Abstract
Human mena (hMENA), a member of the actin cytoskeleton regulators Ena/VASP, is overexpressed in high-risk preneoplastic lesions and in primary breast tumors and has been identified as playing a role in invasiveness and poor prognosis in breast cancers that express HER2. Here we identify a unique isoform, hMENAΔv6, derived from the hMENA alternative splicing program. In an isogenic model of human breast cancer progression, we show that hMENA11a is expressed in premalignant cells, whereas hMENAΔv6 expression is restricted to invasive cancer cells. “Reversion” of the malignant phenotype leads to concurrent down-regulation of all hMENA isoforms. In breast cancer cell lines, isoform-specific hMENA overexpression or knockdown revealed that in the absence of hMENA11a, overexpression of hMENAΔv6 increased cell invasion, whereas overexpression of hMENA11a reduced the migratory and invasive ability of these cells. hMENA11a splicing was shown to be dependent on the epithelial regulator of splicing 1 (ESRP1), and forced expression of ESRP1 in invasive mesenchymal breast cancer cells caused a phenotypic switch reminiscent of a mesenchymal-to-epithelial transition (MET) characterized by changes in the cytoskeletal architecture, reexpression of hMENA11a, and a reduction in cell invasion. hMENA-positive primary breast tumors, which are hMENA11a-negative, are more frequently E-cadherin low in comparison with tumors expressing hMENA11a. These data suggest that polarized and growth-arrested cellular architecture correlates with absence of alternative hMENA isoform expression, and that the hMENA splicing program is relevant to malignant progression in invasive disease.
Keywords: ENAH, EMT, splice variants
MENA along with VASP and EVL comprise the Ena/VASP family of actin regulatory proteins, which modulate cell adhesion and migration by antagonizing actin capping proteins (1, 2), bundling actin filaments, and nucleating and extending filopodia (1–7). The MENA gene encodes the 570-aa hMENA protein and different alternative splicing-derived isoforms, often expressed in a tissue-specific manner, have been reported in human (8, 9) and mouse (1, 10, 11). The neuronal variant is characterized by an extended exon 6 (1, 8), the spleen-specific variant lacks the proline-rich region (10), and an invasion-specific splice variant (MENAINV), with an additional exon just after the EVH1 domain, has been shown to regulate chemotaxis in mouse and rat mammary tumor cells (12). Previously, we characterized hMENA11a, an epithelial-associated hMENA splice variant with an additional exon (exon 11a) (9). hMENA11a, expressed in human pancreatic (13) and breast cancer cells (9), is phosphorylated downstream of HER2 and EGFR following EGF and NRG1 treatment and in turn influences the mitogenic signals of these receptors in luminal breast cancer cell lines (9, 14).
hMENA isoforms, undetectable in normal breast tissue, are progressively expressed in premalignant breast lesions, suggesting that their presence could be used as an early stage marker of breast neoplasia in women at a higher risk of malignancy (15). Recently, Warzecha et al. implicated the epithelial splicing regulatory proteins 1 and 2 (ESRP1 and ESRP2) as central coordinators of an alternative splicing network that underlies the epithelial-to-mesenchymal transition (EMT) in breast cancer through targeting of many genes, including hMENA (16, 17), involving MENA splicing in breast cancer progression.
Here we describe the molecular cloning and characterization of a unique hMENA splice variant lacking the internal exon 6 (hMENAΔv6). Further, we provide evidence that ESRPs regulate epithelial-specific hMENA splicing and also show that alternative splicing of hMENA regulates the morphology and invasion of cancer cells. Using an isogenic model of breast cancer progression, HMT-3522 (18–21), we show that neither hMENA11a nor hMENAΔv6 are detectable in the nonmalignant breast cells (S1), unless treated with EGF. hMENA11a appears however in preneoplastic cells that have self-sufficiency for EGFR activity (22, 23), and hMENAΔv6 is restricted to invasive but nonmetastatic HMT-3522–T4-2 cells. In a panel of breast cancer cell lines we show that the two hMENA isoforms, hMENA11a and hMENAΔv6, are expressed in cells with epithelial or mesenchymal phenotypes, respectively. We further show that these two isoforms have antagonistic roles in cell invasion and migration. Finally and of importance, alternative splicing of hMENA also occurs within primary breast tumors. hMENA11a expressing tumors, have a high proliferation index (Ki67 > 15%), whereas pan-hMENA–positive and hMENA11a-negative tumors have a higher frequency of down-regulated E-cadherin. These results establish that hMENA splicing influences cell morphology and invasion, and that isoform-specific hMENA detection may offer a useful signature for the diagnosis and prognosis of early stage breast cancer.
Results
Molecular Cloning and Characterization of a Unique hMENA Splice Variant.
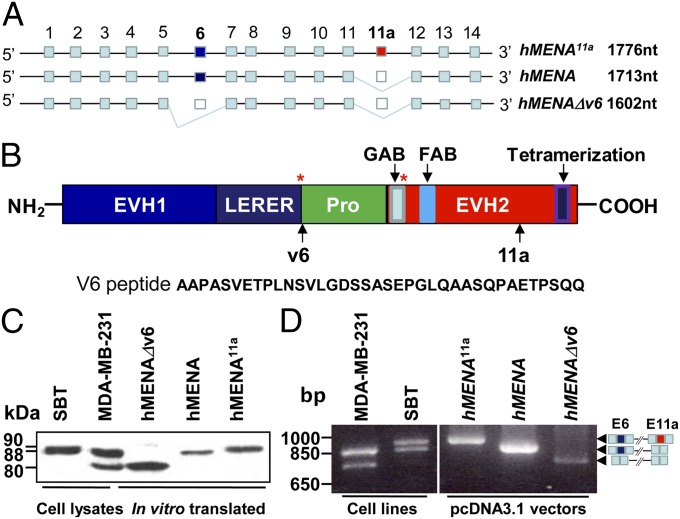
Using mRNA from MDA-MB-231 cells and standard cDNA cloning techniques we identified a unique 1,602 nucleotide variant of hMENA. We named this isoform hMENAΔv6 (GenBank accession no. EU255274), because it lacks the exon 6-encoded 37-aa internal peptide located between the LERER and the proline-rich region of hMENA (Fig. 1 A and B). The absence of this peptide brings the LERER domain and PKA Ser phosphorylation site (Ser-236 in mice) with the proline-rich domain closer together (Fig. 1B). No mouse or human MENA sequences, lacking exon 6, have been described in GenBank; however, a search of the EST database has revealed two mouse Mena sequences with a deleted exon 6, one from embryonic stem cells and one from a mammary infiltrating ductal carcinoma. We found also a human EST from a duodenal adenocarcinoma cell line (Fig. S1A). hMENAΔv6 shares 88% identity with Rattus norvegicus ENAH, with the divergences mainly located in the LERER domain; 87% with the AVENAII from Gallus gallus, and 77% with the ENABLED protein from Xenopus laevis (Fig. S1B). The Western blot (WB) analysis of hMENA11a- (hMENA11a variant includes an exon absent in hMENA), hMENA-, and hMENAΔv6- in vitro translated proteins, using an antibody that recognizes all hMENA isoforms (pan-hMENA) (Fig. 2A), showed that the proteins migrate with apparent molecular weights of 90 kDa (hMENA11a), 88 kDa (hMENA), and 80 kDa (hMENAΔv6) (Fig. 1C). The two in vitro translated hMENA and hMENAΔv6 isoforms appear to correspond to the bands revealed by WB analysis in MDA-MB-231 tumor cell protein extracts (Fig. 1C). RT-PCR experiments performed on SBT and MDA-MB-231, the breast cancer cell lines we used to clone hMENA11a and hMENAΔv6, respectively, revealed that, hMENA was expressed in both. Differently, hMENA11a was expressed only in the luminal breast cancer cell line SBT, whereas hMENAΔv6 only in the basal MDA-MB-231 (Fig. 1D).
Fig. 1.
Molecular cloning and characterization of the unique hMENAΔv6 splice variant. (A) Diagrammatic representation of hMENA splice variants. (B) hMENA protein domains. Asterisks indicate Ser phosphorylation sites. GAB and FAB correspond to the G-actin and F-actin binding sites, respectively. Sites of V6 and 11a peptides are indicated by arrows. Sequence of peptide V6, absent in hMENAΔv6 isoform, is reported. (C) In vitro translated hMENA11a, hMENA, and hMENAΔv6 analyzed by WB analysis using the pan-hMENA Ab (10 μg/mL). Cell lysates (30 μg) of SBT and MDA-MB-231 breast cancer cell lines (used to obtain hMENA11a and hMENAΔv6 cDNAs, respectively) were also tested by WB to identify the corresponding in vitro translated protein bands. (D) RT-PCR performed with primers (MTC1 forward and MTC4 reverse) flanking the region of alternative splicing on MDA-MB-231 and SBT RNA. PCR experiment was also done on the pcDNA3.1 vectors containing the three hMENA variants. PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining. E6, exon 6; E11a, exon 11a.
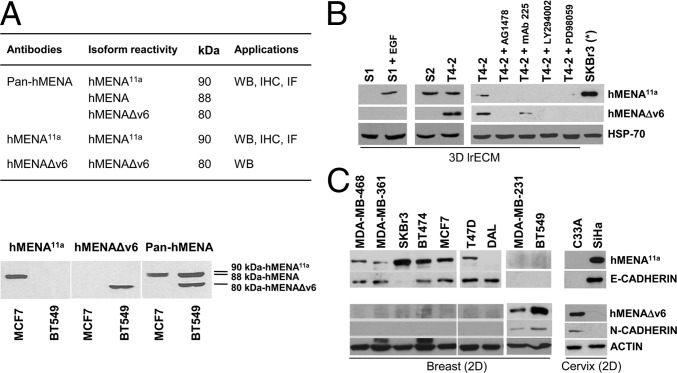
Fig. 2.
hMENAΔv6 is expressed only in malignant T4-2 cells but not in nonmalignant S1 and premalignant S2 cells. hMENA11a and hMENAΔv6 define an epithelial and mesenchymal phenotype, respectively, in breast and cervix cancer cell lines. (A) Description of isoform reactivity and application of anti-hMENA antibodies and representative WB analysis of MCF7 (hMENA/hMENA11a-positive) and BT549 (hMENA/hMENAΔv6-positive) cells with the hMENA11a, hMENA∆v6, and pan-hMENA antibodies. WB, Western blot; IHC, immunohistochemistry; IF, immunofluorescence. (B) hMENA isoform expression in a 3D model of breast cancer progression. WB analysis of lysates of 3D grown HMT-3522 S1, S2, and T4-2 progression series cells with hMENA isoform-specific antibodies is shown. S1 cells grown for 24 h in growth medium depleted of EGF (S1) do not express hMENA11a isoform, which is expressed in the presence of EGF. WB analysis of T4-2 cells and T4-2 cells reverted with AG1478, mAb225, LY294002, and PD98059 treatments, showing a strong reduction of hMENA isoform expression following the reversion of the transformed phenotype. 2D grown SKBr3 (*) cell lysate is reported as a control of hMENA11a expression. (C) WB analysis of lysates of breast and cervix tumor cell lines with hMENA isoform-specific antibodies, anti–E-cadherin and N-cadherin antibodies, indicating a strong correlation between hMENA11a and E-cadherin as well as between hMENAΔv6 and N-cadherin expression.
hMENA Isoforms Are Expressed Differentially in an Organotypic Model of a Breast Cancer Progression Series.
We previously showed that hMENA is absent in normal breast tissue, but becomes expressed in high-risk benign lesions and invasive tumors, suggesting that hMENA expression could be an early marker of breast tumorigenesis (15). We therefore turned to the HMT-3522 isogenic cell strain series, a model of human breast cancer progression where no oncogenes were used for transformation (19–21), to ask whether hMENA alternative splicing is linked to malignant progression. In 3D laminin-rich gel assays (3D lrECM) (24), we found that nonmalignant HMT-3522-S1 mammary epithelial cells do not express hMENA11a unless treated with EGF, whereas the premalignant HMT-3522-S2 cells, which have autocrine EGFR signaling (23), express hMENA11a (Fig. 2B). The tumorigenic and invasive, but not metastatic, HMT-3522–T4-2 cells express both the hMENA11a and hMENAΔv6 isoforms. Targeting the EGFR, MAPK, or PI3K signaling in HMT-3522–T4-2 cells using small molecule inhibitors, (AG1478, PD98059, or LY294002, respectively) or using an anti-EGFR function blocking antibody, mAb-225 “reverts” the phenotype of these cells from malignant to nonmalignant by restoring apicobasal polarity and inducing growth arrest in 3D lrECM (18). We found that reversion leads to concurrent down-regulation of hMENA11a and hMENAΔv6 both at protein (Fig. 2B and Fig. S2A) and RNA levels (Fig. S2B). These data suggest that polarized and growth-arrested cellular architecture correlates inversely with hMENA isoform expression.
hMENA11a and hMENAΔv6 Define an Epithelial and Mesenchymal Phenotype, Respectively.
We found previously that hMENA11a expression is associated with breast and pancreatic cancer cell lines that have an epithelial phenotype in 2D cultures (9, 13). We asked whether expression of hMENA11a and hMENAΔv6 isoforms correlates with either epithelial or mesenchymal markers in a panel of breast cancer cell lines. With the exception of DAL, the only cell line not expressing the hMENA gene (25), we found that all E-cadherin-positive cell lines express hMENA11a (Fig. 2C). However, hMENA11a expression was independent of functional E-cadherin because SKBr3 cells, which have a mutation in the E-cadherin gene (26), retained hMENA11a expression. On the other hand, absence of hMENA11a expression correlated with the expression of hMENAΔv6, absence of E-cadherin and the presence of N-cadherin. This pattern of hMENA isoform expression is found also in cervix cancer cell lines (C33A and SiHa) (Fig. 2C). These data indicate that hMENA11a and hMENAΔv6 expression correlates with epithelial and mesenchymal/basal phenotypes, respectively, but the hMENA itself (88 kDa, using a pan-hMENA Ab) is expressed in all breast cancer cell lines examined (Fig. S2A).
Mesenchymal hMENAΔv6 and Epithelial hMENA11a Isoforms Have Opposite and Antagonistic Roles in Cell Migration and Invasion.
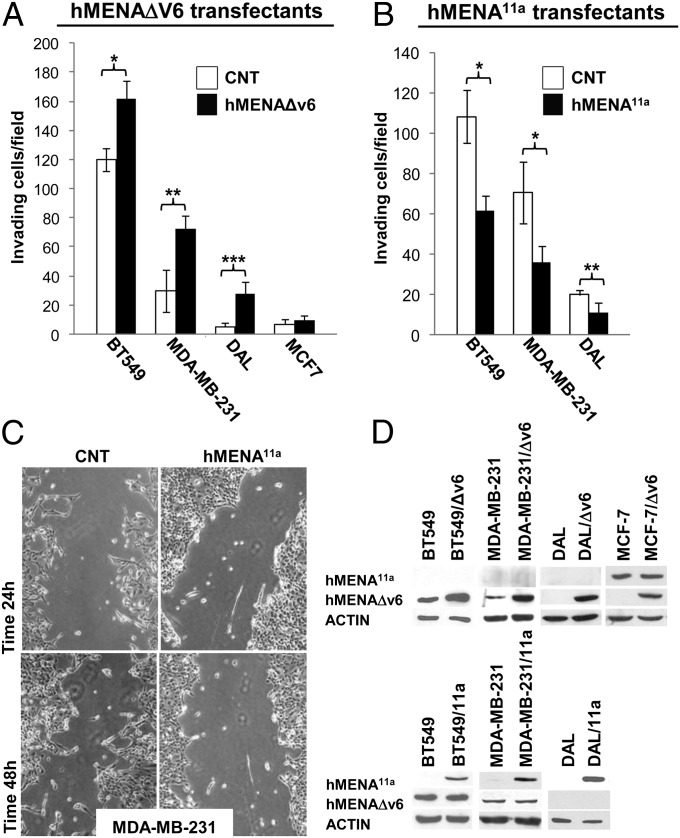
Given the data in rodent models linking Mena splicing with mammary tumor cell invasion and migration (11, 12), we performed isoform-specific overexpression and hMENA siRNA experiments to evaluate the role of alternative splicing of hMENA in human breast cancer invasion and progression. Transfection of hMENAΔv6 into BT549, MDA-MB-231, and DAL cells appreciably increased their invasiveness (Fig. 3A). Our efforts to perform isoform-specific hMENAΔv6 siRNA were not conclusive, but knockdown of hMENA/hMENAΔv6 by siRNA that targets all hMENA mRNA in BT549 cells, significantly reduced their invasion through basement membrane-coated transwell inserts (Fig. S3). However, exogeneous expression of hMENAΔv6 in hMENA11a-positive MCF7 cells did not induce the invasive behavior, suggesting that hMENA11a can overcome the invasive function of hMENAΔv6 (Fig. 3A). Indeed, forced expression of hMENA11a in MDA-MB-231 and BT549 cells decreased their invasiveness significantly (Fig. 3B), and in wound-healing assays, hMENA11a-transfected cells did not scatter or migrate into the wound (Fig. 3C). The low invasive DAL cells completely lost the ability to invade the Matrigel when transfected with hMENA11a (Fig. 3B). These data suggest a proinvasive role for the mesenchymal hMENAΔv6 and a dominant, anti-invasive role for the epithelial hMENA11a.
Fig. 3.
hMENAΔv6 transfection increases the invasive ability of breast cancer cells only in the absence of hMENA11a. (A) Matrigel invasion assay performed on BT549 (50,000 cells; 24 h of invasion) MDA-MB-231 (50,000 cells; 24 h of invasion), DAL (75,000 cells; 72 h of invasion), and MCF7 (75,000 cells; 48 h of invasion) cells stably or transiently (BT549) transfected with the empty vector as control (CNT) or hMENAΔv6, toward serum. The ability of invasion has been measured by the use of Matrigel-coated transwell filters (BD Biosciences), toward serum [RPMI medium with 10% (vol/vol) FCS]. *P < 0.0001, **P = 0.0023, ***P = 0,0003. (B) Matrigel invasion assay perfomed on BT549 (50,000 cells; 24 h of invasion), MDA-MB-231 (50,000 cells; 48 h of invasion), and DAL (150,000 cells; 72 h of invasion) cells stably transfected with the empty vector or hMENA11a, toward serum. *P ≤ 0.0001, **P = 0.009. The assay was repeated three times, performed in triplicate each time. SDs are indicated. (C) Wound-healing assay perfomed on MDA-MB-231 cells stably transfected with the empty vector or hMENA11a. (D) WB analysis of lysates of transfected cells used in the experiments shown in A–C.
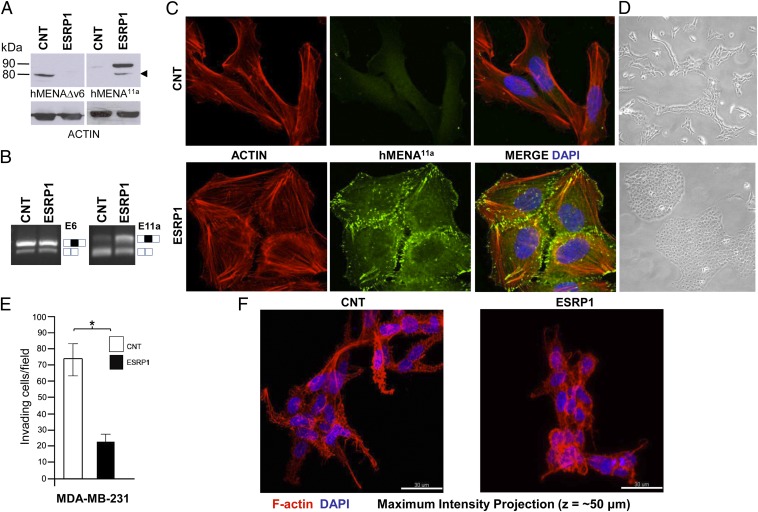
hMENA11a Expression Is Regulated by Epithelial Splicing Regulatory Protein 1 (ESRP1) and Changes the Actin Cytoarchitecture.
ESRP1 and ESRP2, the recently discovered splicing regulatory proteins, have been shown to regulate hMENA11a splicing in cancer cell lines (16, 17). Because ESRP1 is a more robust splicing regulator than ESRP2 (17), we tested whether ESRP1, transduction into the ESRP1 negative MDA-MB-231 (17) and BT549 cells could affect the hMENA splicing program we report here. At protein level, we found that ESRP1 overexpression increased hMENA11a and decreased the hMENAΔv6 expression (Fig. 4A and Fig. S4A). This is compatible with exon 11a inclusion in hMENA and also in hMENA∆v6, as revealed by the additional band recognized either by the anti-hMENA11a mAb or the anti-hMENA∆v6 Ab in BT549 ESRP1 cells (Fig. S4A). To verify that the observed changes of hMENA isoform expression are due to alternative splicing, RT-PCR experiments using oligonucleotides flanking the 11a or 6 exons were done. Results clearly showed the inclusion of exon 11a in the transcripts of ESRP1-transduced cells (as evident in the increase of the upper band and the decrease in the lower band intensity of Fig. 4B and Fig. S4B, Right), but no changes were evident in exon 6 splicing (Fig. 4B and Fig. S4B, Left).
Fig. 4.
Epithelial Splicing Regulatory Protein 1 (ESRP1) transduction induces hMENA11a expression in parallel with changes in cell shape and actin cytoskeleton architecture in MDA-MB-231 cells. (A) WB analysis of MDA-MB-231 cells transduced with the empty vector (CNT) or with ESRP1 (ESRP1), with the hMENA isoform-specific antibodies. hMENA11a is expressed only in the MDA-MB-231 ESRP1 cells. hMENA11a isoform-specific antibody also decorates a band compatible with the insertion of 21 aa of the exon 11a in the hMENAΔv6 isoform (arrowhead). (B) RT-PCR with RNA extracted from MDA-MB-231 cells transduced with the empty vector (CNT) or with ESRP1 (ESRP1), performed either with primers flanking exon 6 (Left), exon 6 inclusion = □■□; exon 6 skipping = □□, or with primers flanking the exon 11a (Right), exon 11a inclusion = □■□; 11a skipping = □□. PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining. E6, exon 6; E11a, exon 11a. (C) Confocal analysis of MDA-MB-231 cells transduced with the empty vector (CNT) or with ESRP1 using hMENA11a mAb (green) and phalloidin (red). Cells were imaged using a Zeiss 710 laser-scanning microscope. Magnification, 63×. (D) Phase-contrast images of MDA-MB-231 cells transduced with the empty vector or with ESRP1. (E) Matrigel invasion assays of MDA-MB-231 cells stably transduced with the empty vector (CNT) or ESRP1 (35,000 cells; 48 h of invasion). *P = 0.002. (F) Phalloidin staining of transduced cells grown in 3D lrECM culture.
Using confocal microscopy, MDA-MB-231 ESRP1 cells showed a dramatic morphological change from a mesenchymal to an epithelium-like phenotype (MET) compared with controls. ESRP1-transduced cells grow in tightly packed, cobblestone-like colonies with decreased cell spreading compared with control cells, hallmark features of epithelial cells in culture (Fig. 4 C, D, and F). This change was associated with the rearrangement of the actin cytoskeletal network and an increase in cell–cell contacts, where hMENA11a was observed to colocalize with F-actin; no hMENA11a staining was observed in vector control cell lines (Fig. 4C). Similar results were also obtained using the pan-hMENA Ab (Fig. S5). Similar to the phenotype obtained by the forced expression of hMENA11a, ESRP1 overexpression significantly reduced the invasive ability of cells (Fig. 4E). ESRP1-transduced cells grown in 3D lrECM have a lower number of shorter F-actin–rich projections into the ECM and lack filopodia-like secondary protrusions, compared with the control cells (Fig. 4F and Movies S1 and S2). Similar results were obtained in BT549 ESRP1 cells (Fig. S4 C–E and Movies S3 and S4). These data suggest a central role for ESRP1 in mediating MET-like phenotypic changes through alternative splicing of genes such as hMENA. hMENA11a appears to be a key regulator of this morphological switch (Fig. S4F).
hMENA Isoform Expression Correlates with Proliferation Index and E-Cadherin in Primary Breast Tumor Tissues.
hMENA isoform expression was evaluated by immunohistochemistry using both a pan-hMENA antibody that recognizes all of the hMENA isoforms and an antibody specific for hMENA11a. The anti-hMENAΔv6 antibody was unsuitable for immunohistochemical evaluation in paraffin-embedded tissues, and we did not succeed in either raising suitable antibodies or finding other sources (Fig. 2A). We tested tissue samples of 162 patients with invasive breast cancer (BC), whose pathological parameters are shown in Table S1. Fifty two cases (32%) were negative for both antibodies, and 41 cases (25%) were positive for both. The remaining 69 tumors (43%) were pan-hMENA positive/hMENA11a negative. Considering the pattern of expression identified in cancer cell lines (Fig. 2), and the reactivity of antibodies used (Fig. 2A), the group of tumors that are pan-hMENA positive/hMENA11a negative would include the tumors that express the hMENA and hMENA∆v6 isoforms, with a mesenchymal-like phenotype. No staining was evident in morphologically normal breast ducts, using either anti pan-hMENA or anti-hMENA11a (Fig. S6). Ki67 staining revealed that only 23% of pan-hMENA negative/hMENA11a negative and 39% of pan-hMENA–positive/hMENA11a-negative tumors presented a high proliferation index (Ki67 > 15%). On the other hand, 63% of hMENA11a-positive tumors showed a high proliferation index (P < 0.001).
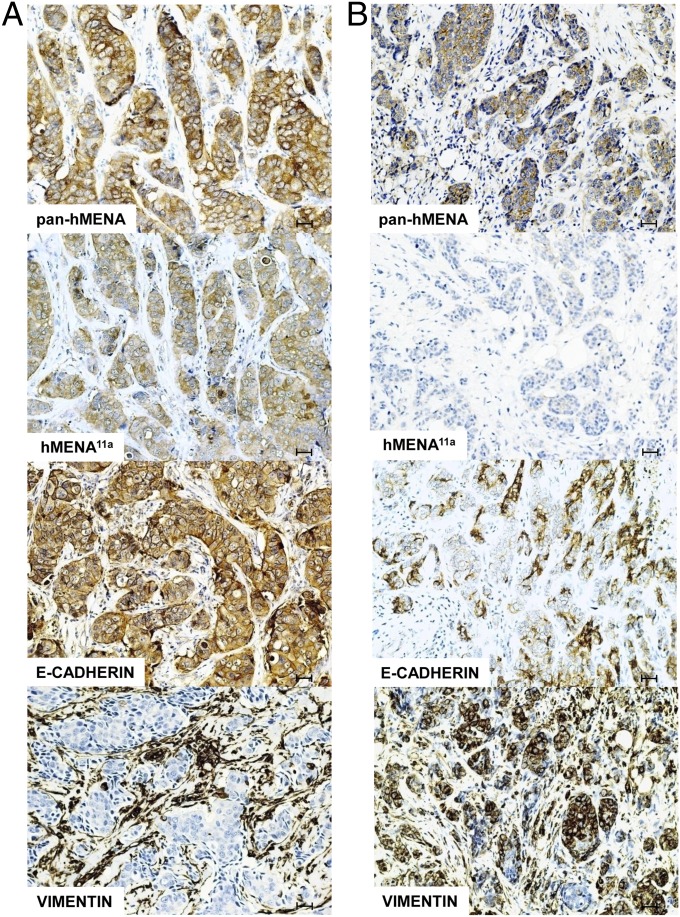
The E-cadherin immunostaining, carried out on the 158 nonlobular carcinomas, revealed normal E-cadherin expression in 56 cases (35%), with the remaining cases showing reduced or no expression of the protein (SI Materials and Methods). The percentage of E-cadherin reduced/negative tumors are significantly more frequent in the subgroup of pan-hMENA–positive/hMENA11a-negative tumors (76%) than in the double negative or double positive cases (57% and 56%, respectively) (P = 0.045). A few representative cases were also stained for vimentin, and the results obtained indicated that tumors expressing hMENA11a and displaying a normal E-cadherin staining, were negative for vimentin, although the antigen was present in the surrounding stroma (Fig. 5A). Conversely, pan-hMENA–positive tumors lacking hMENA11a presented decreased E-cadherin expression and were associated with strong vimentin staining (Fig. 5B).
Fig. 5.
Pan-hMENA, hMENA11a, E-cadherin, and vimentin staining by immunohistochemistry in consecutive sections of two illustrative cases of invasive ductal carcinoma. (A) Immunoreactivity of pan-hMENA and hMENA11a Abs is associated with homogeneous E-cadherin expression and lack of vimentin immunostaining in cancer cells, whereas (B) positive pan-hMENA and negative hMENA11a immunoreactivity is associated with low or absence of E-cadherin and vimentin positivity. Magnification, 20×. (Scale bars, 30 µm.)
In summary, the balance of expression of hMENA splicing isoforms leads to profoundly different phenotypic and functional outcomes in breast cancer cells.
Discussion
Here we report on the molecular cloning of a unique splice variant of human MENA, an actin regulatory protein. The unique isoform, hMENAΔv6, is associated with a mesenchymal phenotype in breast cancer. We provide biochemical, morphological, and functional evidence that alternative splicing generates at least two hMENA isoforms: hMENA11a, which inhibits and hMENAΔv6, which promotes invasive behavior in breast cancer cells. There is no information on this sequence in the literature; the National Center for Biotechnology Information database contains similar splice variants observed in rat, chicken, and Xenopus, and the EST database contains mouse and human sequences lacking exon 6 (Fig. S1). A neuronal-specific isoform generated by alternative splicing and extension of exon 6 has been characterized in mouse (1) and human (8), which in conjunction with our discovery of hMENAΔv6, suggest that exon 6 is a splicing “hotspot” for hMENA.
hMENA has been reported to be an early marker of breast cancer progression expressed in high-risk premalignant lesions and in frank malignancy, but absent in the normal breast (15). Using an isogenic model of breast cancer progression (18–21), here we found evidence to suggest that hMENA alternative splicing is associated with malignant progression (Fig. 2 and Fig. S2). The hMENAΔv6 isoform is expressed only in the tumorigenic and invasive HMT-3522–T4-2 cells, but not in the premalignant (S2) or nonmalignant (S1), as would be expected from nonmalignant “normal” cells. Neither hMENA11a nor hMENAΔv6 isoform are expressed in S1 cells; however when EGF is added to the medium, hMENA11a is induced (Fig. 2 and Fig. S2). Using 3D lrECM cultures, we were able to show that signaling inhibitors that lead to “phenotypic reversion” of T4-2 cells to a nonmalignant morphology (18, 27, 28), concurrently cause down-regulation of both hMENA11a and hMENAΔv6 isoforms. These data corroborate and extend previous results showing that EGF signaling up-regulates hMENA and hMENA11a expression (9), and that reversion of T4-2 cells also “normalizes” the malignant phenotype (for review, ref. 28), and suggest that the activity of splicing factors regulating the hMENA splicing program are controlled by several signaling pathways via changes in expression level as well as posttranslational modifications (29).
Furthermore, we show that hMENA11a expression is usually associated with breast and cervix cancer cell lines that express E-cadherin, whereas hMENAΔv6 is expressed in those cancer cells displaying EMT features and migratory behavior (Fig. 2C). These data suggest that hMENA11a and hMENAΔv6 can identify cancer cells with noninvasive and invasive phenotypes, respectively. Accordingly, the alternative splicing of hMENA yields opposing regulatory functions in tumor cell invasion and migration. Transfection of hMENA11a into cells already expressing hMENAΔv6 suppresses invasion; similarly transfection of hMENAΔv6 into cells already expressing hMENA11a fails to promote invasion, providing compelling evidence that hMENA11a is dominant over hMENAΔv6. The hMENAΔv6 isoform promotes cancer cell invasion in a BM-coated Boyden chamber based on overexpression and knockdown experiments in cell lines lacking hMENA11a. However, it appears that hMENA11a expression can suppress these effects by acting as a dominant-active, antiinvasive isoform.
The mechanistic basis by which the hMENA11a isoform competes with hMENAΔv6 in suppressing invasion is not yet known. However, our data that the presence of hMENA11a (ESRP1 induced) reduces the number and the length of filopodia projecting into the gel in 3D cultures (Fig. 4), is consistent with the hypothesis that the 21-aa insertion of the 11a exon, influences the ability of the Ena/VASP tetramer to promote filopodia formation and extension (11). In a xenograft rodent mammary cancer model, the invasive cancer cells showed a decrease of Mena11a and an increase of MenaINV isoform expression (11), reminiscent of our data. The MENAINV isoform, which has an exon next to the Ena/VASP homology 1 (EVH1) domain, facilitates cell invasion by stabilizing invadopodia (12) and promoting discohesive tumor morphology, whereas MENA11a expression appears to promote epithelial tumor morphology (30). hMENA11a, unlike hMENA, is phosphorylated following activation of the EGFR or its other family members, suggesting that exon 11a may represent a site for regulation of hMENA11a in breast cancer (9). Furthermore, we predict that the exon 6 deletion influences protein function and regulation by bringing two functional regions closer together, with one site containing a PKA Ser phosphorylation residue and the other site containing the regulatory proline-rich region.
To investigate the mechanism of hMENA regulation in cancer, one must understand the intricate balance of lineage- and tumor stage-specific hMENA splicing. The presence of isoforms with antagonistic functions underscores the importance of studying both genes that regulate alternative splicing and the generated spliced variants. Indeed, a major shortcoming of conventional microarray-based gene expression profiling is the lack of alternative spliced forms of transcripts. Splicing regulatory factors process numerous genes, including hMENA, that regulate biological functions, such as morphology, adhesion, migration, and proliferation (17, 31, 32). In particular, the epithelial-specific hMENA11a isoform belongs to a cluster of genes spliced by ESRP1/2, which are involved in actin cytoskeleton organization, cell adhesion, and cell motility (16); and a splicing signature of ESRP-regulated exons has been proposed as a unique biomarker of EMT (17). Consistent with data showing that ESRP1/2 knockdown reduces the hMENA11a expression in epithelial cells (17, 33), our data demonstrate that ectopic expression of ESRP1, in “mesenchymal-like” ESRP1 negative breast cancer cell lines (17, 33), results in up-regulation of hMENA11a, down-regulation of hMENAΔv6 protein isoform, and a dramatic reorganization of the actin cytoskeleton. Even though ESRP1 does not affect the splicing of exon 6 (Fig. 4B and Fig. S4B), hMENA11a and ESRP1expression led to altered cell morphology (from spindle shaped to cobblestone) and suppressed cell invasion in cancer cells. All of the above changes are related to the MET process. This is reminiscent of a study of the Twist-induced EMT-driven alternative splicing program also regulated by ESRP1 (34).
In contrast to cancer cell lines, in tumor tissues only a minority of cells may display characteristics of having entered into or passed through EMT. Thus, pathologists often have the difficult task of classifying a whole tumor with an epithelial or mesenchymal phenotype. The immunostaining of a cohort of primary breast tumors (Table S1 and Fig. 5) suggests that multiple splicing patterns of hMENA may occur also in vivo. The findings that the higher frequency of pan-hMENA–positive/hMENA11a-negative breast cancers occurs when E-cadherin is down-regulated, support the experimental evidence that the hMENA alternative splicing might contribute to a different tumor cell morphology accounting for a different invasive ability. Furthermore, we find that the majority of these pan-hMENA–positive/hMENA11a-negative primary tumors are slow-cycling (Ki67 low) tumors with enriched mesenchymal genes. The hMENA exon 11a splicing is regulated also by Rbfox2 and appears to be an important marker in the newly defined Claudin low subtype (31), described as significantly enriched in EMT cell-like features and related to poor prognosis in breast cancer (35, 36).
As splicing studies evolve, further subclassification of breast tumors is expected and this study paves the way for defining hMENA splicing and its regulation as promising new biomarkers of invasiveness. Combined with other clinicopathologic markers, hMENA and its spliced forms may improve the early diagnosis of breast cancer and efficacious clinical decision for patient treatment.
Materials and Methods
The cell lines used, information relative to patients, and tissue specimens as well as materials and methods relative to antibody production, RT-PCR, in vitro transcription-coupled translation, Western blot analysis, transfections, small-interfering RNA treatment, ESRP1 transduction, 3D culture, confocal analysis, wound-healing assay, cell invasion assay, immunohistochemistry, and statistical analysis are reported in SI Materials and Methods.
Total RNA was extracted from MDA-MB-231 cells using TRIzol reagent (Life Technologies) and 2 µg was used to generate a cDNA library using a first-strand cDNA synthesis kit (Amersham Pharmacia Biotech). cDNA was amplified by PCR using hMENA-specific P1-ATG and P8-stop primers as already reported (9). PCR products were analyzed on a 1% agarose gel, excised from the gel, and purified using a gel extraction kit (Qiagen). The extracted amplicon was incubated with 1 unit of AmpliTaq polymerase and 1 μL of 10 mM dATP (both from Applied Biosystems) to add 3′ adenines, and was then cloned into pcDNA3.1/V5-HIS TOPO following the manufacturer’s protocol (Invitrogen). Plasmid DNA was isolated by Wizard Plus minipreps DNA purification system (Promega) and was sequenced using T7, T3, and internal sequencing primers for hMena (9). The study was reviewed and approved by the ethics committee of the Regina Elena National Cancer Institute, and written informed consent was obtained from all patients.
Supplementary Material
Acknowledgments
This work was supported in part by the Associazione Italiana per la Ricerca sul Cancro (AIRC) Cinque per Mille Grants 12182 and IG 11631 (to P.N.); US Department of Energy, Office of Biological and Environmental Research, and Low Dose Radiation Program Contract DE-AC02-05CH1123 grants; National Cancer Institute Awards R37CA064786, U54CA126552, U54CA112970, U01CA143233, and U54CA143836, Bay Area Physical Sciences–Oncology Center; and US Department of Defense Grant W81XWH0810736 (to M.J.B).
Footnotes
Conflict of interest statement: P.N. and F.D.M. are the inventors on a patent related to the function of hMena variants in tumor cells, which includes Δv6. The Regina Elena National Cancer Institute is a stockholder in Metastat, a company with the exclusive license to the Mena patent suite, but this company does not hold the license for the patent which includes Δv6. Other authors do not have a conflict of interest.
Data deposition: The data reported in this paper has been deposited in the GenBank database (accession no. EU255274).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214394109/-/DCSupplemental.
References
- 1.Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- 2.Bear JE, et al. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 2000;101:717–728. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- 3.Bear JE, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- 4.Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: Regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- 5.Scott JA, et al. Ena/VASP proteins can regulate distinct modes of actin organization at cadherin-adhesive contacts. Mol Biol Cell. 2006;17:1085–1095. doi: 10.1091/mbc.E05-07-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barzik M, et al. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem. 2005;280:28653–28662. doi: 10.1074/jbc.M503957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Applewhite DA, et al. Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol Biol Cell. 2007;18:2579–2591. doi: 10.1091/mbc.E06-11-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urbanelli L, et al. Characterization of human Enah gene. Biochim Biophys Acta. 2006;1759(1–2):99–107. doi: 10.1016/j.bbaexp.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Di Modugno F, et al. Molecular cloning of hMena (ENAH) and its splice variant hMena+11a: Epidermal growth factor increases their expression and stimulates hMena+11a phosphorylation in breast cancer cell lines. Cancer Res. 2007;67:2657–2665. doi: 10.1158/0008-5472.CAN-06-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tani K, et al. Abl interactor 1 promotes tyrosine 296 phosphorylation of mammalian enabled (Mena) by c-Abl kinase. J Biol Chem. 2003;278:21685–21692. doi: 10.1074/jbc.M301447200. [DOI] [PubMed] [Google Scholar]
- 11.Goswami S, et al. Identification of invasion specific splice variants of the cytoskeletal protein Mena present in mammary tumor cells during invasion in vivo. Clin Exp Metastasis. 2009;26:153–159. doi: 10.1007/s10585-008-9225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philippar U, et al. A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis. Dev Cell. 2008;15:813–828. doi: 10.1016/j.devcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pino MS, et al. hMENA+11a isoform serves as a marker of epithelial phenotype and sensitivity to EGFR inhibition in human pancreatic cancer cell lines. Clin Cancer Res. 2008;14:4943–4950. doi: 10.1158/1078-0432.CCR-08-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Modugno F, et al. The cooperation between hMena overexpression and HER2 signalling in breast cancer. PLoS ONE. 2010;5:e15852. doi: 10.1371/journal.pone.0015852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Modugno F, et al. The cytoskeleton regulatory protein hMena (ENAH) is overexpressed in human benign breast lesions with high risk of transformation and human epidermal growth factor receptor-2-positive/hormonal receptor-negative tumors. Clin Cancer Res. 2006;12:1470–1478. doi: 10.1158/1078-0432.CCR-05-2027. [DOI] [PubMed] [Google Scholar]
- 16.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warzecha CC, et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 2010;29:3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, et al. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc Natl Acad Sci USA. 1998;95:14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briand P, Petersen OW, Van Deurs B. A new diploid nontumorigenic human breast epithelial cell line isolated and propagated in chemically defined medium. In Vitro Cell Dev Biol. 1987;23:181–188. doi: 10.1007/BF02623578. [DOI] [PubMed] [Google Scholar]
- 20.Briand P, Nielsen KV, Madsen MW, Petersen OW. Trisomy 7p and malignant transformation of human breast epithelial cells following epidermal growth factor withdrawal. Cancer Res. 1996;56:2039–2044. [PubMed] [Google Scholar]
- 21.Rizki A, et al. A human breast cell model of preinvasive to invasive transition. Cancer Res. 2008;68:1378–1387. doi: 10.1158/0008-5472.CAN-07-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenny PA, Bissell MJ. Targeting TACE-dependent EGFR ligand shedding in breast cancer. J Clin Invest. 2007;117:337–345. doi: 10.1172/JCI29518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madsen MW, Lykkesfeldt AE, Laursen I, Nielsen KV, Briand P. Altered gene expression of c-myc, epidermal growth factor receptor, transforming growth factor-alpha, and c-erb-B2 in an immortalized human breast epithelial cell line, HMT-3522, is associated with decreased growth factor requirements. Cancer Res. 1992;52:1210–1217. [PubMed] [Google Scholar]
- 24.Petersen OW, Rønnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci USA. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Modugno F, et al. Human Mena protein, a serex-defined antigen overexpressed in breast cancer eliciting both humoral and CD8+ T-cell immune response. Int J Cancer. 2004;109:909–918. doi: 10.1002/ijc.20094. [DOI] [PubMed] [Google Scholar]
- 26.Lombaerts M, et al. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br J Cancer. 2006;94:661–671. doi: 10.1038/sj.bjc.6602996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weaver VM, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bissell MJ, Kenny PA, Radisky DC. Microenvironmental regulators of tissue structure and function also regulate tumor induction and progression: The role of extracellular matrix and its degrading enzymes. Cold Spring Harb Symp Quant Biol. 2005;70:343–356. doi: 10.1101/sqb.2005.70.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. J Biol Chem. 2008;283:1223–1227. doi: 10.1074/jbc.R700034200. [DOI] [PubMed] [Google Scholar]
- 30.Roussos ET, et al. Mena invasive (Mena(INV)) and Mena11a isoforms play distinct roles in breast cancer cell cohesion and association with TMEM. Clin Exp Metastasis. 2011;28:515–527. doi: 10.1007/s10585-011-9388-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lapuk A, et al. Exon-level microarray analyses identify alternative splicing programs in breast cancer. Mol Cancer Res. 2010;8:961–974. doi: 10.1158/1541-7786.MCR-09-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venables JP, et al. Identification of alternative splicing markers for breast cancer. Cancer Res. 2008;68:9525–9531. doi: 10.1158/0008-5472.CAN-08-1769. [DOI] [PubMed] [Google Scholar]
- 33.Dittmar KA, et al. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol Cell Biol. 2012;32:1468–1482. doi: 10.1128/MCB.06536-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shapiro IM, et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011;7:e1002218. doi: 10.1371/journal.pgen.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5(1):5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prat A, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.