Abstract
Mammalian voltage-gated sodium channels are composed of four homologous voltage sensor domains (VSDs; DI, DII, DIII, and DIV) in which their S4 segments contain a variable number of positively charged residues. We used single histidine (H) substitutions of these charged residues in the Nav1.4 channel to probe the positions of the S4 segments at hyperpolarized potentials. The substitutions led to the formation of gating pores that were detected as proton leak currents through the VSDs. The leak currents indicated that the mutated residues are accessible from both sides of the membrane. Leak currents of different magnitudes appeared in the DI/R1H, DII/R1H, and DIII/R2H mutants, suggesting that the resting state position of S4 varies depending on the domain. Here, DI/R1H indicates the first arginine R1, in domain DI, has been mutated to histidine. The single R1H, R2H, and R3H mutations in DIV did not produce appreciable proton currents, indicating that the VSDs had different topologies. A structural model of the resting states of the four VSDs of Nav1.4 relaxed in their membrane/solution environment using molecular dynamics simulations is proposed based on the recent NavAb sodium channel X-ray structure. The model shows that the hydrophobic septa that isolate the intracellular and the extracellular media within the DI, DII, and DIII VSDs are ∼2 Å long, similar to those of Kv channels. However, the septum of DIV is longer, which prevents water molecules from hydrating the center of the VSD, thus breaking the proton conduction pathway. This structural model rationalizes the activation sequence of the different VSDs of the Nav1.4 channel.
Keywords: molecular dynamics simulations, omega currents, pH, ion channel, voltage-clamp
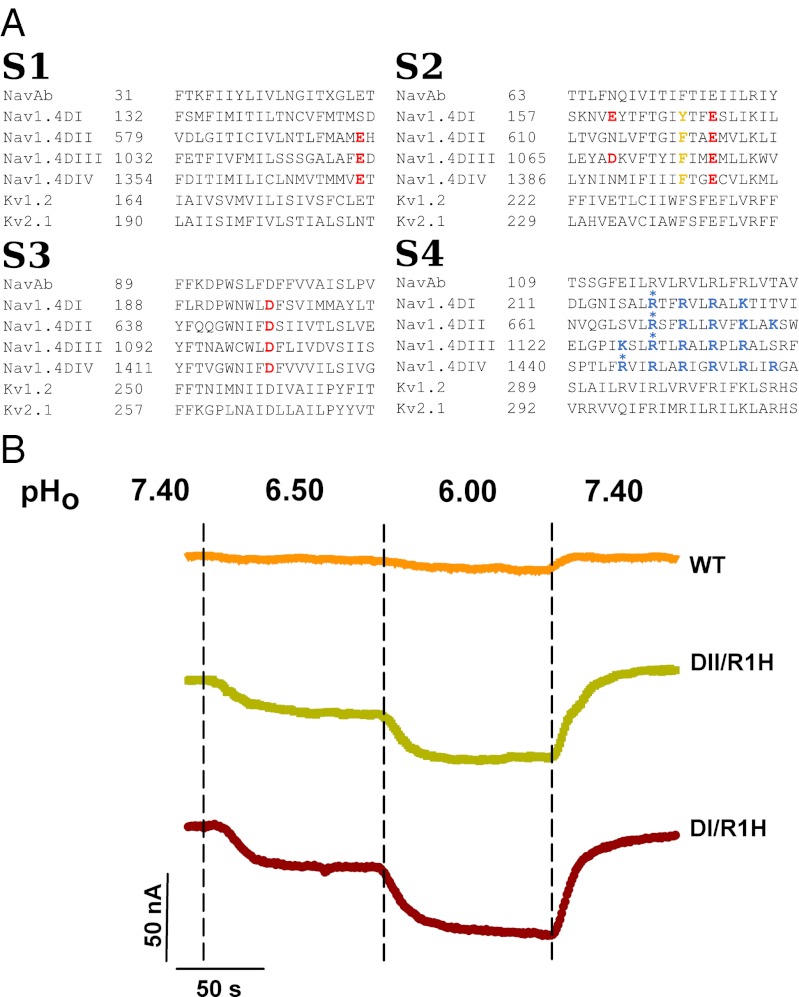
Voltage-gated sodium channels (VGSCs) produce the rapid upstroke of the action potential and are critical elements for maintaining the cell’s electrical activity. VGSCs are composed of four homologous domains, each of which contains six helical transmembrane (TM) segments (S1–S6) divided into two structures: the voltage sensor domain (VSD; S1–S4) and the pore domain (S5 and S6). The S4 segments contain variable numbers of positively charged amino acids, either arginines (R) or lysines (K), at every third position. These charged residues are conserved in several ion channels and are responsible for the voltage sensitivity of the channels (1) (Fig. 1).
Fig. 1.
Sequence alignment of the TM segments (S1–S4) of the voltage sensor domains of various voltage-gated ion channels and proton currents recordings. (A) The different TM segments of the four domains of Nav1.4 were aligned with Kv1.2, Kv2.1, and NavAb. Note the variability in the quantity and position of the positively charged (in blue) and negatively charged residues (in red) in Nav1.4. (B) Lowering the extracellular pH (pHo) induced an inward proton current in oocyte expressing NaV1.4/DI/R1H and NaV1.4/DII/R1H mutant channels but not in oocytes expressing NaV1.4/WT channels. The oocytes were held at –80 mV. Currents were recorded every second in NMDG solution containing 1 μM TTX. The dashed vertical lines indicate when the extracellular solution was changed.
Unlike voltage-gated potassium channels, for which a wealth of structural and functional results is available, other voltage-gated ion channels such as VGSCs have received less attention. Their low level of expression and the fact that they are composed of four different domains make them challenging to investigate. The few studies that have probed the voltage-dependent movement of S4 using cysteine (C) accessibility (2) and fluorescence measurements (3) have provided important information about the structure and function of VGSCs. Moreover, the recent discovery of bacterial VGSCs, which are composed of four identical subunits, has stirred interest in these proteins. The first crystal structure of a VGSC channel (NavAb) revealed molecular details of the closed pore as well as the VSD region (4). This breakthrough made it possible to propose models of the resting state (5) and to start building reliable homology models of mammalian channels (6).
The Nav1.4 VGSC controls the excitability of skeletal muscles and was the first member of the VGSC family shown to be subject to leak currents through the VSD. These currents can give rise to striated muscle channelopathies when specific positively charged residues of the S4 segment of specific Nav channels are mutated (7–11). The discovery of leak currents through the VSD of the Shaker potassium channel dates back to 2004. The first study to observe leak currents through the VSD showed that the substitution of the first R (R1) for a histidine (H) allows the permeation of protons through the VSD (12). The mechanism by which conduction is achieved likely involves Grotthus hopping of the proton along a water wire and the passage of the R1H gate through a shuttle mechanism (10, 12). Subsequent studies confirmed this discovery and showed that the replacement of R1 by an uncharged amino acid, such as serine (S), alanine (A), glutamine (Q), etc., results in the generation of a large cationic current now known as the gating pore current or omega current (7, 13). These studies suggested that the permeation pathway is open when the mutated amino acid on the S4 helix is located in a constriction site of the VSD. The permeation pathway is occluded when the mutated amino acid moves away from the constriction site.
Molecular dynamics (MD) simulations of the effect of such mutations in the mammalian Kv1.2 potassium channel (a Shaker homolog) uncovered the molecular details of the phenomenon (14). These simulations indicate that among all positively charged residues of S4, only the mutation of the amino acid located below the hydrophobic septum in the VSD, i.e., the residue that interacts with the conserved phenylalanine (F) residue of S2 and the conserved glutamate (E) residue of S3, gives rise to a gating pore current (15, 16). If the mutated residue is placed above this constriction site, the negative residue E1 (S2) must also be mutated to give rise to a gating pore current (17). The molecular mechanism of proton transport is different from the one of transport of other ions but the overall characteristics of the VSD topology can likely be extrapolated from the R1Q or R1S to the R1H mutant. The absence of bond breakage and formation in classic MD simulations precludes the description of proton transport. It is noteworthy that methods exist that enable the simulation of such a process, such as ab initio MD (18, 19) or multistate empirical valence bond (20), but are beyond the scope of the present study.
Here, the single histidine substitution approach is used to assess the potential of individual mutations of R in the four homologous VSDs (DI, DII, DIII, DIV) of Nav1.4 to generate proton currents at hyperpolarized voltages. The results are used to probe the location of the charged amino acids of the S4 segments in the VSD at hyperpolarized voltages. We propose a structural model of the resting state of Nav1.4 relaxed in its membrane/solution environment, which accounts for the experimental observations.
Results and Discussion
R1-to-H Substitution Causes a Proton Leak at Hyperpolarized Voltages in Specific Domains of Nav1.4.
Previous studies on the substitution R1-to-H on S4 of the Shaker potassium channel (12) and on S4 of the second domain (DII) of the Nav1.4 VGSC revealed the presence of a specific proton pathway (8). To determine whether the pathway could be observed when R1 of each domain of Nav1.4 was mutated, we prepared four constructs, in which H substituted R1 on each individual domain, and we assessed their potential to generate a proton leak. The experiments were performed in a chloride-free extracellular solution containing N-methyl-d-glucamine (NMDG), which is impermeable to many ion channels. The pH was adjusted using methanesulfonate in the appropriate pH buffers (Materials and Methods). The extracellular solution contained 1 μM TTX to block the ion permeation pathway through the main pore. In oocytes expressing DI/R1H or DII/R1H, a reversible inward current was recorded at a constant potential of –80 mV at varying extracellular pHs (7.4–6.5 and then 6.0) (Fig. 1B). Because the extracellular solution contained impermeable NMDG, this suggested that the inward current is carried by protons. The presence of TTX also ensured that the proton current permeated through the VSD. Similar reversible inward currents could not be recorded in DIII/R1H and DIV/R1H mutants (see the following section).
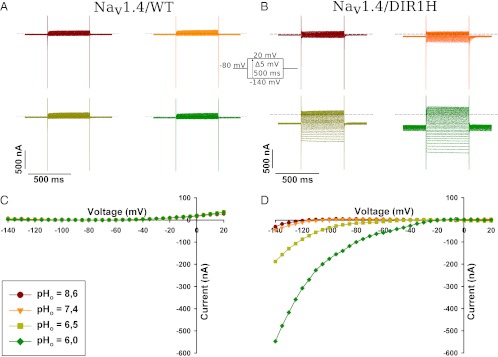
To assess the voltage dependence of the DI/R1H proton current, the current–voltage (I-V) relationship was constructed. The oocyte was held at –80 mV and pulses were applied from –140 mV to +20 mV in 5-mV increments for 500 ms. The proton currents generated exhibited a fast onset of activation and were stable for the duration of the pulses (Fig. 2 A and B). An offline leak subtraction was used to correct for the nonspecific linear leak, and the corresponding I-Vs were plotted (Fig. 2 C and D). The inward currents were larger when the extracellular solution was more acidic, which was likely due to a larger electrochemical gradient. The inward rectification was observed at all pHs (Fig. 2B).
Fig. 2.
I-V relationship of the proton current for NaV1.4/DI/R1H and NaV1.4/WT. Representative current traces recorded in response to an I-V protocol at different extracellular pHs in an oocyte expressing NaV1.4/WT (A) and NaV1.4/DI/R1H (B). The I-V protocol consisted of 500-ms voltage pulses from –140 mV to 20 mV in 5-mV increments while maintaining the oocyte at a holding potential of –80 mV. (Inset) I-V protocol. After offline linear leak subtraction, the values from current traces shown in (A and B) were plotted as a function of the test potential (C and D).
Because the I-V curve of the proton current does not reach a maximum near the V1/2 value of the gating current-voltage (Q-V) curve as it was described for the R365H (R2H) substitution in the Shaker channel, we infer that the protons do not permeate the VSD through a transport mechanism (21). The permeation pathway rather would involve a proton wire similar to what has been proposed for the R362H (R1H) substitution in the Shaker channel and to the permeation mechanism proposed for the gramicidin channel (12, 22).
DIII/R2H Mutation Yields a Proton Current, but Single-Mutation DIII/R1H and Single Mutations in DIV Do Not.
The I-V relationship was built for the constructs (Fig. 3A). The amplitudes of the proton currents generated by each construct were compared with the amplitude of proton current measured in the WT channel (0.07% ± 0.06% of the alpha current). Statistical analysis using Bonferroni’s post hoc test revealed that the DIII/R1H and DIV/R1H mutations did not generate a significant proton current (Fig. 3B). Thus, we sought to identify the R residue involved in the appearance of a leak. We mutated the second R of domains III and IV individually. Proton currents were measured at different extracellular pHs, and the I-V curves of the amplitudes of the proton currents were constructed (Fig. 3A). Although statistical analysis revealed that the mutation of R2 in DIII gave rise to a current, the corresponding mutation in domain IV did not. Furthermore, the DIV/R3H mutation did not give rise to a proton current (Fig. 3), which is in agreement with a recent study showing that a DIV/R1C mutation in a patient with paramyotonia congenital did not generate a gating pore current (23). The absence of a proton current in DIV indicated that the VSD of DIV is probably structurally different from the VSDs of the other domains. This hypothesis is supported by the observations of Capes et al. (24), who observed gating pore currents in DIV only when the first three Rs were mutated. This special DIV structure could explain why DIV plays a pivotal role in linking activation to inactivation (3, 25).
Fig. 3.
I-V relationship of the proton current for R-to-H substitutions in the S4 segments of the four domains of NaV1.4 and a comparison of current amplitudes at –140 mV and pHo 6.0. (A) The I-V curves at various extracellular pHs were measured in oocytes expressing mutant NaV1.4 channels. Offline linear leak subtraction was performed before the proton current amplitudes were normalized. The normalization consisted of dividing the current measured after linear leak subtraction by the maximum sodium current recorded with an I-V protocol for the oocyte in Ringer’s solution in the absence of TTX. The mean I-V relationships of the proton currents for all of the mutants are displayed. The error bars represent the SEM. (B) The magnitude of the proton currents (after leak subtraction and normalization) at –140 mV and pHo 6.0 were plotted as a function of the locations of the R-to-H substitutions. Significant differences were observed between the NaV1.4/WT and NaV1.4 mutant channels (*). When significant differences were found, the probability of the null hypothesis was very low (**P < 0.01, ***P < 0.001).
Proton Current Amplitude Varies Depending on the Domain.
In the Shaker channel, the gating pore current is estimated to represent 1–6% of the alpha current, depending on the nature of the substitution (13). In the present study, the proton current represented 2.2–4.4% of the alpha current, depending on the location of the substitution. The magnitude of the proton current was higher in DI than in DII and DIII (Fig. 3B). This may be an indication that the VSD of DI is more hydrated than the other VSDs, and that DI/R1 is more accessible to the solution than DII/R1 or DIII/R1. Furthermore, the amplitude of current recorded in DIV was not significant compared with the current measured in the WT channel.
Structural Model for the Resting State of Nav1.4 Suggests the VSD of DIV Has a Markedly Different Topology.
The topology of the solvent accessible volume in the VSDs of Kv channels has been inferred from the results of fluorescence neutron diffraction measurements and solid state NMR and MD simulations (26–28). In general, it was found that a network of interactions between basic, acidic and polarizable residues [such as the conserved F (S2)] holds the four-helix bundle together (29). The hour-glass-shaped solution permeates the bundle from both sides of the membrane (28, 30). The constriction site that prevents the solutions from communicating at the center of the domain is located near the gating charge transfer center involving the conserved F residue of S2 (31). Because the position of S4 changes upon activation, a specific positive residue occupies this site at a given voltage. It is the mutation of this residue that leads to the appearance of gating pore currents at this specific voltage.
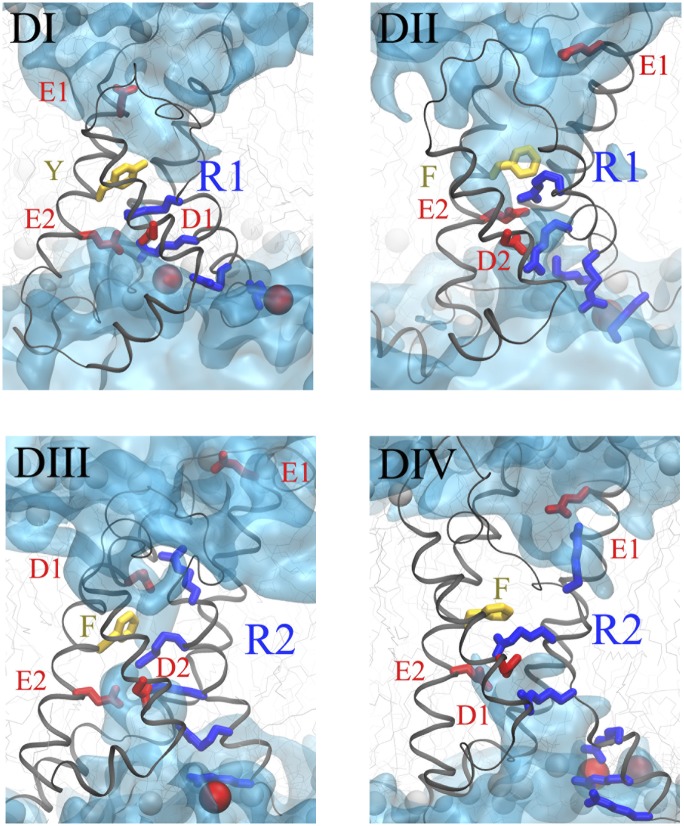
To construct a resting state model of Nav1.4 based on the results presented previously, we generated a homology model of the TM domains of Nav1.4 based on its alignment (Fig. 1A) with the crystal structure of its closest homolog of known structure, NavAb. We then biased the conformation of its VSDs to simultaneously locate DI/R1, DII/R1, and DIII/R2 in their respective constriction sites (Materials and Methods and SI Materials and Methods). Because no conclusive data were produced in this work for DIV, we chose to place DIV/R2 close to the constriction site (see discussion in the following section). The structures were then relaxed in their membrane/solution environment. The interaction network between the positive charges of S4 and negative charges of S2/S3 remained in place over trajectories spanning tens of nanoseconds in each of the subunits (Fig. 4 and Fig. S1). As with Kv1.2, the lipid head groups of the bottom leaflet served as binding sites for the bottom charges of S4 in each subunit. The topology of the solvent accessible volume was markedly different in DIV from the one observed in the first three domains (Fig. 4 and Fig. S2). Although the constriction region was ∼2 Å long in DI–DIII, which is reminiscent of Kv channels (27, 30, 32), it was much longer in DIV, where the amino acid sequence seemed to prevent water molecules from hydrating the center of the VSD.
Fig. 4.
Structural model of the resting state VSDs of the four domains of NaV1.4. The four backbones are represented as gray ribbons, the positively charged residues of S4 as blue sticks, and the negative counter charges in red (sticks for the protein residues of S1, S2, and S3; spheres for the lipid head groups). The conserved residues of S2 defining the hydrophobic seal are shown in yellow. The numbering of the residues corresponds to the one in Fig. S1. The hourglass-like solvent accessible volume is shown as a transparent cyan surface. Note the similarity of the first three domains (thin hydrophilic septum). Domain IV stands out with a several-angstrom-thick hydrophobic volume in the center of the VSD.
The position of S4 in domain DIV remains uncertain because of the inconclusive electrophysiology measurements. Nevertheless, the computed structure seems to agree with previous experimental results. Indeed, it has been shown that mutations of R1, R2, and R3 are required to create a gating pore current through DIV (24). The notion that the hydrophobic septum of DIV is very large compared with the other domains is in agreement with this observation. In addition, placing R2 in the constriction site positions R1 and R3 close enough to the site provides a possible explanation for the observations of Capes et al. (24), who reported that three substitutions in DIV are required to observe a gating pore current. In this position, R1 is located in the extracellular water crevice, whereas R3 is located in the intracellular water crevice. Because no mutated residue has access to both water crevices, no gating pore current is generated. Furthermore, this location of R2 may be in agreement with the results of Yang et al. (2). In fact, R1 might be accessible to water and protons but not to the 2-(trimethylammonium)ethyl ethanethiosulfonate (MTSET) reagent because the technique used by Yang et al. has a lower spatial resolution than the techniques used in the present study (21). Moreover, it seems reasonable to place R1 in the extracellular water crevice because this makes the R1 and R3 residues accessible from one of the sides of the membrane, which is necessary to stabilize the DIV/S4 in a TM position.
The large hydrophobic septum identified in the present study would confer DIV its slow activation kinetics (3). In fact, the free energy barrier for the transfer of a charge across a low dielectric (the hydrophobic septum) is much higher than across a high dielectric (such as a salt solution) (33). Because the energy required to move the S4 may be higher for DIV than for the other domains, the activation of DIV/S4 may be delayed compared with the S4s of DI, DII, and DIII, as reported by Chanda and Bezanilla (3). Because the movement of DIV/S4 is linked to the inactivation of the channel (25), it is important that the conformation of the VSD of DIV changes last (3).
DIII Activates Before DII and DI.
The I-V curves of DI/R1H, DII/R1H, and DIII/R2H displayed statistically significant proton currents. In addition, they all exhibited an inward rectification near –50 mV. Such a rectification should be attributed to the closure of the gating pore because of the movement of the S4 segment rather than to the lowering driving force. Indeed, –50 mV is the value around which the voltage sensor starts moving outward. Moreover, this rectification is not the effect of the test potential neighboring the equilibrium potential because the equilibrium potential is evaluated to be around +71 mV. Taking into account that the oocytes injected with the DI/R1H, DII/R1H, or DIII/R2H mutant channels would undergo acidification during the incubation would only lower the equilibrium potential to values neighboring +53 mV because the internal pH of the oocyte would be around 6.9 rather than 7.2 (10). The rectification potential of the proton current was different for each substitution (Fig. 3A), suggesting that the S4 of each domain starts moving separately. A Student t test was used to assess the values at which the currents recorded at pH = 6 were statistically different from those recorded at pH = 8.6. A –35 mV rectification potential was found for DI/R1H, –45 mV for DII/R1H, and –50 mV for DIII/R2H, respectively. These values indicated that the S4 of DIII activates more promptly than the S4s of DII and DI. As mentioned previously, the rectification potential of the proton current in DIV could not be measured because the amplitude of the current measured was not statistically significant. However, the MD simulations tended to indicate that the activation barrier is higher in this particular domain. We thus inferred that the S4 of DIV is likely activated last. Our results are in agreement with the sequence of activation reported by Chanda and Bezanilla (3), i.e., DIII, DII, DI, and then DIV.
The present study offers a structural model of mammalian Nav1.4 VSDs in their resting state at hyperpolarized voltages. The model also highlights the different topologies of the four domains. Moreover, our results provide a better understanding of the mechanisms that cause pathologies linked to the appearance of gating pore currents such as hypokalemic periodic paralysis, normokalemic periodic paralysis cardiac arrhythmias associated with dilated cardiomyopathy (7–10). Last, other pathologies such as epilepsy and familial hemiplegic migraine may also be linked to gating pore currents (34).
Materials and Methods
Electrophysiology.
Xenopus oocytes, which were handled in accordance with the principles and guidelines of the Université Laval animal protection committee and the Canadian Council on Animal Care, were prepared as described previously (25). Macroscopic currents from mRNA-injected oocytes were recorded using the voltage-clamp technique with two 1-mM KCl-filled microelectrodes. The membrane potential for the two-microelectrode voltage-clamp technique was controlled using a Warner oocyte clamp (Warner Instrument Corp.). The currents were filtered at 5 kHz (–3 dB; four-pole Bessel filter). For experiments using chloride-free solutions, the headstage of the Warner amplifier was attached to a plastic pool containing a 3-M NaCl solution through a silver chloride wire connected to the bath solution using an agar bridge. The bridge contained 3% (wt/vol) agar, 500 mM NMDG, and 10 mM Hepes (pH 7.4) and was threaded with a thin platinum/iridium wire to lower the high-frequency impedance. In all of the experiments using chloride-free solutions, alpha currents were monitored by recording sodium currents in Ringer’s solution before superfusing the oocytes with a sodium-free solution containing 1 μΜ TTX. Because the amplitudes of gating pore currents are closely correlated with the amplitudes of alpha currents (13), the proton currents were normalized to the sodium current (Fig. 3A). This normalization was required to compare the magnitudes of the proton currents measured with each histidine substitution (Fig. 3B). To report only the proton-specific current, the linear leak was subtracted from the I-V curves before normalization. The linear leak substraction consisted in fitting the data points obtained between –20 mV and –40 mV to a linear equation. The value of the linear leak was then calculated for each data point according to the linear equation and subtracted from the I-V curve.
Solutions and Reagents.
The Ringer's bathing solution was composed of 116 mM NaCl, 2 mM KCl, 2 mM CaCl2, 2.9 mM MgCl2, and 5 mM Hepes. The pH was adjusted to 7.6 at 22 °C using 1 M NaOH. The external chloride-free solution was composed of 120 mM NMDG, 2 mM CaCl2, and either 20 mM N-cyclohexyl-2-aminoethanesulfonic acid (pH 8.6), 20 mM Hepes (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (pH 7.4–6.8), or 20 mM MES (pH ≤ 6.5). The NMDG solutions were adjusted to the desired pH using methanesulfonic acid (Sigma). For proton current recordings, 1 μM TTX was added to the NMDG solution. All of the recording solutions had an osmolarity of 240–260 mOsm. All of the chemicals and drugs were purchased from Sigma, except for tetrodotoxin, which was purchased from Latoxan. All of the experiments were carried out at 22 °C.
Homology Modeling and MD.
A homology model of Nav1.4 was constructed based on the NavAb X-ray structure (4), identified as the best template among the protein data bank (pdb) structures by a position-specific iterative Position-Specific Iterated (PSI)-BLAST search. To this end, a ClustalW2 multiple sequence alignment of the first 100 sequences that were homologous to NavAb (identified among the National Center for Biotechnology Information reference protein database using the PSI-BLAST server) was built (35). In agreement with previous work (36), these sequences encompass voltage-dependent calcium channels, voltage-gated sodium channels (Nav1.1–Nav1.9), cation channel sperm-associated proteins, and two pore calcium channel proteins. Resorting to a multiple sequence alignment ensures that the pairwise alignment between NavAb and Nav1.4 is of good quality. The structure obtained with MODELLER (http://salilab.org/modeller/) was inserted into a model 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) membrane and embedded in a 150-mM NaCl solution. The systems were then equilibrated under normal constant temperature and pressure conditions (298 K, 1 atm) after constraining the positions of all of the atoms of the channel during the first nanoseconds to ensure reorganization of the lipids and solution. Details of the homology modeling and MD simulation parameters can be found in SI Materials and Methods.
Because the template corresponds to a state of the NavAb in which the pore is closed but the VSDs are “partially activated” (4), the corresponding Nav1.4 homology model also features the VSDs in a “partially activated” state. Note here that two recent structures of bacterial channels were resolved, confirming the overall structure of the VSD (37) and further proposing a fully activated state of the VSD (38). The resting state was reached by applying external forces using a procedure comparable to the one used in the study of Kv1.2 (15). Based on the patch clamp experiment results, we biased our MD simulation in a stepwise procedure to drag DI/R1, DII/R1, and DIII/R2 toward the constriction site while maintaining a maximum number of basic/acidic residues interactions in the VSD (Fig. S2). Because the data for domain DIV were inconclusive, we chose to bias DIV/R2 toward the constriction site based on other experimental results (3). Details of this procedure can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
This study was supported by grants from the Heart and Stroke Foundation of Quebec and the Canadian Institutes of Health Research (Grant MOP-86564). M.L.K. acknowledges support from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217990109/-/DCSupplemental.
References
- 1.Stühmer W, et al. Structural parts involved in activation and inactivation of the sodium channel. Nature. 1989;339(6226):597–603. doi: 10.1038/339597a0. [DOI] [PubMed] [Google Scholar]
- 2.Yang N, George AL, Jr, Horn R. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 1996;16(1):113–122. doi: 10.1016/s0896-6273(00)80028-8. [DOI] [PubMed] [Google Scholar]
- 3.Chanda B, Bezanilla F. Tracking voltage-dependent conformational changes in skeletal muscle sodium channel during activation. J Gen Physiol. 2002;120(5):629–645. doi: 10.1085/jgp.20028679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475(7356):353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yarov-Yarovoy V, et al. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc Natl Acad Sci USA. 2012;109(2):E93–E102. doi: 10.1073/pnas.1118434109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tikhonov DB, Zhorov BS. Architecture and pore block of eukaryotic voltage-gated sodium channels in view of NavAb bacterial sodium channel structure. Mol Pharmacol. 2012;82(1):97–104. doi: 10.1124/mol.112.078212. [DOI] [PubMed] [Google Scholar]
- 7.Sokolov S, Scheuer T, Catterall WA. Gating pore current in an inherited ion channelopathy. Nature. 2007;446(7131):76–78. doi: 10.1038/nature05598. [DOI] [PubMed] [Google Scholar]
- 8.Struyk AF, Cannon SC. A Na+ channel mutation linked to hypokalemic periodic paralysis exposes a proton-selective gating pore. J Gen Physiol. 2007;130(1):11–20. doi: 10.1085/jgp.200709755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sokolov S, Scheuer T, Catterall WA. Depolarization-activated gating pore current conducted by mutant sodium channels in potassium-sensitive normokalemic periodic paralysis. Proc Natl Acad Sci USA. 2008;105(50):19980–19985. doi: 10.1073/pnas.0810562105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gosselin-Badaroudine P, et al. A proton leak current through the cardiac sodium channel is linked to mixed arrhythmia and the dilated cardiomyopathy phenotype. PLoS ONE. 2012;7(5):e38331. doi: 10.1371/journal.pone.0038331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sokolov S, Scheuer T, Catterall WA. Ion permeation through a voltage-sensitive gating pore in brain sodium channels having voltage sensor mutations. Neuron. 2005;47(2):183–189. doi: 10.1016/j.neuron.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 12.Starace DM, Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 2004;427(6974):548–553. doi: 10.1038/nature02270. [DOI] [PubMed] [Google Scholar]
- 13.Tombola F, Pathak MM, Isacoff EY. Voltage-sensing arginines in a potassium channel permeate and occlude cation-selective pores. Neuron. 2005;45(3):379–388. doi: 10.1016/j.neuron.2004.12.047. [DOI] [PubMed] [Google Scholar]
- 14.Delemotte L, Treptow W, Klein ML, Tarek M. Effect of sensor domain mutations on the properties of voltage-gated ion channels: Molecular dynamics studies of the potassium channel Kv1.2. Biophys J. 2010;99(2):L72–L74. doi: 10.1016/j.bpj.2010.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delemotte L, Tarek M, Klein ML, Amaral C, Treptow W. Intermediate states of the Kv1.2 voltage sensor from atomistic molecular dynamics simulations. Proc Natl Acad Sci USA. 2011;108(15):6109–6114. doi: 10.1073/pnas.1102724108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen MØ, et al. Mechanism of voltage gating in potassium channels. Science. 2012;336(6078):229–233. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- 17.Khalili-Araghi F, Tajkhorshid E, Roux B, Schulten K. Molecular dynamics investigation of the ω-current in the Kv1.2 voltage sensor domains. Biophys J. 2012;102(2):258–267. doi: 10.1016/j.bpj.2011.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geissler PL, Dellago C, Chandler D, Hutter J, Parrinello M. Autoionization in liquid water. Science. 2001;291(5511):2121–2124. doi: 10.1126/science.1056991. [DOI] [PubMed] [Google Scholar]
- 19.Hassanali A, Prakash MK, Eshet H, Parrinello M. On the recombination of hydronium and hydroxide ions in water. Proc Natl Acad Sci USA. 2011;108(51):20410–20415. doi: 10.1073/pnas.1112486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Wu Y, Voth GA. Origins of proton transport behavior from selectivity domain mutations of the aquaporin-1 channel. Biophys J. 2006;90(10):L73–L75. doi: 10.1529/biophysj.106.084061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Starace DM, Bezanilla F. Histidine scanning mutagenesis of basic residues of the S4 segment of the shaker k+ channel. J Gen Physiol. 2001;117(5):469–490. doi: 10.1085/jgp.117.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pomès R, Roux B. Molecular mechanism of H+ conduction in the single-file water chain of the gramicidin channel. Biophys J. 2002;82(5):2304–2316. doi: 10.1016/S0006-3495(02)75576-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis DG, Rybalchenko V, Struyk A, Cannon SC. Leaky sodium channels from voltage sensor mutations in periodic paralysis, but not paramyotonia. Neurology. 2011;76(19):1635–1641. doi: 10.1212/WNL.0b013e318219fb57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capes DL, Arcisio-Miranda M, Jarecki BW, French RJ, Chanda B. Gating transitions in the selectivity filter region of a sodium channel are coupled to the domain IV voltage sensor. Proc Natl Acad Sci USA. 2012;109(7):2648–2653. doi: 10.1073/pnas.1115575109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chahine M, et al. Sodium channel mutations in paramyotonia congenita uncouple inactivation from activation. Neuron. 1994;12(2):281–294. doi: 10.1016/0896-6273(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 26.Bezanilla F. The voltage-sensor structure in a voltage-gated channel. Trends Biochem Sci. 2005;30(4):166–168. doi: 10.1016/j.tibs.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Treptow W, Tarek M. Environment of the gating charges in the Kv1.2 Shaker potassium channel. Biophys J. 2006;90(9):L64–L66. doi: 10.1529/biophysj.106.080754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krepkiy D, et al. Structure and hydration of membranes embedded with voltage-sensing domains. Nature. 2009;462(7272):473–479. doi: 10.1038/nature08542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pless SA, Galpin JD, Niciforovic AP, Ahern CA. Contributions of counter-charge in a potassium channel voltage-sensor domain. Nat Chem Biol. 2011;7(9):617–623. doi: 10.1038/nchembio.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jogini V, Roux B. Dynamics of the Kv1.2 voltage-gated K+ channel in a membrane environment. Biophys J. 2007;93(9):3070–3082. doi: 10.1529/biophysj.107.112540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328(5974):67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freites JA, Tobias DJ, White SH. A voltage-sensor water pore. Biophys J. 2006;91(11):L90–L92. doi: 10.1529/biophysj.106.096065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacCallum JL, Bennett WFD, Tieleman DP. Partitioning of amino acid side chains into lipid bilayers: Results from computer simulations and comparison to experiment. J Gen Physiol. 2007;129(5):371–377. doi: 10.1085/jgp.200709745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jurkat-Rott K, Groome J, Lehmann-Horn F. Pathophysiological role of omega pore current in channelopathies. Front Pharmacol. 2012;3:112. doi: 10.3389/fphar.2012.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Pavlov E, Bladen C, Diao C, French RJ. In: Bacterial Ion Channels and Their Eukaryotic Homologs. Kubalski A, Martinac B, editors. Washington, DC: ASM Press; 2005. pp. 191–207. [Google Scholar]
- 37.Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486(7401):135–139. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486(7401):130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.