Abstract
The activation of eukaryotic SLO K+ channels by intracellular cues, mediated by a cytoplasmic structure called the gating ring, is central to their physiological roles. SLO3 channels are exclusively expressed in mammalian sperm, where variations of intracellular pH are critical to cellular function. Previous studies primarily focused on the mouse SLO3 orthologue and revealed that, in murine sperm, SLO3 mediates a voltage- and alkalization-activated K+ current essential to male fertility. Here we investigate the activation of the human SLO3 channel by intracellular pH at the functional and structural level. By using electrophysiology in a heterologous system, we show that human SLO3 opens upon intracellular pH increase and that its expression and functional properties are modulated by LRRC52, a testis-specific accessory subunit. We next present the crystal structure of the human SLO3 gating ring. Comparison with the known structures of the corresponding domain from SLO1, a Ca2+-activated homologue, suggests that the SLO3 gating ring structure may represent an open state. Together, these results present insights into the function of a protein expected to be critical for human reproduction and provide a framework to study the mechanism of pH gating in SLO3 channels.
Keywords: membrane protein, protein structure, ion channel
Eukaryotic SLO1 and SLO3 channels share a unique property among the extended family of voltage-gated K+ channels in that their opening requires the synergistic action of membrane depolarization and specific intracellular cues (1). This is mirrored by the distinctive architecture of SLO1 and SLO3, which includes a transmembrane voltage sensor as well as a large cytoplasmic structure called the gating ring, responsible for regulation by intracellular signals. The two homologues are closely related in amino acid sequence (42% identity between human channels), and, in fact, sequence analysis suggests that SLO3, which exists only in mammals, evolved from a duplication of the Slo1 gene, which is found across metazoans (1, 2). However, SLO1 and SLO3 diverge in one critical aspect: whereas SLO1 opens upon binding of intracellular Ca2+, SLO3 is activated by an increase in intracellular pH (3–5). This difference underlies fundamentally distinct physiological roles for the two channels. Whereas SLO1 is expressed in excitable cells such as neurons or muscles, where Ca2+ entry into the cytoplasm is critical for function, SLO3 is found exclusively in sperm cells and is essential for male fertility in animal models (3, 6, 7). This exclusive expression pattern reflects the key importance of intracellular [H+] signaling in the physiology of sperm, in which critical steps such as the initiation of motility or the acrosome reaction depend on intracellular alkalization (8, 9).
Hence, SLO1 and SLO3 are closely related in sequence and structure but evolved to serve very divergent functions. What is the molecular basis for the activation of these two channels by different intracellular signals? Studies of chimeras between SLO1 and SLO3 revealed that the specificity for Ca2+ or pH originates solely from variations within the intracellular gating rings: channels containing a SLO1 transmembrane region and a SLO3 gating ring are alkalization-activated but Ca2+-insensitive, and vice versa (10). These results also showed that the allosteric regulation of SLO3 and SLO1 by pH or Ca2+ must be similar enough to allow for the interchangeability of the gating rings. Recent crystal structures of the SLO1 gating ring in the Ca2+-free (i.e., closed) and Ca2+-bound (i.e., open) states have shown how, upon Ca2+ binding, the conformation of the SLO1 gating ring changes in a way that can directly explain the opening of the transmembrane pore (11–13). Determining the extent to which the same principles apply to pH gating in SLO3 channels would provide a better understanding of the gating ring-mediated opening of an important class of eukaryotic ion channels.
So far, studies of SLO3 function have primarily focused on the mouse orthologue, mSLO3 (3). Electrophysiological analysis and genetic deletion studies established that mSLO3 is a voltage- and pH-gated channel that mediates most (if not all) K+ current in murine sperm (4, 6, 7, 14). mSLO3 function is essential to male fertility: Slo3 KO mice are infertile, and KO sperm cells exhibit an array of functional defects (6, 7). Here we study the human SLO3 orthologue, hSLO3. hSLO3 RNA is specifically found in sperm (3), and, based on the results in mouse models, hSLO3 is expected to be the major K+ channel of human spermatozoa (8, 9). However, the functional properties of hSLO3 have not been characterized. Proteins specialized in reproduction, such as SLO3, are known to evolve extremely rapidly, and orthologues can exhibit significant functional variation even between closely related species (15, 16). Therefore, whether the human hSLO3 channel is pH-sensitive is still an open question (8, 9). Here we demonstrate first that hSLO3 functions as a pH-gated channel when expressed in Xenopus oocytes. We next show that the functional expression and gating properties of hSLO3 are critically regulated by LRRC52, an associated subunit first described in mouse sperm (17). Finally, we describe the crystal structure of the hSLO3 gating ring and show how a comparison with the open and closed conformations of the SLO1 gating ring suggests that the hSLO3 structure might represent an open state.
Results
hSLO3 Currents Recorded in Xenopus Oocytes Are pH-Sensitive.
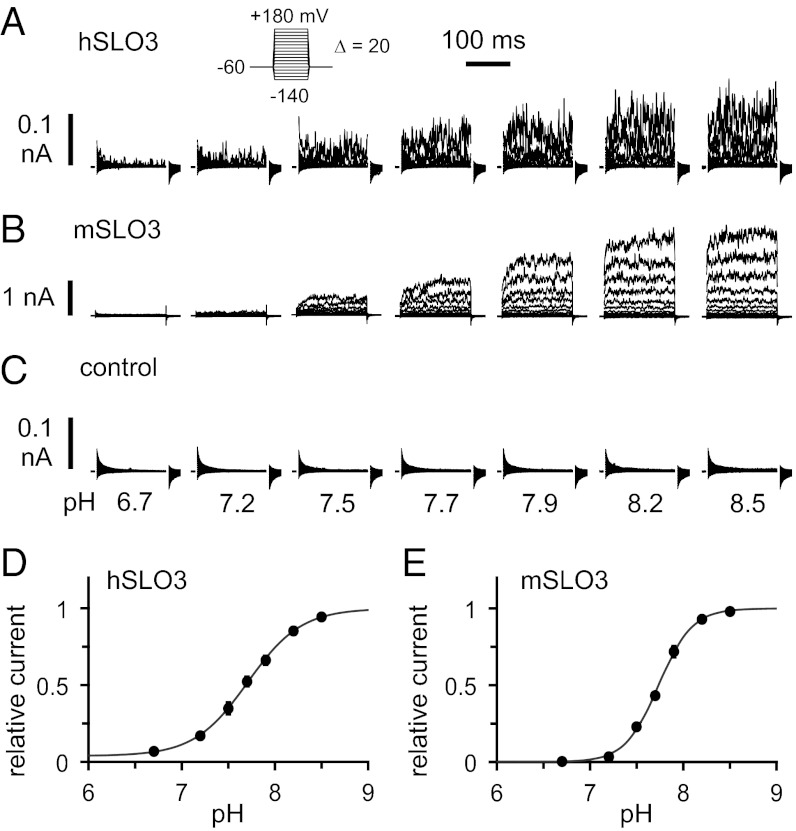
We compared the functional properties of hSLO3 and mSLO3 currents by using heterologous expression in Xenopus oocytes, in which channels are expressed after injection of RNA. The pH sensitivity of SLO3 channels was examined in inside-out patch-clamp electrophysiology experiments, in which the intracellular solution can be precisely controlled. hSLO3 currents from a representative inside-out patch bathed in solutions of increasing pH are shown in Fig. 1A (channel activation is elicited by a series of voltage steps from −140 to +180 mV): hSLO3 currents are clearly pH-sensitive. For comparison, the same experiment performed with mSLO3 is shown in Fig. 1B. Under the same conditions, no channel activity is seen in oocytes injected with water as a negative control (Fig. 1C). The pH sensitivities of hSLO3 and mSLO3 can be compared by plotting the maximum current in these recordings (i.e., at +180 mV) as a function of intracellular pH. This analysis, shown in Fig. 1 D and E, reveals that the pH dependences of hSLO3 and mSLO3 channels are very similar. In particular, the measured pH for half-maximal activation (pH50; at the given voltage of +180 mV) is the same for the two channels (pH50 7.7). However, a significant difference between hSLO3 and mSLO3 is apparent when the total amount of current is compared between Fig. 1 A and B: under similar conditions of pH and voltage, the amplitude of hSLO3 currents is substantially smaller than for mSLO3 currents. This is most likely a result of lower levels of functional expression of hSLO3 in Xenopus oocytes.
Fig. 1.
pH dependence of SLO3 channels. (A) Representative inside-out patch currents recorded from a Xenopus oocyte expressing human hSLO3 channels, bathed in solutions of increasing pH. Voltage pulses are −140 to +180 mV in 20-mV increments (holding voltage, −60 mV). The capacitive transients are masked for clarity. (B) Same as in A from an oocyte expressing mouse mSLO3 channels. (C) Same as in A from a control oocyte injected with water. (D and E) pH dependence of SLO3 currents. Plots of current at +180 mV as a function of intracellular pH, normalized to current at pH 8.5. Data shown as mean ± SEM (hSLO3, n = 6; mSLO3, n = 7; error bars indicate SEM, and some are smaller than the symbols). Solid lines are fits to the Hill equation: I(pH) = 1 + (Imin – 1)/{1 + [10^(−pH50) / 10^(−pH)]n}. For hSLO3, Imin is 0.038 ± 0.011, pH50 is 7.7 ± 0.011, and n is 1.5 ± 0.046. For mSLO3, Imin is 0.0013 ± 0.013, pH50 is 7.7 ± 0.0063, and n is 2.4 ± 0.085.
Coexpression with LRRC52 Increases hSLO3 Expression and Modulates hSLO3 Gating Properties.
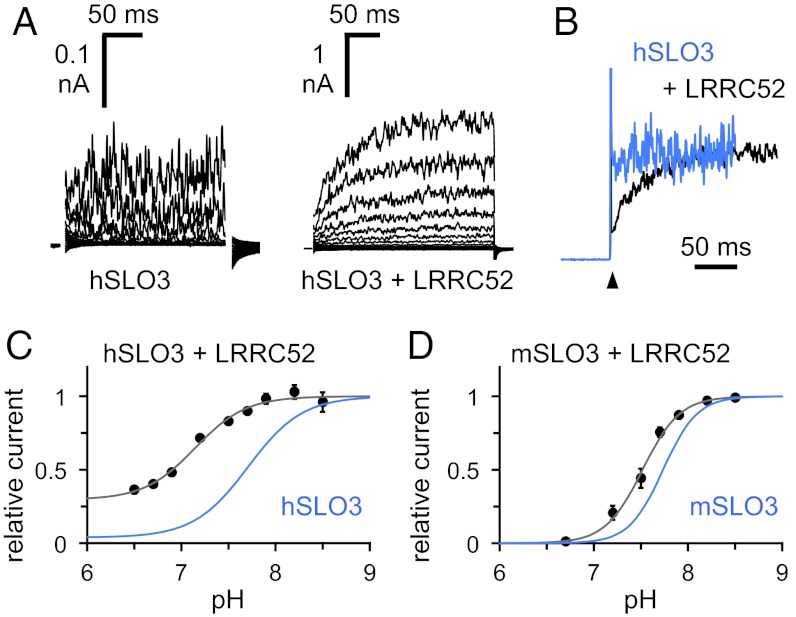
The very limited expression of hSLO3 in Xenopus oocytes makes the functional analysis of hSLO3 currents technically challenging. We hypothesized that the low functional expression of hSLO3 might stem from poor folding and/or trafficking of the hSLO3 protein. Recent studies in murine spermatozoa have led to the identification of a testis-specific mSLO3 auxiliary subunit, LRRC52, a transmembrane protein member of the leucine-rich repeat containing family (17). LRRC52 transcript is specifically enriched in human sperm (18), so we reasoned that LRRC52 might also serve as an auxiliary SLO3 subunit in human, and could facilitate hSLO3 expression. Indeed, coexpression with human LRRC52 (hLRRC52) had several significant effects on hSLO3 currents recorded from Xenopus oocytes. First, the amount of current was greatly increased (Fig. 2A). Second, the activation rate of the current was reduced (Fig. 2B). Third, the pH dependence of hSLO3 activation was substantially shifted, significant channel opening being observed even at low pH (Fig. 2C). The analysis of hSLO3+hLRRC52 currents was complicated by a fast run-down of channel activity upon patch excision, independent of pH (Fig. S1A). To circumvent this issue, we used a strategy in which the same patch is recorded for an extensive period, and we could correct for the effect of run-down by using a given pH (pH 7.2) as a reference point during each pH titration experiment (Fig. S1B). The data presented in Fig. 2C show the result of such analysis. Interestingly, the way LRRC52 affects the pH dependence of hSLO3 is in significant contrast to what is observed with mSLO3 (Fig. 2D). A comprehensive investigation of the functional consequences of LRRC52 coexpression on hSLO3 currents is beyond the scope of the present study, however, two conclusions seem apparent. First, the high levels of K+ currents achieved through coexpression show the presence of pH-dependent gating with an improved signal-to-background ratio, and second, as hSLO3 and LRRC52 are both specifically enriched in human sperm, the pronounced effect of LRRC52 coexpression on hSLO3 function suggests that their interaction could be important for sperm biology in humans.
Fig. 2.
Coexpression of SLO3 channels with LRRC52. (A) Representative inside-out patch currents at pH 8.5 recorded from Xenopus oocytes expressing hSLO3 alone (Left) or hSLO3 and hLRRC52 (Right). Voltage pulses of −140 to +180 mV in 20-mV increments (holding voltage, −60 mV). The capacitive transients are masked for clarity. (B) Normalized current traces from A corresponding to a voltage step from −60 to +180 mV, showing hSLO3 in light blue and hSLO3+hLRRC52 in black. The spikes of current at the voltage step (arrowhead) correspond to the capacitive transients. (C) pH dependence of hSLO3+hLRRC52 currents (at +180 mV). Data shown as mean ± SEM (n = 4–10). Further details are provided in the text. The solid line is a fit to the Hill equation: I(pH) = 1 + (Imin − 1) / {1 + [10^(−pH50) / 10^(−pH)]n}, with Imin of 0.30 ± 0.058 (pH50 7.1 ± 0.069) and n of 1.6 ± 0.16. The pH-dependence of hSLO3 currents from Fig. 1D, in light blue, is shown as a comparison. (D) pH dependence of mSLO3+mLRRC52 currents (at +180 mV). Data shown as mean ± SEM (n = 4). The solid line is a fit to the Hill equation: I(pH) = 1 + (Imin – 1) / {1 + [10^(−pH50) / 10^(−pH)]n}, with Imin of 0.012 ± 0.037 (pH50 7.5 ± 0.027) and n of 2.2 ± 0.024. The pH dependence of mSLO3 currents from Fig. 1E, in light blue, is shown as a comparison. Error bars indicate SEM in C and D, and can be smaller than the symbols.
Structure of hSLO3 Gating Ring.
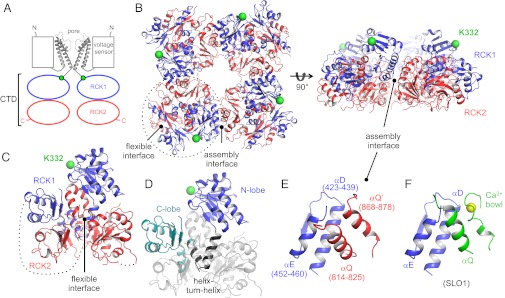
To obtain the best diffracting crystals of the hSLO3 gating ring, we expressed a construct corresponding to the cytoplasmic domain (CTD) of hSLO3 (residues 330–1062; Fig. 3A and Fig. S2) in which two regions predicted to be unstructured (the last 87 C-terminal amino acids and a large internal loop, residues 831–851) were deleted. Similar deletions were used for the crystallization of the Ca2+-bound SLO1 gating ring, for which the deletions facilitated crystallization without affecting the functional properties of the channel (13). The hSLO3 CTD crystallized in space group I222 and diffracted X rays to 3.4-Å resolution. Initial phases were calculated by molecular replacement by using the Ca2+-bound SLO1 monomer structure as a search model (11), and the final model was refined to Rwork/Rfree of 0.248/0.267, where Rwork is the working R-factor and Rfree is the free R-factor. In our crystals, the asymmetric unit contains two CTD subunits. Application of twofold crystallographic symmetry operators generates the structure of the tetrameric hSLO3 gating ring (Fig. 3B).
Fig. 3.
hSLO3 gating ring structure. (A) Domain topology of the hSLO3 channel. Only two opposing subunits are shown for clarity. (B) Crystal structure of the hSLO3 gating ring, showing RCK1 in blue and RCK2 in red. The gating ring is viewed from the top, down the fourfold axis (Left) and from the side (Right). The RCK1 N-terminal residues (K332) that connect to the transmembrane pore are shown as green spheres. A single subunit is indicated by the dotted line. (C) Single subunit from the hSLO3 gating ring structure with the same coloring as in B. Large disordered segments are indicated by dotted lines. (D) Subdomain organization of the hSLO3 gating ring subunit, shown for RCK1. (E) Close-up view of the hSLO3 assembly interface. Amino acid numbers are indicated in parenthesis. (F) Close-up view of the assembly interface from the Ca2+-bound SLO1 gating ring structure (PDB ID code 3U6N), in the same orientation as in E. Ca2+ is shown as a yellow sphere.
The basic structural features of the hSLO3 gating ring are very similar to what is seen in SLO1, as expected given the significant conservation of amino acid sequence between the two channels (Fig. S2). Each hSLO3 CTD subunit is formed by a tandem of regulator of K+ conductance (RCK) domains interacting through a large surface called the flexible interface (Fig. 3C; RCK1 in blue and RCK2 in red). RCK domains, which are ubiquitous in prokaryotic K+ channels and transporters, are a structural signature of eukaryotic channels from the SLO family (19). In hSLO3, as in SLO1 or in the prokaryotic MthK channel (20), each RCK domain can be further divided into three subdomains, called (from N to C termini) the N-lobe, helix–turn–helix motif, and C-lobe (Fig. 3D). To form a gating ring, four CTD protomers assemble via four so-called assembly interfaces bringing together the RCK1 domain from a given subunit and the RCK2 domain from the adjacent subunit. In SLO1, the assembly interface is critical for function because it contains the main Ca2+ binding site of each SLO1 subunit, the Ca2+ bowl, which forms a loop located in the RCK2 domain (11–13). Between hSLO3 and SLO1, the secondary structure elements forming the assembly interface are conserved (helices αD and αE in the RCK1 domain of one subunit contact helix αQ in the RCK2 domain of an adjacent subunit; Fig. 3E). There is, however, a structural difference between hSLO3 and SLO1 in this region: the hSLO3 residues corresponding to the Ca2+ bowl in SLO1 form an extended α helix, which we annotated αQ′ (Fig. 3 E and F).
Structural Comparison of hSLO3 and SLO1 Gating Rings.
For the SLO1 gating ring, structures of both the Ca2+-free (i.e., closed) and Ca2+-bound (i.e., open) states are known (12, 13). Upon Ca2+ binding, the main conformational change occurs in a layer of the SLO1 gating ring proximal to the membrane, formed by the RCK1 N-lobes of all four subunits (hereafter the “RCK1 N-lobe layer”). Ca2+ binding causes the RCK1 N-lobe layer of SLO1 to “open up” like the petals of a flower, leading to an expansion of the gating ring diameter in a way that can directly explain the opening of the transmembrane pore (13) (Fig. S3). How does the hSLO3 gating ring compare with the closed or open conformation of the SLO1 gating ring?
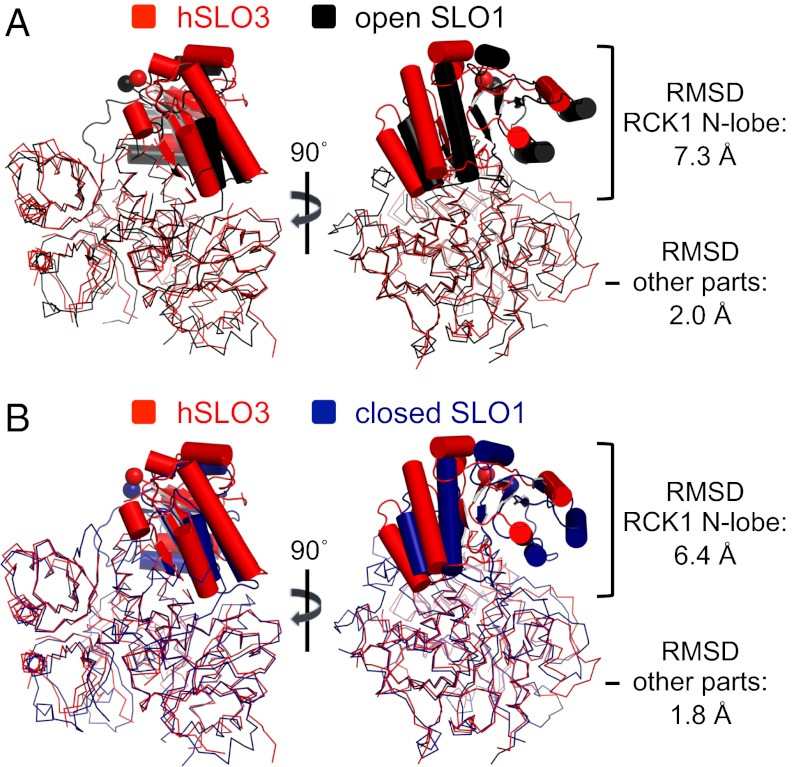
Before we examine the structures of the tetrameric gating rings of hSLO3 and SLO1, let us first compare the structure of isolated subunits from each of these gating rings. Although the overall subdomain organization is conserved between the hSLO3 and SLO1 subunits, the relative orientation of these subdomains in the hSLO3 subunit is quite different from in the closed or open SLO1 structures. A structural alignment reveals that, in the hSLO3 subunit, the RCK1 N-lobe subdomain changes its orientation with respect to the rest of the structure compared with the closed or open conformation of the SLO1 subunit. This is shown in Fig. 4, in which the SLO1 subunits are aligned onto a hSLO3 subunit by superimposing the Cα atoms of all residues except the RCK1 N-lobes. The regions excluding the RCK1 N-lobes align well, but the orientation of the hSLO3 RCK1 N-lobe matches neither conformation of the SLO1 subunit (rmsd values in Fig. 4).
Fig. 4.
Structural comparison of isolated subunits from the hSLO3 and SLO1 gating rings. (A) Superposition of subunits from the hSLO3 (red) and open SLO1 (black; PDB ID code 3U6N) gating rings. The subunits were superimposed by using all Cα atoms excluding the RCK1 N-lobe region. The RCK1 N-lobe subdomains are highlighted. (B) Same as in A, showing superposition of subunits from the hSLO3 and closed SLO1 (deep blue; PDB ID code 3NAF) gating rings.
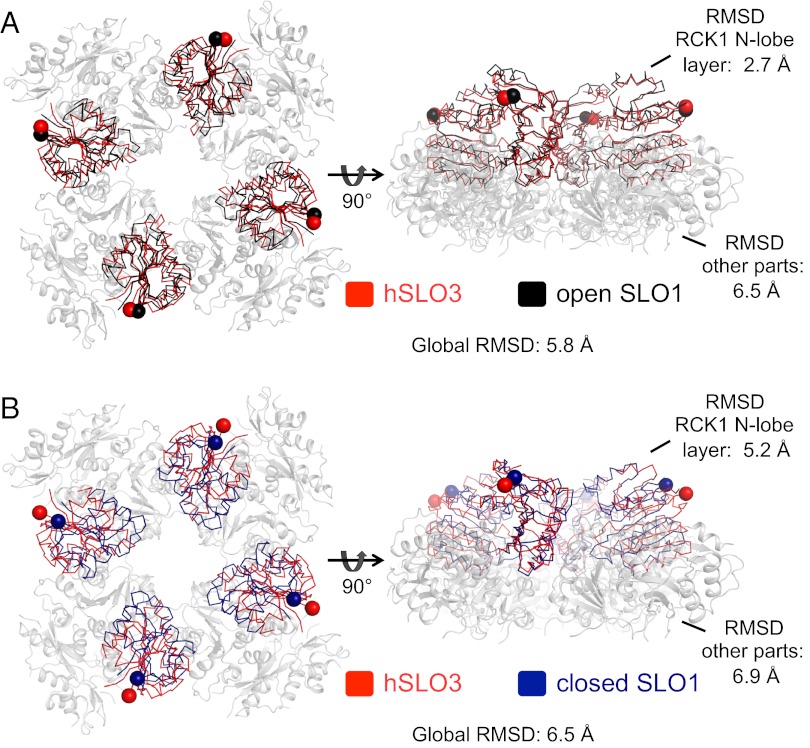
If we now compare the structures of the entire gating rings between hSLO3 and the two conformations of SLO1, the direct result of the different orientation of the RCK1 N-lobes in the isolated hSLO3 subunits is that the hSLO3 gating ring does not align with the open or closed SLO1 gating rings. An objective comparison of these structures is achieved by superimposing all Cα atoms of the tetrameric rings (Fig. S4). The total Cα rmsds (calculated by using all Cα atoms from the tetrameric structures) of these superpositions are very high (4.7 Å and 5.9 Å for the superpositions of hSLO3/open SLO1 and hSLO3/closed SLO1, respectively). This means that, overall, the hSLO3 gating ring structure matches neither the open nor closed conformation of the SLO1 ring. However, an interesting difference is apparent at the level of the RCK1 N-lobe layer. In the superposition using all Cα atoms, the rmsd in the RCK1 N-lobe layer (rmsd calculated using Cα belonging to the RCK1 N-lobes from all four subunits) between the hSLO3 and open SLO1 gating rings is 3.9 Å, vs. 6.0 Å when hSLO3 is compared with the closed SLO1 conformation (Fig. S4). Another feature, independent of specific superposition, can be seen in Fig. S4. Measured from equivalent N-terminal residues (K332 in hSLO3, K343 in SLO1; Fig. S4, spheres), the diameter of the hSLO3 gating ring (94 Å) matches almost exactly the diameter of the open SLO1 gating ring (93 Å vs. 81 Å in the closed SLO1 gating ring).
To further compare the structure of the RCK1 N-lobe layer between the tetrameric hSLO3 and SLO1 gating rings, we performed another structural alignment by superimposing the Cα atoms in this region (using all four subunits; Fig. 5). This analysis reveals that, in the hSLO3 gating ring, the structure of the RCK1 N-lobe layer matches closely the corresponding region in the open conformation of the SLO1 gating ring (rmsd measured using Cα atoms from all four subunits, 2.7 Å), whereas the rest of the structure aligns quite poorly (rmsd measured using Cα atoms from all four subunits, 6.5 Å; Fig. 5A and Fig. S5 A and B). When the closed conformation of the SLO1 gating ring is used in the same superposition, no significant alignment is observed in any part of the structures (Fig. 5B and Fig. S5 C and D).
Fig. 5.
hSLO3 gating ring vs. open and closed conformations of SLO1. (A) Superposition of the hSLO3 and open SLO1 (PDB ID code 3U6N) gating ring structures by aligning the RCK1 N-lobes from all four subunits. The RCK1 N-lobe regions from all four subunits (i.e., RCK1 N-lobe layer) of the hSLO3 (red) and open SLO1 (black) gating rings are shown as a ribbon. The rest of the structure, where the superposition is poor and is not shown, is drawn as a gray cartoon. RCK1 N-terminal residues (K332 in hSLO3, K343 in SLO1) are shown as spheres. (B) Same as in A but showing the superposition of the hSLO3 and closed SLO1 (deep blue; PDB ID code 3NAF) gating rings.
hSLO3 Gating Ring Structure Might Represent an Open Conformation.
The hSLO3 crystals were grown in a solution with initial pH ∼6.8, at which hSLO3 currents recorded in oocytes are mostly closed. However, the pH dependence of the isolated hSLO3 gating ring is not necessarily the same as for the entire channel. Furthermore, the intracellular surface of the cell membrane, where the gating ring is located, is rich in negative charges that concentrate positively charged protons. As a result, the local pH in the immediate vicinity of the hSLO3 “pH sensor” might be significantly lower than in the bulk intracellular solution. For all these reasons, the pH at which crystals were grown might not correspond well to the bulk pH at which membrane-bound channels are activated.
In fact, the hSLO3 gating ring structure most resembles the open conformation of the SLO1 gating ring. Most importantly, the architecture of the RCK1 N-lobe layer in hSLO3 coincides with the architecture observed in the open SLO1 gating ring, and the overall diameters of the hSLO3 and open SLO1 gating rings (measured at the N-terminal residues of the RCK1 domains) are nearly identical. Because, in SLO1 channels, the orientation, and thus the diameter, of the RCK1 N-lobe layer is the main structural determinant of the open vs. closed state of the gating ring, it is likely that the hSLO3 structure represents the open conformation of the hSLO3 gating ring. The fact that hSLO3 and SLO1 channels are similar in amino acid sequence reinforces this hypothesis. In particular, the length of the linkers connecting the CTDs to the transmembrane pore is absolutely conserved, so that the identical diameters of the hSLO3 and open SLO1 gating rings could drive opening of the pore in a similar manner.
Discussion
Altogether, the implications of our results are twofold. First, our data provide insight into the physiological function of the human SLO3 channel. We show that hSLO3 is indeed pH-sensitive. When expressed in Xenopus oocytes, hSLO3 currents open in the same range of pH as mSLO3. However, upon coexpression with the associated subunit LRRC52, the pH dependences of mSLO3 and hSLO3 differ markedly: hSLO3+LRRC52 currents show significant activation at pH < 6.7, a pH at which mSLO3+LRRC52 activity is essentially suppressed. This is interesting because preliminary reports in the literature suggest that the K+ current in human sperm is much less sensitive to changes in intracellular pH than the K+ current of mouse sperm (8, 9). This led to an uncertainty about hSLO3 being responsible for K+ currents in human sperm. If LRRC52 is a physiological partner of hSLO3 in human sperm, its significant effect on the pH dependence of hSLO3 might explain this discrepancy. Ultimately, a precise understanding of the role and functional properties of K+ channels in human sperm will require direct electrophysiological and biochemical characterizations in human sperm cells. In this context, specific pharmacological agents (e.g., hSLO3-blocking toxins) would be very valuable to characterize the exact physiological function of hSLO3. The expression strategy we describe, in which hSLO3 currents can be robustly expressed in a heterologous system in the presence of LRRC52, could be used to identify such modulators.
Second, our results reveal significant similarities in the structures of the gating rings of SLO3 and SLO1 channels that could indicate a convergence in the molecular mechanisms of pH or Ca2+ sensing between the two homologues. Overall, the basic organization of the hSLO3 and SLO1 gating rings is the same: four subunits each contribute a tandem pair of RCK domains that assemble into a tetrameric ring-like structure through interfaces involving equivalent secondary structure elements in the two homologues. The details of how the subdomains are arranged with respect to each other are different in the hSLO3 and SLO1 gating rings, and the end result is a global architecture of the hSLO3 gating ring that does not match the closed or open conformation of the SLO1 gating ring. However, the spatial organization of one critical region (the RCK1 N-lobe layer) is the same in hSLO3 and SLO1 in its open conformation. Because, in SLO1, the conformation of the RCK1 N-lobe layer is precisely what determines the closed vs. open state of the gating ring, we propose that the hSLO3 gating ring structure represents an open state.
What else does the structural conservation of the RCK1 N-lobe layer between hSLO3 and SLO1 imply? In a simple description, the gating ring can be viewed as a module that simply transduces chemical energy (e.g., binding of Ca2+) into mechanical work used to pull open the channel’s transmembrane pore. In this context, only the overall diameter of the gating ring (measured at the N-terminal positions connecting to the pore), but not the details of the gating ring structure, determines channel opening. Therefore, the fact that not only the gating ring diameter, but also the architecture of the entire RCK1 N-lobe layer, appears to be conserved between the open states of hSLO3 and SLO1 suggests the presence of a specific functional or structural constraint. One possibility is that the conformational change in the RCK1 N-lobe layer during the closed-to-open transition of the gating ring is conserved between SLO1 and SLO3 channels, in the same way that the geometry of the active site (but not necessarily the remainder of the structure) is conserved within a given class of enzymes. Another (nonexclusive) possibility is that, in a full channel, the RCK1 N-lobe layer interacts with other parts of the protein (namely, the transmembrane domain) and that this interaction constrains structurally the position of the RCK1 N-lobes. The conserved architecture of the transmembrane domains between SLO1 and SLO3 (expected based on sequence homology) would result in a conserved positioning of the RCK1 N-lobes.
The structure of the hSLO3 gating ring and its close relationship to the structure of the SLO1 gating ring provides a molecular framework with which to understand the molecular mechanisms of pH sensing in SLO3 channels. Understanding how pH affects the conformation and function of proteins is challenging because variations in pH can affect the protonation state of a large number of amino acid residues and as a consequence the pH sensitivity of proteins is often controlled by multiple titratable groups. In particular, the molecular identity of the putative pH sensor in SLO3 channels is still elusive. The hSLO3 gating ring structure will hopefully pave the way for further experimental and computational analyses to address this important question.
Materials and Methods
hSLO3 Construct.
A synthetic gene encoding the human hSLO3 channel (KCNU1, NCBI GeneID: 157074213) was purchased from BioBasics and served as a template for subcloning.
Electrophysiology.
For mRNA production, the full-length hSLO3 channel was subcloned into the pGEM-HE vector under the control of a T7 promoter for transcription (21). Plasmids of mSLO3 in pOX (T3 promoter) and of human and mouse LRRC52 in pXMX (SP6 promoter) were gifts from L. Salkoff and C. Lingle (Washington University, St. Louis, MO), respectively. After linearization of the corresponding plasmids, capped mRNA was produced by in vitro transcription by using the mMessage mMachine reagents (Ambion). mRNA was purified by using phenol/chloroform extraction and ethanol precipitation, dissolved in DEPC-treated H2O and stored at −80 °C. Defolliculated Xenopus laevis oocytes were prepared as described previously (11) and injected with ∼10 ng mRNA. For SLO3/LRRC52 coexpression, the two constructs were injected at a 1:1 wt/wt ratio. Electrophysiological measurements were made 3 to 6 d after injection.
For recordings, inside-out patches were excised from freshly devitellinized oocytes by using fire-polished glass pipettes with a typical resistance of 0.7 to 1.2 MΩ. Large pipettes were used to increase the amplitude of SLO3 currents in a patch. The pipette solution contained (in mM) 140 K-gluconate, 20 Hepes, and 2 MgCl2, with pH adjusted to 7.0 with K-OH. Bath solutions contained (in mM) 140 K-gluconate, 20 Hepes, and 5 EGTA (acid form), with pH carefully adjusted with K-OH to the desired value. The intracellular solution was exchanged by using a custom-built gravity-flow perfusion system, with individual patches being exposed to solutions flowing out of small glass capillaries (Microcaps 1; Drummond).
Analog signals were filtered (2 kHz) by using the built-in low-pass filter of an Axopatch 200B patch clamp amplifier (Molecular Devices), digitized at 10 kHz (Digidata 1440A; Molecular Devices), and stored on a computer hard disk by using Clampex 10.2 software (Molecular Devices).
For recordings of hSLO3 currents or control oocytes injected with DEPC-H2O, the contribution of the nonspecific linear leak was calculated by extrapolating the values of current at negative voltages, at which the channels (if any) are closed, and subtracted from the data (because of the low open probability and the limited number of hSLO3 channels in a patch, the current recorded at negative voltages corresponds almost entirely to the linear leak). For recordings of mSLO3 or hSLO3/mSLO3 coexpression with LRRC52, currents are of large enough amplitude that the contribution of linear leak was insignificant, and no subtraction was performed. For all recordings, the level of current at a given voltage in an individual patch was measured as the average of current in the last 60 ms of a single voltage pulse.
Protein Purification and Structure Determination.
A CTD construct containing hSLO3 residues 330 to 1062 with a loop deletion (residues 831–851) was expressed in Sf9 insect cells and purified following methods described previously (11). Briefly, the hSLO3 CTD construct was expressed fused to a C-terminal GFP with a deca-histidine tag, following a recognition site for the PreScission protease (PreScission-GFP-His10; GE Life Sciences). Cells were disrupted by sonication in buffer containing (in mM) 500 KCl, 50 K-phosphate, 10 imidazole, and 5 β-mercaptoethanol, at final pH 8.0 (K-OH), containing DNase and protease inhibitors. The fusion construct was purified by using a Co2+ affinity resin (TALON; Clontech), supplemented with 10 mM DTT and incubated overnight with PreScission protease (1:40 wt/wt) to remove the C-terminal GFP-His10. The hSLO3 CTD was then isolated by size-exclusion chromatography on a Superose 6 column (GE Life Sciences) in (in mM) 500 KCl, 20 K-phosphate, 20 DTT, and 1.5 Tris (2-carboxyethyl) phosphine, at final pH 8.5 with K-OH. The protein was concentrated to ∼5 mg/mL for crystallization experiments. All purification steps were carried out at 4 °C.
Crystals were grown at 20 °C by using hanging-drop vapor diffusion by mixing equal volumes of protein and a reservoir solution containing 50 mM K-phosphate, 3% (wt/vol) PEG12000, and 1 M ammonium formate, at final pH 6.3 with K-OH. The pH of the final solution obtained by mixing equal amounts of protein and reservoir solutions was measured to be 6.8. The crystals grew to a maximum size of approximately 0.4 mm × 0.15 mm × 0.05 mm within 3 d. Crystals were sequentially transferred into cryoprotective solutions [all containing 500 mM KCl, 70 mM K-phosphate, 4% (wt/vol) PEG12000, and 1 M ammonium formate, at final pH 6.4] containing increasing concentrations of ethylene glycol [5%, 10%, 20%, final ethylene glycol concentration of 30% (vol/vol)] before being flash-frozen in liquid N2. The crystals belong to space group I222 and the unit cell has the following dimensions: a, 124.54 Å; b, 157.94 Å; and c, 249.00 Å; with α, β, and γ equal to 90°. Each asymmetric unit contains two CTD subunits. Diffraction data were measured at beamline X29 of the National Synchrotron Light Source and were processed with the HKL2000 program suite (22). Diffraction of X rays by these crystals was very anisotropic, and analysis revealed that diffraction in the a* and b* directions of the reciprocal cell are the weakest. In the final dataset used for structure determination, diffraction data collected from two independent crystals that varied in their degree of diffraction anisotropy were merged. These reflections were scaled together in SCALEPACK (22) to produce a unique, final dataset. The scaled data set was anisotropically corrected to resolution limits of 3.8, 3.4, and 3.3 Å along the reciprocal cell directions a*, b*, and c*, respectively, using the diffraction anisotropy server at the University of California, Los Angeles (23). An isotropic B factor of −105.89 Å2 was applied to restore the magnitude of the high-resolution reflections diminished by anisotropic scaling. The structure was determined by molecular replacement by using the monomeric Ca2+-bound human SLO1 CTD [Protein Data Bank (PDB) ID code 3MT5] as the search model. Structure refinement was carried out in REFMAC (24) by using strong twofold noncrystallographic symmetry restraints. Iterative model building was carried out in COOT (25). The final model was refined to a resolution of 3.4 Å with an Rwork of 0.248 and Rfree of 0.267. A few disordered regions were not modeled as a result of weak electron density, and the final refined model includes residues 331 to 352, 361 to 371, 378 to 559, 567 to 601, 681 to 706, 710 to 806, 812 to 829, 858 to 931, 956 to 1006, and 1013 to 1043. Residues for which side-chain density was poorly defined were modeled as alanines. The majority (96.5%) of the residues lie in the most favored region in a Ramachandran plot, with the remaining 3.5% in the additionally allowed regions [using MolProbity (26)]. Data collection and structure refinement statistics are shown in Tables S1 and S2. All structural illustrations were prepared with PYMOL (www.pymol.org).
Supplementary Material
Acknowledgments
We thank staff members at NSLS X29, Brookhaven National Laboratory, for beamline assistance; P. Hoff and members of the Gadsby laboratory for help with oocyte preparation; L. Salkoff for the mSLO3 plasmid; and C. Lingle for sharing the LRRC52 constructs. M.D.L. thanks J. Avalos and L. Feng for training and support. R.M. is an investigator in the Howard Hughes Medical Institute. This work was supported by American Asthma Foundation Grant 07-0127.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4HPF).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215078109/-/DCSupplemental.
References
- 1.Salkoff L, Butler A, Ferreira G, Santi C, Wei A. High-conductance potassium channels of the SLO family. Nat Rev Neurosci. 2006;7(12):921–931. doi: 10.1038/nrn1992. [DOI] [PubMed] [Google Scholar]
- 2.Santi CM, Butler A, Kuhn J, Wei A, Salkoff L. Bovine and mouse SLO3 K+ channels: Evolutionary divergence points to an RCK1 region of critical function. J Biol Chem. 2009;284(32):21589–21598. doi: 10.1074/jbc.M109.015040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schreiber M, et al. Slo3, a novel pH-sensitive K+ channel from mammalian spermatocytes. J Biol Chem. 1998;273(6):3509–3516. doi: 10.1074/jbc.273.6.3509. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Zeng X, Lingle CJ. Slo3 K+ channels: Voltage and pH dependence of macroscopic currents. J Gen Physiol. 2006;128(3):317–336. doi: 10.1085/jgp.200609552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Zeng X, Xia X-M, Lingle CJ. pH-regulated Slo3 K+ channels: Properties of unitary currents. J Gen Physiol. 2006;128(3):301–315. doi: 10.1085/jgp.200609551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santi CM, et al. The SLO3 sperm-specific potassium channel plays a vital role in male fertility. FEBS Lett. 2010;584(5):1041–1046. doi: 10.1016/j.febslet.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng X-H, Yang C, Kim ST, Lingle CJ, Xia X-M. Deletion of the Slo3 gene abolishes alkalization-activated K+ current in mouse spermatozoa. Proc Natl Acad Sci USA. 2011;108(14):5879–5884. doi: 10.1073/pnas.1100240108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lishko PV, et al. The control of male fertility by spermatozoan ion channels. Annu Rev Physiol. 2012;74:453–475. doi: 10.1146/annurev-physiol-020911-153258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirichok Y, Lishko PV. Rediscovering sperm ion channels with the patch-clamp technique. Mol Hum Reprod. 2011;17(8):478–499. doi: 10.1093/molehr/gar044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia X-M, Zhang X, Lingle CJ. Ligand-dependent activation of Slo family channels is defined by interchangeable cytosolic domains. J Neurosci. 2004;24(24):5585–5591. doi: 10.1523/JNEUROSCI.1296-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan P, Leonetti MD, Pico AR, Hsiung Y, MacKinnon R. Structure of the human BK channel Ca2+-activation apparatus at 3.0 A resolution. Science. 2010;329(5988):182–186. doi: 10.1126/science.1190414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Y, Yang Y, Ye S, Jiang Y. Structure of the gating ring from the human large-conductance Ca(2+)-gated K(+) channel. Nature. 2010;466(7304):393–397. doi: 10.1038/nature09252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan P, Leonetti MD, Hsiung Y, MacKinnon R. Open structure of the Ca2+ gating ring in the high-conductance Ca2+-activated K+ channel. Nature. 2012;481(7379):94–97. doi: 10.1038/nature10670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navarro B, Kirichok Y, Clapham DE. KSper, a pH-sensitive K+ current that controls sperm membrane potential. Proc Natl Acad Sci USA. 2007;104(18):7688–7692. doi: 10.1073/pnas.0702018104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3(2):137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 16.Lishko PV, Botchkina IL, Kirichok Y. Progesterone activates the principal Ca2+ channel of human sperm. Nature. 2011;471(7338):387–391. doi: 10.1038/nature09767. [DOI] [PubMed] [Google Scholar]
- 17.Yang C, Zeng X-H, Zhou Y, Xia X-M, Lingle CJ. LRRC52 (leucine-rich-repeat-containing protein 52), a testis-specific auxiliary subunit of the alkalization-activated Slo3 channel. Proc Natl Acad Sci USA. 2011;108(48):19419–19424. doi: 10.1073/pnas.1111104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan J, Aldrich RW. BK potassium channel modulation by leucine-rich repeat-containing proteins. Proc Natl Acad Sci USA. 2012;109(20):7917–7922. doi: 10.1073/pnas.1205435109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Y, Pico A, Cadene M, Chait BT, MacKinnon R. Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel. Neuron. 2001;29(3):593–601. doi: 10.1016/s0896-6273(01)00236-7. [DOI] [PubMed] [Google Scholar]
- 20.Jiang Y, et al. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417(6888):515–522. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 21.Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9(5):861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Strong M, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103(21):8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.