Abstract
The gaseous phytohormone ethylene C2H4 mediates numerous aspects of growth and development. Genetic analysis has identified a number of critical elements in ethylene signaling, but how these elements interact biochemically to transduce the signal from the ethylene receptor complex at the endoplasmic reticulum (ER) membrane to transcription factors in the nucleus is unknown. To close this gap in our understanding of the ethylene signaling pathway, the challenge has been to identify the target of the CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) Raf-like protein kinase, as well as the molecular events surrounding ETHYLENE-INSENSITIVE2 (EIN2), an ER membrane-localized Nramp homolog that positively regulates ethylene responses. Here we demonstrate that CTR1 interacts with and directly phosphorylates the cytosolic C-terminal domain of EIN2. Mutations that block the EIN2 phosphorylation sites result in constitutive nuclear localization of the EIN2 C terminus, concomitant with constitutive activation of ethylene responses in Arabidopsis. Our results suggest that phosphorylation of EIN2 by CTR1 prevents EIN2 from signaling in the absence of ethylene, whereas inhibition of CTR1 upon ethylene perception is a signal for cleavage and nuclear localization of the EIN2 C terminus, allowing the ethylene signal to reach the downstream transcription factors. These findings significantly advance our understanding of the mechanisms underlying ethylene signal transduction.
Keywords: mass spectrometry, serine
Ethylene is a plant hormone that plays important roles in growth and development (1–3). Responses to ethylene include fruit ripening, abscission, senescence, and adaptive responses to a wide range of biotic and abiotic stresses (1, 2). Molecular genetic dissection of the ethylene response pathway in Arabidopsis thaliana has led to the identification of key components in ethylene signal transduction (3), but little is known regarding the biochemical mechanisms that transduce the ethylene signal. A major gap in our understanding of the ethylene signaling pathway is how the signal is transduced from CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) to ETHYLENE-INSENSITIVE2 (EIN2) at the endoplasmic reticulum (ER) membrane and then to the nucleus where gene expression is regulated.
The ethylene receptors, which are related to the receptor histidine kinases of the prokaryotic two-component signaling system (4, 5), reside at the ER membrane where they associate with and signal to the CTR1 serine/threonine protein kinase (6–8); in the absence of ethylene, the receptors promote CTR1 kinase activity, which represses ethylene responses, whereas in the presence of ethylene, the receptors, and therefore CTR1, are inactive. CTR1 consists of a unique N-terminal regulatory domain and a C-terminal serine/threonine kinase domain (Fig. 1A). Because CTR1 is most similar in sequence to the Raf protein kinase family (8), CTR1 has long been presumed to function, like Raf, as a mitogen-activated protein kinase kinase kinase (MAPKKK) in a typical MAPK cascade. However, the existence of such a MAPK cascade in ethylene signaling is controversial (9–11), and no authenticated substrate of CTR1 has been identified.
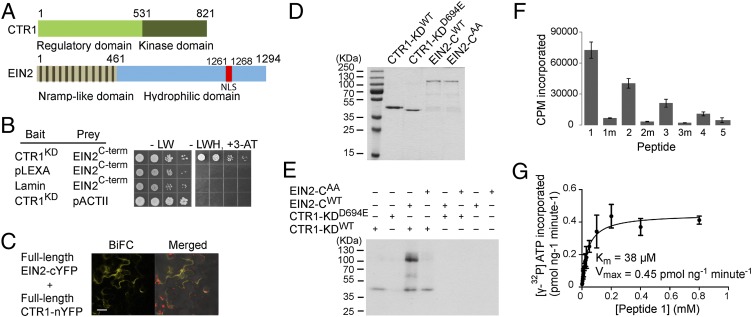
Fig. 1.
CTR1 phosphorylates specific serine/threonine residues in the EIN2 C-terminal domain in vitro. (A) Cartoon of CTR1 and EIN2 protein domain structure. Position of the predicted NLS in EIN2 is shown (19). (B) Yeast two-hybrid assay showing that the CTR1 kinase domain (residues 551–821) interacts with the EIN2 soluble domain (residues 516–1294). Bait vector (pLEXA), prey vector (pACTII), and lamin were negative controls. (C) BiFC interaction of full-length EIN2 and CTR1 in tobacco leaf epidermal cells. Merged image shows BiFC, DIC, and chlorophyll. (Scale bar: 20 µM.) (D) Coomassie-stained SDS/PAGE gel of purified WT and mutant versions of CTR1 kinase domain (KD) and soluble C-terminal domain of EIN2 (EIN2-C). Molecular weight markers are shown on Left. (E) In vitro kinase assay of purified CTR1 kinase domain (residues 531–821) with the EIN2 C-terminal domain (residues 479–1294). The indicated proteins (WT or mutant) were incubated together in kinase reaction buffer, separated by SDS/PAGE, and the incorporated radiolabel detected with a phosphorimager. (F) The relative phosphorylation of various peptides by CTR1-KDWT. Peptides used: 1: KAAPTSNFTVGSDGPPS645FRSLSGK; 1m: KAAPTSNFTVGSDGPPA645FRSLSGK; 2. KAAVANEKKYSS924MPDISGLSMSAR; 2m: KAAVANEKKYSA924MPDISGLSMSAR; 3: KPVGMNQDGPGS1283RKNVTAYG; 3m: KPVGMNQDGPGA1283RKNVTAYG; 4:KQQRTPGS757IDSLYGLQR; 5: KKGMDS739QMTSSLYDSLKQQRT. (G) Kinetic analysis of CTR1 phosphorylation of peptide 1. A total of 20 ng of His6-CTR-KDWT protein was incubated with increasing concentrations of EIN2 peptide1 in kinase buffer with γ-labeled [32P]ATP, and the amount of radioactivity incorporated determined.
EIN2 is a positive regulator of ethylene responses that acts downstream of CTR1 based on genetic analyses (12, 13). No physical or biochemical connection has been reported between EIN2 and CTR1, however, and the mechanism of EIN2 signaling is unknown. EIN2 consists of an N-terminal integral membrane domain of 12 predicted transmembrane helices (residues 1–461) with sequence similarity to Nramp metal ion transporters, followed by a hydrophilic C-terminal domain (residues 462–1294) believed to be cytosolic (12) (Fig. 1A). EIN2 resides at the ER (14) associated with the ethylene receptors (14, 15). In the absence of ethylene, EIN2 protein levels are decreased by protein turnover involving F-box proteins and degradation by the Ub/26S proteasome (16). Acting downstream of EIN2 are several nuclear-localized transcription factors (e.g., EIN3 and ERF1) that mediate the transcriptional response to ethylene (17, 18). Interestingly, the EIN2 C-terminal domain carries a conserved nuclear localization signal (NLS) (19) (Fig. 1A).
Recently, phosphorylation sites in the C-terminal domain of EIN2 were identified by mass spectrometry of microsomal membrane proteins isolated from ethylene-treated and untreated dark-grown Arabidopsis seedlings (20). Interestingly, the data suggested the possibility of differential phosphorylation of EIN2 in vivo in response to ethylene, with phosphorylation occurring primarily in the absence of ethylene (20). Based on this finding, we proposed that the protein kinase responsible for phosphorylating EIN2 could be CTR1. Here, we demonstrate that EIN2 is a direct target of the CTR1 kinase in the absence of ethylene, and that alanine substitutions preventing phosphorylation result in activation of ethylene responses by a mechanism involving translocation of the EIN2 C terminus to the nucleus.
Results
CTR1 Phosphorylates EIN2.
Consistent with the possibility that CTR1 phosphorylates EIN2, the CTR1 kinase domain (residues 551–821) interacted with the EIN2 C-terminal domain (residues 516–1294) in the yeast two-hybrid assay (Fig. 1B), and full-length CTR1 and EIN2 associated in plant cells using bimolecular fluorescence complementation (BiFC; Fig. 1C).
We examined the ability of CTR1 to phosphorylate EIN2 in vitro using purified proteins. The kinase domain of CTR1 (residues 531–821; CTR1-KDWT) was expressed in insect cells, and the hydrophilic, C-terminal domain of EIN2 (residues 479–1294; EIN2-CWT) was expressed in Escherichia coli, and both were purified using 6×-His affinity tags (Fig. 1D). We similarly expressed and purified the catalytically inactive CTR1-1 mutant version (CTR1-KDD694E) (7) as well as EIN2-C carrying Ala substitutions at both Ser645 and Ser924 (EIN2-CAA; Fig. 1D). These two Ser residues were of particular interest, because both had displayed apparent differential phosphorylation in air vs. ethylene-treated seedlings in vivo, and both are conserved in EIN2 homologs in higher plants (20). In an in vitro kinase assay, CTR1-KDWT, but not CTR1-KDD694E, was capable of phosphorylating EIN2-CWT (Fig. 1E). Mass spectrometry analysis of the kinase reaction identified six phosphorylation sites in EIN2 corresponding to the four differentially phosphorylated sites (20) plus two additional sites (Ser659 and Thr819; Fig. S1; Table S1). Notably, the phosphorylation by CTR1- KDWT was substantially reduced (>85%) when EIN2-CAA was used in the reaction (Fig. 1E).
Next, we examined the ability of CTR1 to phosphorylate individual EIN2 peptides corresponding to the four sites found in common in vivo and in vitro. We included a fifth peptide corresponding to a site observed to be phosphorylated both with and without ethylene in vivo (20). The peptides corresponding to Ser645 and Ser924 were robustly phosphorylated by CTR1, and the other two peptides were phosphorylated to a lesser extent, whereas the fifth peptide was only weakly phosphorylated (Fig. 1F). When the target Ser residues in these peptides were substituted with Ala, phosphorylation was again eliminated (Fig. 1F). Kinetic analysis of phosphorylation on Ser645 revealed a Km of 38 μM (Fig. 1G). Taken together, these results indicate that CTR1 phosphorylates EIN2 in vivo on at least four sites.
Preventing EIN2 Phosphorylation on Ser645 and Ser924 Results in Constitutive Activation of Ethylene Responses.
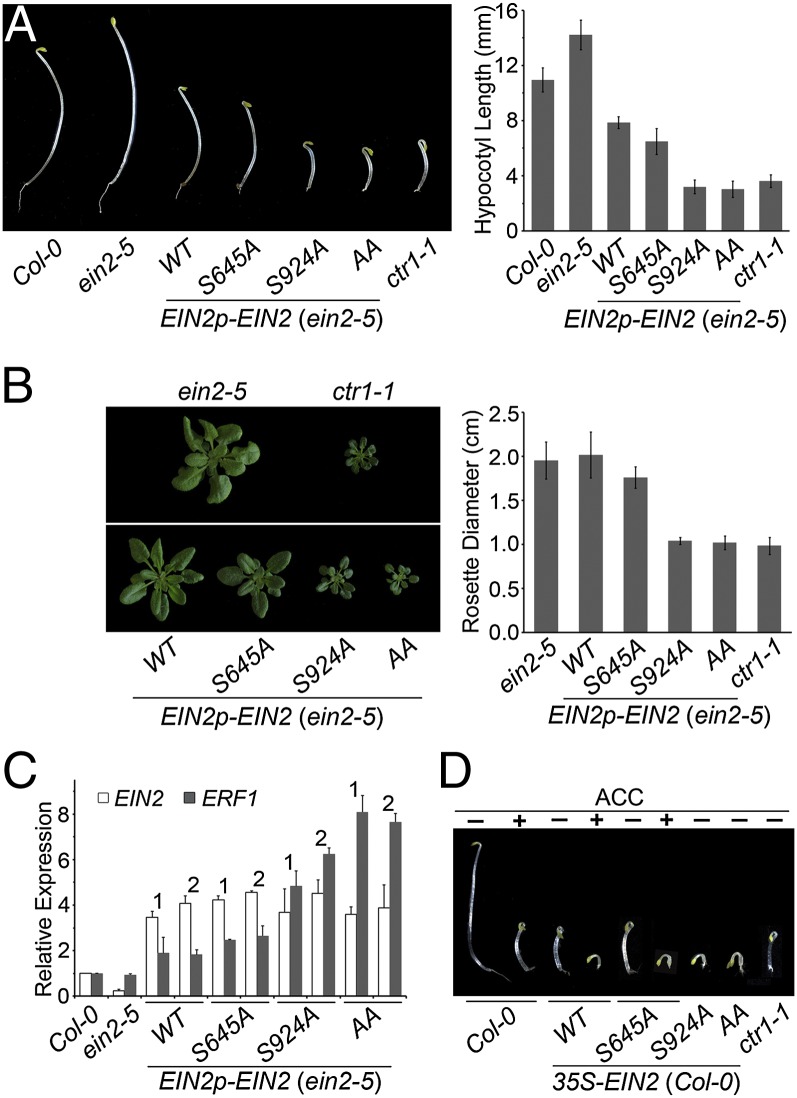
To test the relevance of phosphorylation on Ser645 and Ser924 to ethylene signaling in planta, we constructed EIN2 transgenes encoding Ser-to-Ala substitutions to block phosphorylation at these sites. Mutations encoding the alanine substitutions were introduced into a 9.4-kb genomic EIN2 transgene (EIN2p-EIN2), which included the EIN2 promoter region (2.86 kb upstream of the 5′ UTR) as well as 629 bp downstream of the 3′ UTR. We first confirmed that the wild-type version of this transgene (EIN2WT) could rescue the strong mutant allele, ein2-5, which has a 7-bp deletion (nucleotides 939–945 in the EIN2 coding sequence; Fig. S2). When we introduced transgenes carrying S645A (EIN2S645A), S924A (EIN2S924A), or both substitutions (EIN2AA) into the ein2-5 mutant, constitutive ethylene-response phenotypes were obtained. The EIN2p-EIN2S645A construct conferred only a slight constitutive phenotype (seen in 6 of 11 independent lines), whereas EIN2p- EIN2S924A conferred a stronger phenotype (observed in 7 of 11 independent lines) similar to the constitutive ethylene-response phenotype of the ctr1-1 mutant (Fig. 2 A and B; Fig. S3). EIN2p-EIN2AA gave a strong phenotype (observed in 15 of 32 lines) consistent with an additive effect of EIN2S645A and EIN2S924A (Fig. 2 A and B). For all phenotypic comparisons, we used transgenic lines expressing similar levels of EIN2 (Fig. 2C). Consistent with the above, the EIN2S924A and EIN2AA lines showed elevated expression of the ethylene-responsive transcription factor gene ERF1 (Fig. 2C). Taken together, our results indicate that phosphorylation of Ser645 and Ser924 is involved in repressing EIN2 signaling, and that Ser924 plays a more prominent role in repressing EIN2 function compared with Ser645.
Fig. 2.
Ser645Ala and Ser924Ala substitutions in EIN2 confer constitutive ethylene responses in Arabidopsis. (A) The ethylene-response phenotype in 4-d-old dark-grown seedlings in the absence of ethylene treatment is shown for ein2-5 transformed with WT and mutant versions of genomic transgene EIN2p-EIN2. Representative seedlings are shown in comparison with the WT (Col-0), ein2-5, and ctr1-1. Mean hypocotyl length ± SD is averaged for two independent lines, 20–30 seedlings per line. (B) Inhibition of leaf cell expansion in representative rosettes of 5-wk-old soil-grown plants of the same transgenic lines as in A. Mean rosette diameter ± SD at 4 wk old is averaged for two independent lines, 20–35 rosettes per line. (C) Real-time quantitative PCR expression analysis for EIN2 and the ethylene-responsive transcription factor gene ERF1. Two independent lines (1 and 2) are shown for each transgene. (D) Four-d-old dark-grown Col-0 seedlings stably transformed with WT and mutant versions of 35S-EIN2 treated with or without 20 μM ACC. Representative seedlings are shown in comparison with Col-0 and ctr1-1.
Overexpression of EIN2 in wild-type plants provided additional evidence that Ser924 plays a larger role in ethylene signaling compared with Ser645. Most of the resulting lines displayed ethylene insensitivity (possibly due to EIN2 cosuppression), but 25–40% of the lines for each construct showed a constitutive ethylene-response phenotype. Without ethylene treatment, transgenic lines carrying 35S-EIN2WT displayed a constitutive ethylene-response phenotype similar to ctr1-1, and when grown on the ethylene biosynthesis precursor 1-aminocyclopropane carboxylic acid (ACC), they displayed a distinct stunted phenotype (Fig. 2D) identical to that of ACC-treated seedlings that overexpress the EIN3 transcription factor (17). The 35S-EIN2S645A lines conferred a similar phenotype as 35S-EIN2WT in both the presence and absence of ACC. In contrast, 35S-EIN2S924A and 35S-EIN2AA conferred the distinct stunted phenotype even when grown without ACC treatment (Fig. 2D). Therefore, EIN2S924A and EIN2AA appear to be activated to a greater extent than EIN2WT and EIN2S645A.
Preventing EIN2 Phosphorylation on Ser645 and Ser924 Results in Constitutive Localization of the EIN2 C Terminus in the Nucleus.
We next addressed how the EIN2 protein in its unphosphorylated state is connected to downstream signaling. The putative nuclear localization sequence in EIN2 (19) raised the possibility that the EIN2 C terminus could move into the nucleus if cleaved from the N-terminal, integral-membrane portion of EIN2; we examined this using full-length EIN2 fused to GFP or YFP. Interestingly, the amount of EIN2-GFP in the microsomal fraction was greatly reduced when stably transformed seedlings were treated with ethylene (Fig. 3A), perhaps due to cleavage or degradation of the C terminus. EIN2-GFP protein was found predominantly in the nucleus after ethylene treatment both in onion cells and stably transformed Arabidopsis, whereas without ethylene treatment, the signal was at the ER (Fig. 3 B and C). In contrast, a YFP tag at the N terminus of EIN2 remained at the ER membrane with or without ethylene treatment (Fig. 3B). These data suggest that perception of ethylene leads to a proteolytic cleavage of EIN2, allowing the C-terminal domain to localize to the nucleus while the N terminus remains at the ER membrane.
Fig. 3.
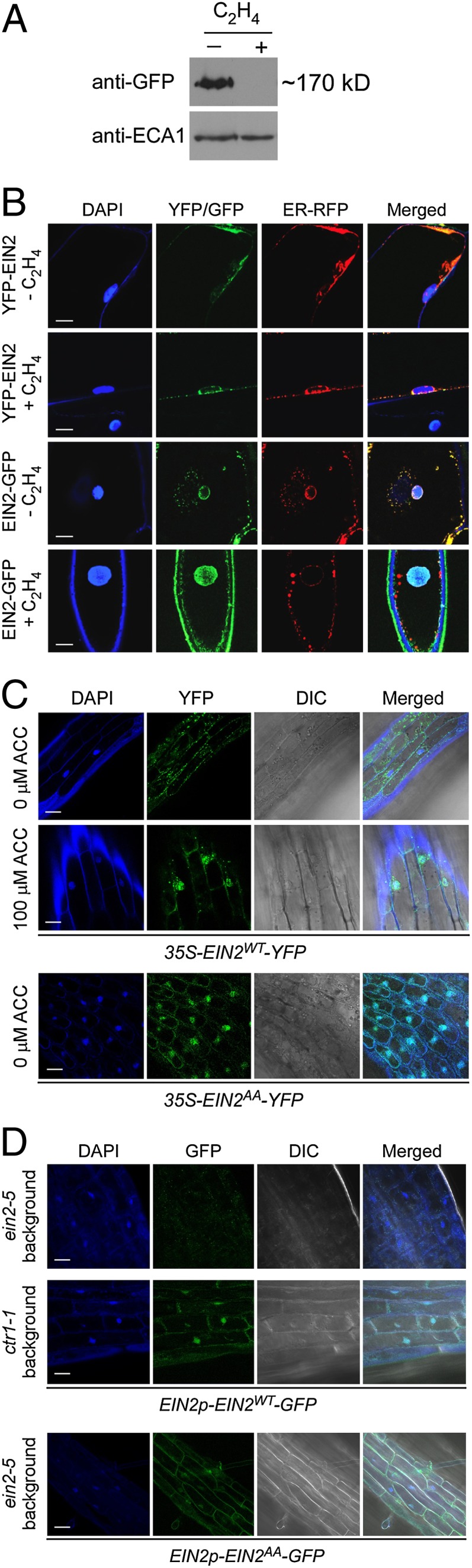
EIN2AA results in constitutive nuclear localization of the EIN2 C-terminal domain. (A) Western blot of EIN2-GFP in the microsomal fraction. Four-d-old dark-grown ein2-5 seedlings stably transformed with EINp-EIN2-GFP were treated without or with ethylene gas for 3 h. The microsomal fraction was analyzed by Western blotting using an anti-GFP antibody. EIN2 (without GFP) is 141 kDa. ECA1 (41) was a loading control. (B) Differential localization of the N and C termini of EIN2 in onion epidermal cells. 35S-YFP-EIN2 and 35S-EIN2-GFP were delivered into onion cells by particle bombardment and fluorescence was visualized by confocal microscopy. The YFP tag and the ER marker (ER-RFP) colocalized both without and with 3–6 h ethylene treatment. The GFP tag colocalized with the ER marker without ethylene treatment, but predominantly colocalized with the nuclear DAPI stain after ethylene treatment. (C) Ethylene-responsive nuclear localization of EIN2WT-YFP and constitutive nuclear localization of EIN2AA-YFP in Arabidopsis hypocotyl cells. Four-d-old dark-grown Col-0 seedlings stably transformed with 35S-EIN2WT-YFP were treated for 3 h with H2O (0 µM ACC) or 100 µM ACC, and then examined by confocal microscopy. Col-0 seedlings transformed with 35S-EIN2AA-YFP were treated for 3 h with only H2O (0 µM ACC). (D) Constitutive nuclear localization of EIN2WT-GFP and EIN2AA-GFP in hypocotyl cells of ctr1-1 and ein2-5, respectively. Four-d-old dark-grown seedlings stably transformed with EIN2p-EIN2WT-GFP or EIN2p-EIN2AA-GFP without ethylene treatment were examined by confocal microscopy. (Scale bar: B–D, 20 μm.)
In wild-type Arabidopsis stably transformed with 35S-EIN2AA-YFP, we detected the YFP signal in the nucleus even without ethylene treatment (Fig. 3C), indicating that the constitutive phenotypes conferred by EIN2AA are correlated with constitutive nuclear localization of the EIN2 C terminus. When we transformed ein2-5 and ctr1-1 mutants with EIN2p-EIN2WT-GFP and localized the GFP signal without ethylene treatment, there was a nuclear signal for ctr1-1, indicating that the loss of CTR1 phosphorylation results in nuclear localization of the C terminus of wild-type EIN2 (Fig. 3D). In the ein2-5 background, there was weak fluorescence with no obvious signal in the nucleus, whereas expression of EIN2p-EIN2AA-GFP produced a GFP signal in the nucleus even without ethylene treatment (Fig. 3D).
Discussion
Since the genetic discovery of CTR1 and EIN2 as critical regulators of ethylene hormone signaling, a major unanswered question has been how their encoded proteins mediate ethylene signaling. This has left a significant gap in our understanding of the ethylene signaling pathway from the ethylene receptor–CTR1 complex at the ER membrane to EIN2, and from EIN2 to the nucleus. Recent mass spectrometry analysis provided evidence of four in vivo phosphorylation sites in the EIN2 C-terminal domain (20). The phosphopeptides were observed only in the absence of ethylene treatment, leading us to hypothesize that CTR1 is responsible for phosphorylating EIN2 in vivo, because this is consistent with CTR1 being an active kinase in the absence but not the presence of ethylene. Here, we have demonstrated that CTR1 directly phosphorylates EIN2 in vitro on the same four sites that were identified in vivo, plus two additional sites. Of particular interest were Ser645 and Ser924, because both had displayed differential phosphorylation in untreated vs. ethylene-treated seedlings in vivo (20), both are conserved in EIN2 homologs (20), both were efficiently phosphorylated by CTR1 in vitro, and Ala substitutions at these residues substantially reduced CTR1 phosphorylation of EIN2 in vitro. The large reduction in the in vitro phosphorylation of EIN2-CAA compared with EIN2-CWT indicated that Ser645 and Ser924 are major sites in EIN2 that are phosphorylated by CTR1. In stably transformed Arabidopsis, the Ser645Ala and Ser924Ala substitutions conferred constitutive ethylene responses and constitutive nuclear localization of the EIN2 C terminus. Our finding that both N-terminal and C-terminal tags on EIN2 localize to the ER without ethylene treatment, whereas only the C-terminal tag appears in the nucleus after ethylene treatment (Fig. 3B), suggests that there is a proteolytic cleavage of the EIN2 C-terminal domain.
Our functional analyses in planta indicated that Ser924 plays a larger role in ethylene signaling compared with Ser645 based on side-by-side analyses of transformants expressing similar transgene levels. The absence of Ser924 phosphorylation might promote proteolytic cleavage of EIN2 or play a role in movement of the EIN2 C terminus to the nucleus, whereas other sites phosphorylated by CTR1 might play supporting roles or regulate EIN2 turnover.
Overexpression of the EIN2 genomic sequence from start to stop codon in wild-type Arabidopsis yielded a majority (60–75%) of transgenic lines that were insensitive to ethylene, suggesting cosuppression of EIN2, whereas the remaining transformed lines yielded different degrees of constitutive ethylene response depending on the construct. Conceivably, high levels of EIN2 expression result in constitutive response phenotypes due to excess EIN2 escaping phosphorylation by CTR1 and consequently activating downstream responses. This activation might occur by enhancing EIN3 activity and/or increasing EIN3 protein levels, because the phenotype conferred by 35S-EIN2WT shown in Fig. 2D is very similar to that conferred by 35S-EIN3 (17).
Based on our findings, we propose a model in which CTR1 phosphorylates EIN2 at Ser645 and Ser924 (and other sites) at the ER membrane when ethylene is absent, causing EIN2 to be inactive. In the presence of ethylene, CTR1 is inactive, and the subsequent lack of phosphorylation on EIN2 results in cleavage of EIN2 and movement of the EIN2 C terminus into the nucleus (Fig. 4). The absence of phosphorylation on the EIN2 C-terminal domain may be the signal for its proteolytic release from the membrane-bound N-terminal domain, freeing the C terminus to migrate into the nucleus. An alternative but not mutually exclusive model is that phosphorylation acts as a signal to target EIN2 for degradation by the 26S proteasome (16), in which case the absence of phosphorylation could inhibit degradation, indirectly resulting in nuclear localization. Once in the nucleus, the EIN2 C terminus possibly regulates the downstream transcription factors, EIN3 and EIL1, either directly or via other components. Several well-known ER membrane- or plasma membrane-localized signaling proteins in animals also undergo cleavage and translocation to the nucleus, such as sterol regulatory element binding proteins (21), Notch (22), and Tra2 (23), and in these cases, the nuclear-localized forms are transcription factors themselves or control other proteins involved in transcription.
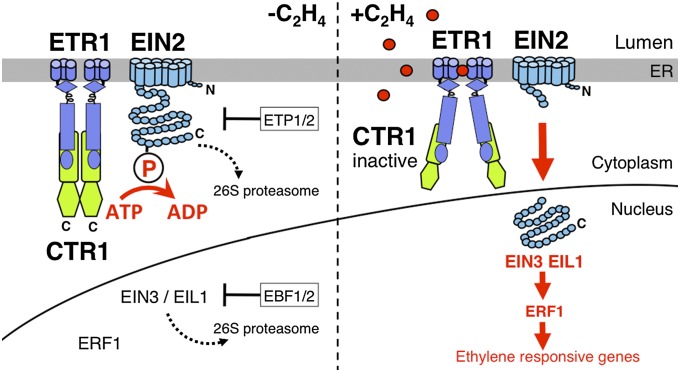
Fig. 4.
Model of ethylene signaling. In the absence of ethylene (Left), the ethylene receptors (e.g., ETR1) at the ER membrane activate the CTR1 protein kinase, a dimer (42), which phosphorylates the C-terminal domain of EIN2, preventing its nuclear localization. Without ethylene, EIN2 is targeted for 26S proteasomal degradation by F-box proteins ETP1/2 (16). Transcription factors EIN3/EIL1 are also targeted for degradation by F-box proteins EBF1/2 (17). In the presence of ethylene (Right), the receptors are inactivated and therefore the CTR1 kinase is no longer active. The absence of phosphorylation on EIN2 results in EIN2 C terminus being cleaved and localizing to the nucleus where it can activate the downstream transcriptional cascade.
Although the molecular functions of the N-terminal and C-terminal domains of EIN2 have yet to be elucidated, the biochemical connection between EIN2 and the so-called Raf-like kinase CTR1, together with insight into how the ethylene signal is relayed from the ER to the nucleus, fills a long-standing gap in our understanding of the ethylene signaling pathway. Though the possibility of a MAPK cascade in ethylene signaling still remains, our findings demonstrate that the regulation of ethylene responses by CTR1 can occur without such a cascade, although we have not ruled out that CTR1 phosphorylates other targets, as proposed by Yoo et al. (24).
While this paper was under review, a paper on the same topic was published by Qiao et al. (25), who similarly demonstrated that EIN2 is differentially phosphorylated at Ser645, and that the EIN2 C terminus migrates to the nucleus after ethylene treatment. In contrast to the results presented here, Qiao et al. (25) reported a strong constitutive ethylene-response phenotype conferred by EIN2S645A. This discrepancy could be the result of a high-level expression of the EIN2 transgene in their studies. Qiao et al. (25) did not uncover or examine phosphorylation of Ser924 of EIN2. Nevertheless, both papers together advance a mechanistic understanding of key steps in the ethylene signal transduction pathway.
Materials and Methods
Plant Materials and Growth Conditions.
The A. thaliana Columbia ecotype (Col-0) was used as the wild type. White onions were obtained from a grocery store. Arabidopsis and Nicotiana benthamiana plants were grown in soil under 16-h light/8-h dark in controlled environment chambers at 22 °C under white fluorescent light. For the seedling assay, seeds were sown on Murashige and Skoog (MS) medium containing 0.8% (wt/vol) agar supplemented with either 5 µM aminoethoxyvinylglycine (AVG) (Sigma Aldrich) or 20 µM ACC (Sigma Aldrich). Following a 3D stratification at 4 °C, the plates were placed in light for 5–6 h and then germinated in the dark for 4 d at 20 °C. For treatment with ethylene gas (Specialty Gases of America), the sown seeds were placed in custom-made light- and gas-tight Plexiglas chambers (Plas-Labs, Inc.) into which ethylene gas was injected to the indicated concentration. Measurements of hypocotyl length and rosette diameter were made using ImageJ software (http://rsbweb.nih.gov/ij/) on digital photographs.
Purification of His6-CTR1-KDWT and His6-CTR1-KDD694E Using Recombinant Virus.
The generation of recombinant virus is described in SI Materials and Methods. Sf9 insect cells grown as an adherent culture in Grace’s insect media were transfected with a P3 stock of recombinant virus expressing His6-CTR1-KDWT and His6-CTR1-KDD694E and incubated for 72 h at 28 °C. Cells were harvested and washed briefly with 1× PBS and resuspended in lysis buffer [20 mM Tris (pH 7.5), 150 mM NaCl, 10 mM NaF, 1% Triton X-100, 1 mM PMSF, 1× Roche Complete Protease Inhibitor mixture]. Supernatant was collected, cleared by centrifugation (16,000 × g × 20 min) and loaded onto a nickel resin (Ni-NTA His⋅Bind Superflow Resin; Novagen). The column was washed with 10 column-volumes of washing buffer I [20 mM Tris (pH 7.5), 300 mM NaCl, 10 mM NaF, 10% (vol/vol) glycerol, 1 mM PMSF, 1× Roche Complete Protease Inhibitor mixture] followed by 20 column-volumes of washing buffer II [20 mM Tris (pH 7.5), 300 mM NaCl, 10 mM NaF, 10% (vol/vol) glycerol, 10 mM imidazole, 1 mM PMSF, 1× Roche Complete Protease Inhibitor mixture]. Recombinant proteins were eluted using two column-volumes of elution buffer [20 mM Tris (pH 7.5), 300 mM NaCl, 10 mM NaF, 10% (vol/vol) glycerol, 200 mM imidazole, 1 mM PMSF, 1× Roche Complete Protease Inhibitor mixture]. Protein concentration was measured using a Bradford assay.
Expression and Purification of Recombinant EIN2 Proteins.
The mutagenesis of EIN2 is described in SI Materials and Methods. The EIN2WT (14) and EIN2AA expression plasmids were transformed into the E. coli strain BL21 for protein expression. Protein expression was induced in cultures grown at 30 °C to an A600 of 0.6 by the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside and then incubated for an additional 48 h at 18 °C. Cells were harvested and resuspended in extraction buffer [20 mM Tris (pH 7.5), 150 mM NaCl, 10 mM NaF, 1 mM PMSF, 0.5 mM DTT, 1× Roche Complete Protease Inhibitor mixture], followed by sonication and centrifugation (16,000 × g × 30 min). The His-tagged proteins were purified as described above.
In Vitro Kinase Assay.
A total of 20 ng purified His6-CTR1-KDWT or His6-CTR1-KDD694E protein was incubated with 100 ng of His6-EIN2WT-His6 or His6-EIN2AA-His6 in kinase reaction buffer [50 mM Tris (pH 7.5), 10 mM MgCl2, 1× Roche Complete Protease Inhibitor mixture, 1 µCi [γ-32P]ATP] for 30 min at room temperature. After incubation, reactions were terminated by boiling in 6× Laemmli SDS sample buffer for 3 min. Samples were subjected to SDS/PAGE, dried, and visualized by autoradiography.
Phosphorylation of EIN2 Peptides by His6-CTR1-KDWT.
The kinase assay described above was carried out using 20 ng of purified His6-CTR1-KDWT and 400 µM of each EIN2 peptide (University of North Carolina High-Throughput Peptide Synthesis and Array Facility, Chapel Hill, NC). Reactions were terminated by adding 2 vol of ice-cold 75-mM phosphoric acid, and the samples were then spotted onto a 96-well unifilter microplate (Whatman). The unifilter microplate was then washed several times with ice-cold 75-mM phosphoric acid to remove unincorporated [γ-32P]ATP. The plate was washed with acetone and dried, and the incorporated radioactivity determined with a scintillation counter. The reactions were performed in triplicate. For analysis of enzyme kinetics, 20 ng of His6-CTR1-KDWT protein was incubated with increasing amounts of EIN2 peptide1 in kinase reaction buffer and analyzed as above.
Evaluation of In Vitro Phosphorylation of EIN2 by Multi-Stage Mass Spectrometry (MS2 and MS3).
His6-EIN2WT-His6 was reacted with His6-CTR1-KDWT and ATP (referred to as EIN2_CTR1) and separated by SDS/PAGE (Fig. S1A). As controls, His6-CTR1-KDWT and His6-EIN2WT-His6 were reacted by themselves. Coomassie-stained His6-EIN2WT-His6 bands were excised and destained, and the proteins were reduced, carboxyamidomethylated, and digested with trypsin as previously described (26). Peptides from each sample were separated on a C18 reverse-phase column (100 × 0.18 mm BioBasic-18) using a linear gradient from 5% to 40% acetonitrile/0.1% formic acid at a flow rate of 3 mL/min, which was controlled by an Accela HPLC pump (Thermo Fisher Scientific). The eluent was electrosprayed at 3.5 kV directly into the orifice of an LTQ-Orbitrap XL mass spectrometer (27) (Thermo Fisher Scientific) controlled by Xcalibur 2.0.7 software (Thermo Fisher Scientific). A parent-ion scan was performed in the Orbitrap over the range of 400 to 1,600 m/z at 30,000 resolution, 1 million automatic gain control (AGC), 750-ms ion injection time, and one microscan. Lock mass was enabled (28). Data-dependent MS2 and MS3 were performed in the linear ion trap with 10,000 AGC and 100-ms ion injection times with three microscans. MS2 was performed on the five most intense MS ions, and MS3 was triggered if one of the top three MS2 ions corresponded with neutral loss of 98.0, 49.0, and 32.7 Da for +1, +2, and +3 charged ions, respectively (29). Minimum signals were 5,000 and 500 respectively. An isolation width of 2 m/z and normalized collision energy of 35% were used for MS2 and MS3. Dynamic exclusion was used with a repeat count of one 30-s repeat duration, a list of 50, list duration of 3 min, and exclusion mass width of ±0.7 Da.
MS2 and MS3 spectrum data files were separately extracted from the raw data with BioWorks 3.3.1 (Thermo Fisher Scientific) using the parameters 600−4,500 mass range, zero group scan, one minimum group count, and five minimum ion counts. There were 2,317 MS2 spectra from the EIN2 sample and 2,506 MS2 and 65 MS3 spectra from the EIN2_CTR1 sample. Sets of MS2 and MS3 spectra were searched independently with Mascot 2.4.0 (30). For MS2 spectra, search parameters were for tryptic digests, one possible missed cleavage, fixed amino acid modification [+57, C], variable amino acid modifications for phosphorylation [+80, S, T], monoisotopic mass values, ±10 ppm parent ion mass tolerance, ±0.8 Da fragment ion mass tolerance, and 13C = 1 enabled. For MS3 spectra, search parameters were for tryptic digests, one possible missed cleavage, fixed amino acid modification [+57, C], variable amino acid modifications [−18, S, T] and [+80, S, T], monoisotopic mass values, ±1.5 Da parent ion mass tolerance, and ±0.8 Da fragment ion mass tolerance. The searched database consisted of version 8.0 of the A. thaliana genome protein reference database (ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/; 32,825 records) appended with a list of common contaminants (32,997 records total). Peptide-spectrum matches with Expect values less than 0.05 were accepted. Phosphorylation of S and T were evaluated based on the Mascot peptide-spectrum match assignments. It was discovered by manual examination that BioWorks assigned an incorrect parent ion mass to the MS2 spectrum EIN2_CTR1_B5.1556.1556.2. Therefore, Mascot Distiller was used to reextract the peak list from the raw data, which resulted in correct parent ion mass assignment for the EIN2_CTR1_B5.1556.1556.2 spectrum reported here.
To evaluate phosphorylated peptides and phosphosite localization, we used the Mascot Delta score (31) and a decision tree (20) that considered the number of moieties and the number of potential sites for phosphorylation, the Mascot Ions score magnitude and the Expect value, the peptide charge state (32), +80 Da mass shifts (meaning no neutral loss and phosphorylation mass gain at an amino acid position), phosphoric acid neutral losses (leading to −18 Da mass loss at S/T), and corroborating neutral loss-generated MS3 spectra (33). This information was used to assign high, moderate, or low confidence for phosphorylation positioning to the specific peptides listed in Table S1.
Generation of Stably Transformed Arabidopsis.
Cloning of all EIN2 binary plasmid constructs and site-directed in vitro mutagenesis are described in SI Materials and Methods. The binary plasmids were transformed into Agrobacterium tumefaciens strain GV3101, and then Arabidopsis plants were transformed using the floral dip method (34). Transformants were selected with the herbicide Basta (Bayer Crop Science) or hygromycin. Homozygous lines were obtained in the T3 generation for the wild-type EIN2p-EIN2 and EIN2p-EIN2-GFP in ein2-5 and mutant versions of EIN2p-EIN2 in ein2-5. The T2 generation was analyzed for transformants carrying 35S-EIN2WT-GFP, 35S-EIN2AA-GFP, EIN2p-EIN2WT-GFP, EIN2p-EIN2AA-GFP, and wild-type and mutant versions of 35S-EIN2 without GFP.
Yeast Two-Hybrid Assay.
Cloning of the EIN2 C-terminal domain in pACTII is described in SI Materials and Methods. The resulting EIN2 prey clone was paired with an existing CTR1 kinase domain clone (in bait vector pLexA) and tested along with negative controls as in Clark et al. (35). To select for interaction, the medium lacked histidine and contained 2.5 mM 3-aminotriazole (3-AT).
BiFC in Tobacco Leaf Epidermal Cells.
BiFC constructs are described in SI Materials and Methods . For Agrobacterium infiltration of tobacco leaves, we followed the protocol of Schütze et al. (36) using Agrobacterium strain C58C1. As a negative control, EIN2-cYFP was paired with ECA1 (an ER-localized Ca2+-ATPase)-nYFP described in Dong et al. (37).
Subcellular Localization of EIN2 in Onion Epidermal Cells.
Plasmid constructs are described in SI Materials and Methods. For particle bombardment, we used a Helios Gene Gun (BioRad) as described (38). Bullets were prepared for each of the above constructs together with an ER marker, ER-rb (Arabidopsis Biological Resource Center) (39).
Real-Time Quantitative PCR Analysis of EIN2 and ERF1 mRNA Abundance.
Total RNA was prepared from 4-wk-old soil-grown plants or 4-d-old seedlings using RNeasy Plant Mini Kit (QIAGEN) and reverse transcribed using RevertAid First Strand cDNA Synthesis Kit (Fermentas) according to the manufacturers’ instructions. Quantitative PCR was performed with SsoFast EvaGreen Supermix (Bio-Rad) on a CFX96 Real-Time System (Bio-Rad) using the gene-specific primers listed in Table S2. Three technical replicates were carried out for each sample. Relative expression levels in each cDNA sample were obtained by normalization to the reference gene GAPDH.
Western Blotting.
Four-day-old dark-grown seedlings (homozygous EINp-EIN2-GFP in ein2-5) grown on MS agar containing AVG were treated with or without 100 ppm ethylene gas for 3 h. The microsomal fraction was prepared as described (40), separated by 7.5% SDS/PAGE, transferred into PVDF membrane (BioRad) by wet-tank transfer, and subjected to immunoblotting with a 1:1,500 dilution of anti-GFP antibody (Santa Cruz Biotechnology). The same membrane was stripped and immunoblotted with a 1:3,000 dilution of anti-ECA1 antibody (kindly provided by H. Sze, University of Maryland, College Park, MD) as a loading control.
Fluorescence Microscopy.
Imaging of GFP, YFP, and RFP was conducted under a laser-scanning confocal microscope (Zeiss LSM710). Samples were directly mounted on glass slides in water. For BiFC, three replicate experiments were carried out using 15–20 tobacco leaves per experiment. For onion epidermal cells, Petri dishes containing onion peels were placed in the light- and gas-tight chambers described above, and for ethylene treatment, ethylene gas was injected into the chamber at a final concentration of 167 ppm. Onion peels were stained with 20 µg/mL DAPI solution for 30 min. Three replicates were carried out for each construct using 12 onion pieces per replicate. For Arabidopsis transformed with 35S-EIN2-YFP, 4-d-old dark-grown seedlings were treated with 0 µM or 100 µM ACC (dissolved in water) for 3 h in combination with DAPI [5 µg/mL in H2O plus 0.2% (vol/vol) Triton X-100] for at least 1 h and rinsed with water before examination. Five independent transgenic lines were examined using five or more seedlings per construct, and ACC treatment experiments were repeated at least three times. For EIN2p-EIN2-GFP transformants, 4-d-old dark-grown seedlings were treated only with DAPI; for EIN2p-EIN2WT, 15 and 12 independent lines in ein2-5 and ctr1-1, respectively. For EIN2p-EIN2AA, we examined 28 independent lines in ein2-5. At least five seedlings were examined per line.
Supplementary Material
Acknowledgments
We thank Jason Shockey and Chris McClellan for yeast two-hybrid constructs; Mandy Kendrick for the CTR1 cDNA in pDONR221; Ruiqiang Chen for primer design; Klaus Harter for BiFC vectors; Amy Beaven for microscopy assistance; Heven Sze for the anti-ECA1 antibody; and Plant Bioscience Limited for Agrobacterium strain C58C1 (pCH32) and p19 plasmid. This work was supported by National Science Foundation Grants MCB-0923796 (to C.C.) and MCB-1021704 (to J.K.); a Specific Cooperative Agreement between the University of Maryland and the Agricultural Research Service of the US Department of Agriculture; the Deutsche Forschungsgemeinschaft (G.G.); and the Maryland Agricultural Experiment Station (C.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214848109/-/DCSupplemental.
References
- 1.Bleecker AB, Kende H. Ethylene: A gaseous signal molecule in plants. Annu Rev Cell Dev Biol. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Abeles FB, Morgan PW, Saltveit ME., Jr . Ethylene in Plant Biology. 2nd Ed. San Diego: Academic; 1992. [Google Scholar]
- 3.Alonso JM, Ecker JR. The ethylene pathway: A paradigm for plant hormone signaling and interaction. Sci STKE. 2001;2001(70):re1. doi: 10.1126/stke.2001.70.re1. [DOI] [PubMed] [Google Scholar]
- 4.Schaller GE, Kieber JJ, Shiu SH. Two-component signaling elements and histidyl-aspartyl phosphorelays. Arabidopsis Book. 2008;6:e0112. doi: 10.1199/tab.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 6.Gao Z, et al. Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J Biol Chem. 2003;278(36):34725–34732. doi: 10.1074/jbc.M305548200. [DOI] [PubMed] [Google Scholar]
- 7.Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ. Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J. 2003;33(2):221–233. doi: 10.1046/j.1365-313x.2003.01620.x. [DOI] [PubMed] [Google Scholar]
- 8.Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72(3):427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Q, Guo HW. Paradigms and paradox in the ethylene signaling pathway and interaction network. Mol Plant. 2011;4(4):626–634. doi: 10.1093/mp/ssr042. [DOI] [PubMed] [Google Scholar]
- 10.Hahn A, Harter K. Mitogen-activated protein kinase cascades and ethylene: Signaling, biosynthesis, or both? Plant Physiol. 2009;149(3):1207–1210. doi: 10.1104/pp.108.132241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ecker JR. Reentry of the ethylene MPK6 module. Plant Cell. 2004;16(12):3169–3173. [Google Scholar]
- 12.Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284(5423):2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 13.Roman G, Lubarsky B, Kieber JJ, Rothenberg M, Ecker JR. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics. 1995;139(3):1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bisson MMA, Bleckmann A, Allekotte S, Groth G. EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem J. 2009;424(1):1–6. doi: 10.1042/BJ20091102. [DOI] [PubMed] [Google Scholar]
- 15.Bisson MMA, Groth G. New insight in ethylene signaling: Autokinase activity of ETR1 modulates the interaction of receptors and EIN2. Mol Plant. 2010;3(5):882–889. doi: 10.1093/mp/ssq036. [DOI] [PubMed] [Google Scholar]
- 16.Qiao H, Chang KN, Yazaki J, Ecker JR. Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 2009;23(4):512–521. doi: 10.1101/gad.1765709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.An F, et al. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell. 2010;22(7):2384–2401. doi: 10.1105/tpc.110.076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solano R, Stepanova A, Chao Q, Ecker JR. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998;12(23):3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bisson MMA, Groth G. New paradigm in ethylene signaling: EIN2, the central regulator of the signaling pathway, interacts directly with the upstream receptors. Plant Signal Behav. 2011;6(1):164–166. doi: 10.4161/psb.6.1.14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen RQ, et al. Proteomic responses in Arabidopsis thaliana seedlings treated with ethylene. Mol Biosyst. 2011;7(9):2637–2650. doi: 10.1039/c1mb05159h. [DOI] [PubMed] [Google Scholar]
- 21.Sakai J, et al. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85(7):1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 22.Fortini ME. Notch signaling: The core pathway and its posttranslational regulation. Dev Cell. 2009;16(5):633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Lum DH, Kuwabara PE, Zarkower D, Spence AM. Direct protein-protein interaction between the intracellular domain of TRA-2 and the transcription factor TRA-1A modulates feminizing activity in C. elegans. Genes Dev. 2000;14(24):3153–3165. [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature. 2008;451(7180):789–795. doi: 10.1038/nature06543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao H, et al. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science. 2012;338(6105):390–393. doi: 10.1126/science.1225974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper B, Eckert D, Andon NL, Yates JR, Haynes PA. Investigative proteomics: Identification of an unknown plant virus from infected plants using mass spectrometry. J Am Soc Mass Spectrom. 2003;14(7):736–741. doi: 10.1016/S1044-0305(03)00125-9. [DOI] [PubMed] [Google Scholar]
- 27.Makarov A, et al. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem. 2006;78(7):2113–2120. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- 28.Olsen JV, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4(12):2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Beausoleil SA, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA. 2004;101(33):12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 31.Savitski MM, et al. Confident phosphorylation site localization using the Mascot Delta Score. Mol Cell Proteomics. 2011;10(2) doi: 10.1074/mcp.M110.003830. M110.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aguiar M, Haas W, Beausoleil SA, Rush J, Gygi SP. Gas-phase rearrangements do not affect site localization reliability in phosphoproteomics data sets. J Proteome Res. 2010;9(6):3103–3107. doi: 10.1021/pr1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng J, Garrett WM, Naiman DQ, Cooper B. Correlation of multiple peptide mass spectra for phosphoprotein identification. J Proteome Res. 2009;8(11):5396–5405. doi: 10.1021/pr900596u. [DOI] [PubMed] [Google Scholar]
- 34.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 35.Clark KL, Larsen PB, Wang X, Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA. 1998;95(9):5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schütze K, Harter K, Chaban C. Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Methods Mol Biol. 2009;479:189–202. doi: 10.1007/978-1-59745-289-2_12. [DOI] [PubMed] [Google Scholar]
- 37.Dong CH, et al. Molecular association of the Arabidopsis ETR1 ethylene receptor and a regulator of ethylene signaling, RTE1. J Biol Chem. 2010;285(52):40706–40713. doi: 10.1074/jbc.M110.146605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hollender CA, Liu Z. Bimolecular fluorescence complementation (BiFC) assay for protein-protein interaction in onion cells using the helios gene gun. J Vis Exp. 2010;40(40):1963. doi: 10.3791/1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51(6):1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 40.Dong CH, Rivarola M, Resnick JS, Maggin BD, Chang C. Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. Plant J. 2008;53(2):275–286. doi: 10.1111/j.1365-313X.2007.03339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang F, Cunningham KW, Harper JF, Sze H. ECA1 complements yeast mutants defective in Ca2+ pumps and encodes an endoplasmic reticulum-type Ca2+-ATPase in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94(16):8579–8584. doi: 10.1073/pnas.94.16.8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayerhofer H, Panneerselvam S, Mueller-Dieckmann J. Protein kinase domain of CTR1 from Arabidopsis thaliana promotes ethylene receptor cross talk. J Mol Biol. 2012;415(4):768–779. doi: 10.1016/j.jmb.2011.11.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.