Abstract
The human malaria parasite Plasmodium falciparum is auxotrophic for most amino acids. Its amino acid needs are met largely through the degradation of host erythrocyte hemoglobin; however the parasite must acquire isoleucine exogenously, because this amino acid is not present in adult human hemoglobin. We report that when isoleucine is withdrawn from the culture medium of intraerythrocytic P. falciparum, the parasite slows its metabolism and progresses through its developmental cycle at a reduced rate. Isoleucine-starved parasites remain viable for 72 h and resume rapid growth upon resupplementation. Protein degradation during starvation is important for maintenance of this hibernatory state. Microarray analysis of starved parasites revealed a 60% decrease in the rate of progression through the normal transcriptional program but no other apparent stress response. Plasmodium parasites do not possess a TOR nutrient-sensing pathway and have only a rudimentary amino acid starvation-sensing eukaryotic initiation factor 2α (eIF2α) stress response. Isoleucine deprivation results in GCN2-mediated phosphorylation of eIF2α, but kinase-knockout clones still are able to hibernate and recover, indicating that this pathway does not directly promote survival during isoleucine starvation. We conclude that P. falciparum, in the absence of canonical eukaryotic nutrient stress-response pathways, can cope with an inconsistent bloodstream amino acid supply by hibernating and waiting for more nutrient to be provided.
Keywords: protease, artemesinin, autophagy
Human red blood cells (RBCs) provide the intraerythrocytic malaria parasite Plasmodium falciparum with an abundant nutrient supply in the form of hemoglobin. However, human hemoglobin lacks the amino acid isoleucine (1). Isoleucine is present in more than 99% of the proteins encoded by P. falciparum (2), and because Plasmodium is unable to synthesize this amino acid de novo (3), the parasite must obtain isoleucine from human serum (4, 5). Humans also cannot make isoleucine and must acquire this essential amino acid through the diet (3, 6). In endemic regions, malaria patients often are severely malnourished, drastically limiting the availability of free amino acids in the plasma (7, 8). Normal plasma isoleucine levels are in the 100-μM range but can be less than one-tenth this concentration in malnourished children (7). During in vitro culturing, P. falciparum growth is optimal above 20 μM isoleucine, but the parasite is unable to grow in medium devoid of isoleucine (4). This observation raises the question of how P. falciparum responds to low isoleucine conditions that may exist during human infection.
Eukaryotes have canonical mechanisms for responding to amino acid deprivation. The target of rapamycin (TOR) complex, which functions as a master regulator of cell growth (9), is repressed during amino acid starvation. Conversely, the eukaryotic initiation factor 2 alpha (eIF2α) kinase, GCN2, is activated by amino acid-limiting conditions (10, 11). Activated GCN2 mediates a reduction in translation efficiency, allowing resource conservation, metabolic readjustment, and promotion of an adaptive transcriptional program, inducing GCN4 in yeast and ATF4 in mammals. These transcription factors control the response to amino acid deprivation by turning on pathways for amino acid biosynthesis, among others (12, 13). Plasmodium does not have a TOR complex (14) and lacks the downstream transcription factors and biosynthetic pathways that mediate GCN2 action. An ortholog of eIF2α and three putative eIF2α kinases have been identified previously in the P. falciparum genome (15–17). One of them, PfeIK2, controls latency in sporozoite development in the mosquito (17). Another, PfPK4, is involved in intraerythrocytic schizogony (18). The third, PfeIK1, recently has been confirmed as the amino acid-sensing GCN2 ortholog, active in the blood stages of the parasite and able to phosphorylate eIF2α in response to amino acid starvation (15). A knockout of GCN2 in the related apicomplexan parasite Toxoplasma gondii has an extracellular tachyzoite fitness defect (19), but the biological role of the Plasmodium ortholog has not been defined and is tenuous, given the lack of GCN2-responsive transcription factors and amino acid biosynthesis pathways.
To understand how P. falciparum responds to and survives amino acid limitation, we monitored the growth recovery, metabolic activity, and gene expression of cultured parasites exposed to isoleucine-free medium. We show that parasites slow their metabolism and cell-cycle progression in a hibernatory fashion that allows them to survive prolonged isoleucine starvation. Notably, its GCN2 amino acid-sensing pathway is active but does not play a role in starvation survival. We conclude that P. falciparum hibernates upon exposure to amino acid limitation to allow its long-term survival.
Results
P. falciparum Growth Is Recoverable After Prolonged Isoleucine Starvation.
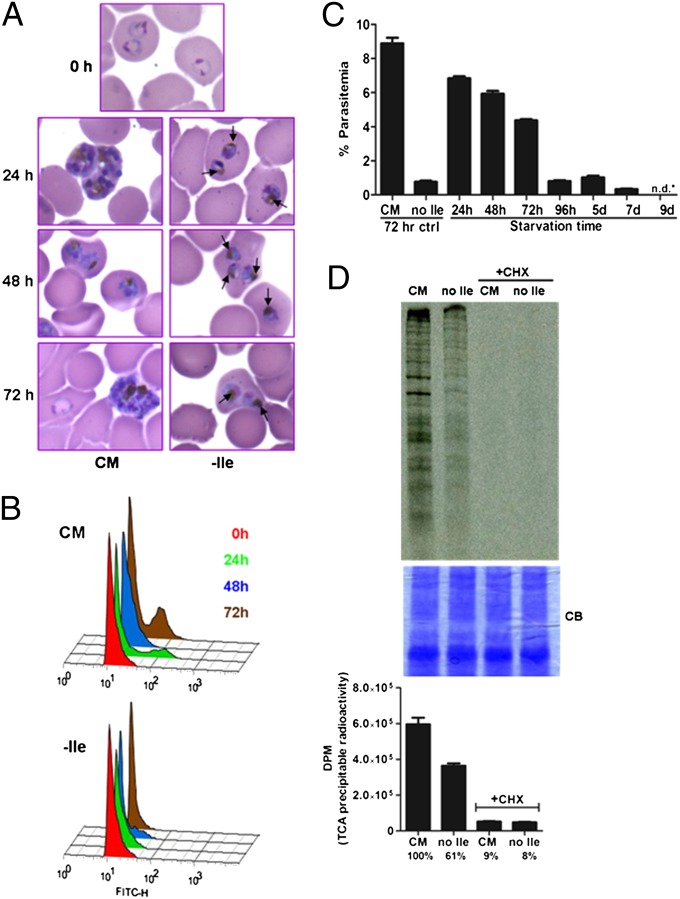
We monitored the growth of synchronized ring-stage P. falciparum parasites (Fig. 1). In complete medium (CM), parasites progressed normally through the asexual cycle, reinvaded fresh RBCs, and continued growth. However, in the absence of isoleucine (−Ile), parasites progressed slowly to the trophozoite stage (Fig. 1A) but did not enter S-phase, as indicated by the absence of a high DNA content peak at 24 to 72 h postincubation in flow cytometry traces (Fig. 1B). Surprisingly, parasite morphology remained essentially normal during isoleucine starvation (Fig. 1A). In contrast, within hours of glucose starvation, P. falciparum parasites appeared as shrunken, rounded bodies with pyknotic nuclei and failed to recover (Fig. S1). To determine whether isoleucine-starved parasites maintain viability, we incubated synchronized ring-stage parasites in isoleucine-free RPMI medium for varying periods of time and then supplemented each starved culture with isoleucine at the concentration found in complete RPMI (382 μM). Parasite growth was followed for an additional 72 h, and parasitemia was measured by flow cytometry. When parasites were starved for isoleucine for 24, 48, and 72 h, a large fraction of the parasite population was able to recover (Fig. 1C). However, parasites starved for longer periods (up to 9 d) were no longer detectable on Giemsa-stained blood smears, and recovery of growth after isoleucine supplementation dropped precipitously (Fig. 1C). In cultures with lower recovery (i.e., those that had been starved for 4 d or more), gametocytes were undetectable, and asexual forms remained prevalent, suggesting that the vast reductions in the recovery of growth by parasites subjected to extended starvation were caused by decreased viability.
Fig. 1.
P. falciparum growth during and recovery from isoleucine starvation. (A) Representative images of Giemsa-stained thin blood smears prepared from parasites at 0, 24, 48, and 72 h of incubation in CM or Ile-free (−Ile) RPMI. Arrows in images from isoleucine-starved cultures indicate hemozoin pigmentation. (B) Flow cytometry assessment of DNA content. Synchronous 3D7 parasites were grown in CM or Ile-free RPMI, and samples were harvested at 0 (red), 24 (green), 48 (blue), and 72 (brown) h. Offset overlaid histograms of FITC-H channel fluorescence for the indicated time points and medium conditions are shown. Samples were treated with RNase, allowing haploid ring and trophozoite populations (left-most peak) to be better distinguished from polyploid schizonts (right-most peaks). The gated uninfected RBC population was removed for clarity. (C) Growth recovery following isoleucine resupplementation of parasites starved for the indicated times. A control set of parasites were fed (CM) or isoleucine-starved (no Ile) for 72 h. Synchronized 3D7 parasites were starved for up 9 d, followed by supplementation with isoleucine. Parasitemia of all cultures was measured by flow cytometry after 72 h of recovery. Data shown represent the mean parasitemia ± SEM; n = 3; n.d.*, none detected. (D) Protein synthesis in starved parasites. Parasites were fed or starved for 6 h and labeled with [35S]methionine/cysteine for the last hour while incubated in CM or isoleucine-free (no Ile) labeling RPMI medium in the presence or absence of the protein synthesis inhibitor cycloheximide (CHX). Parasite proteins were resolved by SDS/PAGE for autoradiography (Upper) or were TCA-precipitated to determine incorporated radioactivity through scintillation counting (Lower). The SDS/PAGE gel was stained with Coomassie Brilliant Blue (CB) to ensure even protein loading. Data shown represent the mean disintegrations per minute (DPM) of incorporated radioactivity ± SEM; n = 6.
To determine whether protein translation is affected in isoleucine-starved parasites, we incubated synchronized parasites in complete or isoleucine-free labeling medium containing [35S] methionine and cysteine. Starved parasites incorporated the radiolabel into protein, but at a reduced rate, indicating a slowed metabolism (Fig. 1D). Recovery from starvation was similar whether parasites were adapted to low (20 μM) or high (200 μM) concentrations of exogenous isoleucine before starvation (Fig. S2), suggesting that parasite survival during starvation does not require an abundant preexisting isoleucine pool.
Impairment of Proteolysis During Isoleucine Starvation Abrogates Recovery.
Members of the aspartic and cysteine protease families, plasmepsins and falcipains, respectively, reside in the digestive vacuole (DV) of P. falciparum, where they degrade massive amounts of host-cell hemoglobin, a process that supplies the parasite with amino acids (4). The bulk of hemoglobin degradation takes place during the trophozoite stage, when the metabolic activity of P. falciparum is at its highest (20). During long-term isoleucine starvation, parasites display evidence that hemoglobin degradation remains active, in that hemozoin (the sequestered heme byproduct of catabolism) becomes visible in the DV within 24 h (Fig. 1A, arrows).
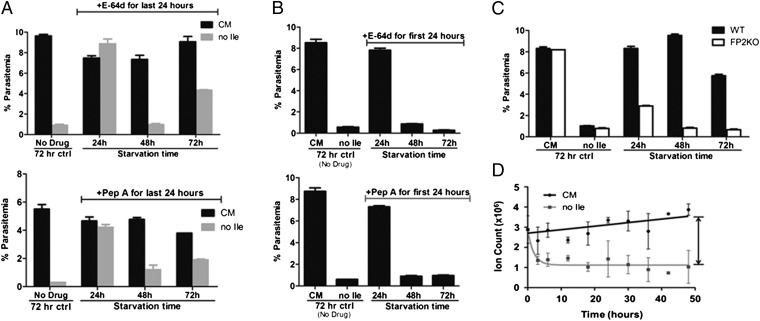
To determine whether proteolytic activity is required to maintain viability, we incubated ring-stage parasites in isoleucine-free RPMI and exposed them to E-64d, a membrane-permeable cysteine protease inhibitor, for a 24-h window at different times during the starvation period. Under the conditions used, growth could be restored in a fed control (i.e., cultured in CM) after drug removal (Fig. 2A). Parasites starved for 24 h in the presence of E-64d recovered well after washout of the drug and resupplementation with isoleucine. Even parasites starved for 72 h and exposed to E-64d for the last 24 h of the incubation recovered nearly half of the growth seen in controls, in line with the decrease in recovery seen previously under similar starvation conditions without drug (Fig. 1C). However, parasites starved for 48 h with E-64d present during the last half of the incubation (the trophozoite stage with peak hemoglobin degradation) were not viable (Fig. 2A).
Fig. 2.
Protease activity is required to maintain viability during isoleucine starvation. (A) Synchronous 3D7 parasites were fed (black bars) or starved for isoleucine (gray bars) for the indicated times in the presence of 10 μM E-64d (Upper) or 5 μM pepstatin A (Pep A, Lower) for the last 24 h of the incubation. After drug removal, each culture was replated in CM for recovery. Parasitemia of all cultures was measured by flow cytometry after 72 h of recovery. A control set of parasites, shown on the far left of each graph, was grown in the absence of drug for 72 h. Data shown represent the mean parasitemia ± SEM; n = 3. (B) Synchronous 3D7 parasites were starved for isoleucine for the indicated times in the presence of 10 μM E-64d (Upper) or 5 μM pepstatin A (Pep A, Lower) for the first 24 h of the incubation. After drug removal (and extended starvation for the 48-h and 72-h samples), parasites were replated in CM for recovery. Parasitemia of all cultures was measured by flow cytometry after 72 h of recovery. A control set of parasites was fed (CM) or isoleucine-starved (no Ile) in the absence of drug for 72 h. Data shown represent the mean parasitemia ± SEM; n = 3. (C) Growth recovery after isoleucine resupplementation of synchronous 3D7 (WT, black bars) and fp2 knockout (FP2KO, white bars) parasites incubated in Ile-free RPMI for the indicated times. Control parasites were fed (CM) or starved for isoleucine without refeeding (no Ile) for 72 h. Parasitemia of all cultures was measured by flow cytometry after 72 h of recovery. Data shown represent the mean parasitemia ± SEM; n = 3. Each experiment was repeated at least three times. Representative experiments are shown. (D) Intracellular isoleucine content of fed (CM) and isoleucine-starved (no Ile) parasites over time. Extracts of 3D7 infected-erythrocyte cultures were analyzed via LC/MS. Signal from uninfected erythrocyte (cultured under the same conditions) extracts was subtracted from the infected-erythrocyte signal. The data are presented as the total ion count detected for derivatized isoleucine. Curves were fitted using the equation Y = (Y0 - P)*exp(-K*X) + P, where P is the Y value at infinite times and K is the rate constant. Data shown represent the mean ± SD, n = 2.
We performed a similar experiment with the aspartic protease inhibitor pepstatin A. As with E-64d, parasites cultured in CM and treated with pepstatin A maintained viability (Fig. 2A). Again, parasites starved for 48 h and treated with inhibitor for the last 24 h were not viable, whereas parasites starved and treated for 24 h recovered well. Although recovery was notably lower in the pepstatin A-treated 72 h-starved cultures than in the corresponding E-64d–treated cultures, the surviving parasites consistently outgrew those from the 48 h-starved condition. These data suggest that proteolytic activity is most critical between 24–48 h of starvation, soon after which the parasite appears to reach its developmental limit.
The morphology of starved parasites over time indicates that they transition slowly through the trophozoite stage, displaying early characteristics of this stage at 24 h of starvation (Fig. 1A). As noted above, when parasites were starved for 24 h in the presence of E-64d or pepstatin A, recovery of growth was relatively unaffected. However, when starvation time was extended beyond the withdrawal of either drug, recovery was attenuated (Fig. 2B, compare with Fig. 2A, gray bars). It appears that continuous protein degradation is essential for surviving the second day of starvation, and protease inhibition throughout the first or second day of isoleucine starvation is lethal to the parasite.
The cysteine protease falcipain 2 (FP2) plays a pivotal role in hemoglobin degradation of P. falciparum, in that its genetic disruption leads to the accumulation of undigested hemoglobin and swelling of the DV (21), a phenotype similarly seen with E-64d treatment. We incubated a clonal line of fp2− parasites (4) in isoleucine-free RPMI and then supplemented each starved culture with isoleucine to allow recovery. Parasites lacking FP2 exhibited significant defects in recovery after starvation, with only minimal growth restored in mutants starved for 24 h and virtually no recovery of the mutant parasites starved for 48 or 72 h (Fig. 2C). These data complement the protease inhibitor studies and emphasize the requirement for DV proteolysis in surviving isoleucine starvation.
We also used mass spectrometry-based metabolite detection to measure isoleucine levels in starved and control parasites over a 48-h period. Control parasites maintained a consistently high level of isoleucine. Surprisingly, starved parasites contained low but detectable levels of free isoleucine throughout the starvation period (Fig. 2D), despite the complete removal of exogenous isoleucine from the culture medium. This observation is consistent with the requirement for proteolysis to maintain viability.
Starvation-Induced Hibernation Differs from Artemisinin-Associated Quiescence.
In patients with falciparum malaria, monotherapy with artemisinin frequently results in parasite recrudescence (22). Parasites recovered posttherapy remain sensitive to drug. A similar phenomenon has been observed in vitro with parasites cultured in the presence of artemisinin and has given rise to the concept of parasite dormancy, a state that presumably confers drug tolerance (22–24). To determine whether isoleucine-starved parasites are in a similar dormant state, we cultured parasites in isoleucine-free medium, exposed them to artemisinin, and then assessed viability. There was no increased resistance to artemisinin in starved parasites; if anything, there was a slight increase in sensitivity (Fig. S3). Therefore isoleucine starvation status does not appear to be equivalent to artemisinin-associated dormancy.
Expression Profile of Starved Parasites Reveals a Delayed Growth Phenotype.
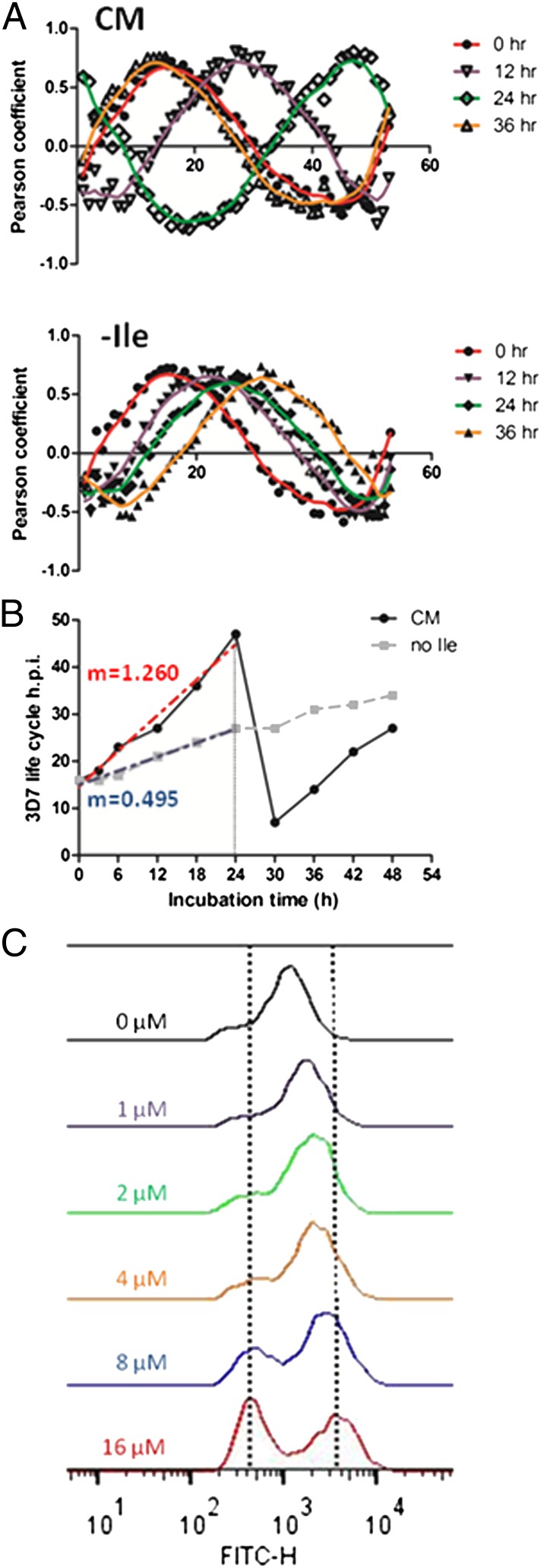
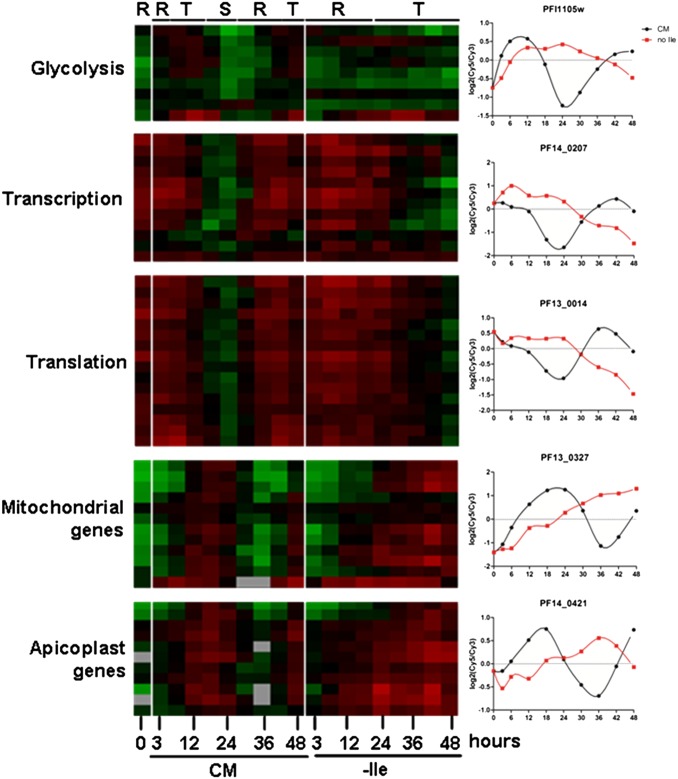
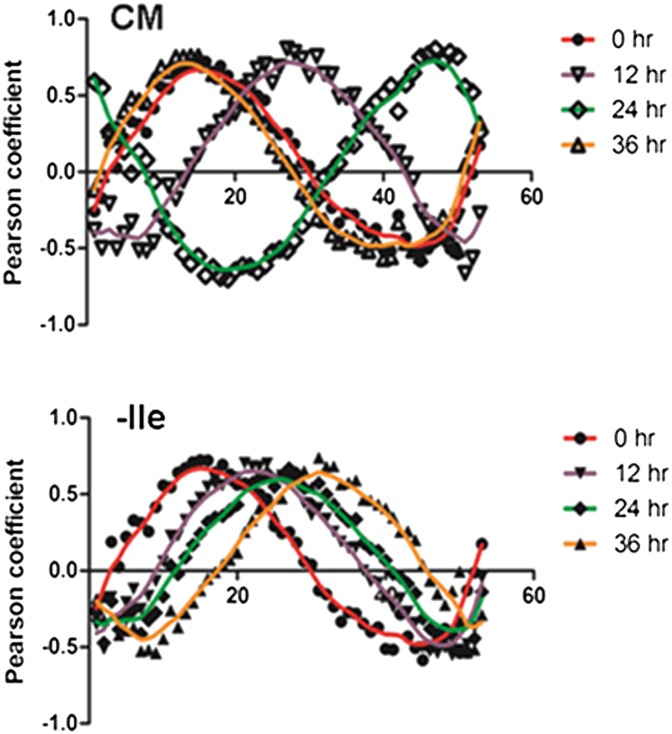
Global transcription in P. falciparum is characterized by successive waves of gene activation tightly coordinated with the parasite’s developmental progression (25, 26). To examine mRNA abundance in isoleucine-starved parasites, we isolated RNA from synchronized parasites incubated in isoleucine-free RPMI, harvesting samples after 3 and 6 h of incubation and every 6 h thereafter up to 48 h total. In parallel, RNA was isolated from a control set of synchronized parasites that were maintained in CM and harvested along with the starved samples. The expression level of nearly 4,800 genes was analyzed from both conditions at each time point by DNA microarray (Dataset S1). We observed that gene expression of control parasites (CM) best correlated with isoleucine-starved parasites (−Ile) that had been grown for 2.5 to threefold longer periods [e.g., the 18-h −Ile sample correlated best with the 6-h CM sample, and the 30-h −Ile sample correlated best with the 12-h CM sample] (Table S1). The starvation-associated growth retardation occurred quickly, with significant transcriptional deviation between the fed and starved samples apparent within 6 h of incubation (Table S1). Using Pearson correlation, the expression profiles generated from this dataset were compared with those from a high-resolution transcriptional array study that sampled the complete intraerythrocytic developmental cycle (IDC) of in vitro-cultured P. falciparum every hour (27). These results demonstrate the remarkably retarded progression of the isoleucine-starved parasites through the trophozoite stage (Fig. 3 A and B and Dataset S2). The developmental rate of the starved parasites through one life cycle decreased by 60%, ultimately ending at the midtrophozoite stage, whereas the CM-fed control parasites progressed normally through the IDC, transitioning from late rings to trophozoites to schizonts and continuing on to initiate another round of invasion (Fig. 3B and Dataset S2). Parasites cultured in suboptimal but nonzero concentrations of isoleucine had intermediate rates of developmental progression (Fig. 3C). In other eukaryotes, amino acid starvation activates a distinct shift in transcriptional activity, in which genes that support adaptation and cell viability are up-regulated selectively (9, 13). However, in P. falciparum, transcription remained inextricably coupled with parasite development during starvation, with both gene expression and parasite morphology remaining in phase but displaying slowed progression through the IDC. Most notably, there were no obvious transcriptional alterations indicative of a conventional starvation-related stress response (Fig. 4 and Dataset S3). For instance, genes involved in glycolysis, translation, transcription, and apicoplast and mitochondrial function appear to follow their normal course of regulation, albeit at a slowed rate, as in the general transcription program.
Fig. 3.
Developmental progression of hibernating parasites. RNA was isolated from synchronous 3D7 parasites that were fed (CM) or starved for isoleucine (−Ile), with samples harvested at 3- or 6-h intervals over the course of 48 h. (A) Pearson coefficient values were calculated by comparing the global transcriptional data generated from parasites maintained in the two medium conditions at the indicated time points against corresponding data from each time point generated in the high-resolution 3D7 IDC transcriptome from ref. 27. The y-axis shows Pearson coefficient; the x-axis shows hours post invasion (h.p.i.) in the IDC data set. The apex of the peak in each graph corresponds to the approximate point in the IDC to which the fed (CM; open symbols) or starved (−Ile; filled symbols) parasites best correlate at the indicated incubation time. Plots are shown with a Loess fit of the data: 0 h, red; 12 h, purple; 24 h, green; 36 h, orange. (B) Summary plot of progress through the IDC (based on Pearson coefficient) of parasites that were fed (CM, black circles) or starved (no Ile, gray squares, dashed line) for the indicated incubation times. The red and blue dashed lines indicate the slope (m) of the best-fit curve for the CM and no-Ile points up to 24 h of incubation, respectively. (C) Histograms of nucleic acid content of parasites grown in varying concentrations of isoleucine for 41 h from the early ring stage. Live cells were stained with Acridine Orange and analyzed by flow cytometry. Vertical lines mark peaks of rings and schizonts. Parasites at higher isoleucine concentrations progressed to schizonts, and some reinvaded, forming new rings. Parasites at lower isoleucine concentrations progressed at slower rates. Blood smears correlated well with flow cytometry results. Shown is a representative experiment, one of three similar determinations.
Fig. 4.
Expression of metabolic, organellar, and functional pathway genes in starved parasites. Cy5-labeled cDNA from synchronous 3D7 parasites that were fed (CM) or starved for isoleucine (−Ile) over a 48-h period was hybridized against a Cy3-labeled parasite cDNA reference pool. The expression profiles of representative genes involved in the indicated biological pathways are shown. The labels at the top denote the parasite stage of each sample at the indicated time point: R, ring; T, trophozoite; S, schizont. The panels on the right are of plots of the log2(Cy5/Cy3) expression values over time for representative genes from each of the indicated pathways. CM, black circles; no Ile, red squares. PFI1105w, phosphoglycerate kinase; PF14_0207, DNA-directed RNA polymerase III subunit; PF13_0014, 40S ribosomal protein S7; PF13_0327, cytochrome c oxidase subunit 2; PF14_0421, apicoplast acyltransferase. On heat map, scale [log2(Cy5/Cy3)] is from −3 (green) to +3 (red).
Isoleucine-Starved Parasites Have Reduced Levels of Central-Carbon Metabolism Intermediates.
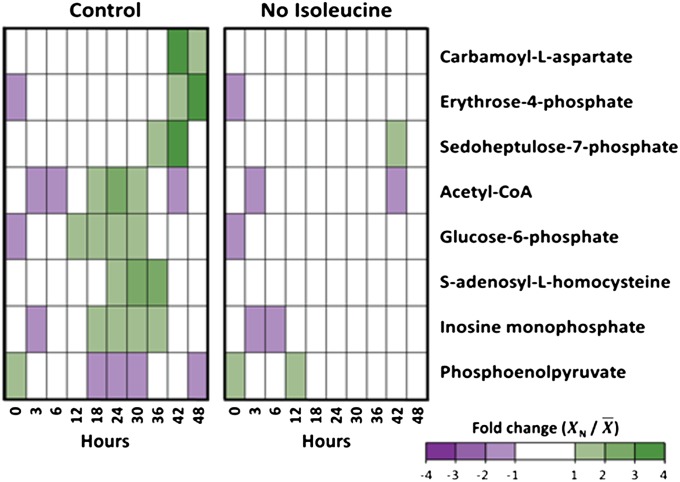
The metabolite profile of parasites was monitored over a 48-h period using liquid chromatography mass spectrometry (LC/MS). The overall metabolomic profile (87 metabolites) of isoleucine-starved parasites was similar to that of the control throughout the time course (Fig. S4). However, several intermediates of glycolysis and the pentose phosphate pathway were suppressed under isoleucine starvation (Fig. 5). These included glucose-6-phosphate, acetyl-CoA, erythrose-4-phosphate, and sedoheptulose-7-phosphate. The overall trend toward lower levels of energy-related metabolites was accompanied by reductions in intermediates of pyrimidine biosynthesis (carbamoyl-aspartate) and methyl-group transfer reactions, such as membrane biogenesis and DNA methylation (S-adenosyl homocysteine), under isoleucine-starved conditions. These intermediates normally increase during late-stage development (28); in starved parasites their suppression was persistent throughout the time course and did not follow the delayed periodicity of the transcriptional expression.
Fig. 5.
Metabolite profile of infected-erythrocyte cultures under standard and isoleucine-starved conditions. Profiles of eight metabolites from 3D7 P. falciparum-infected erythrocytes over 48 h. Infected erythrocytes were cultured under standard conditions (Control) and isoleucine-depleted conditions (No Isoleucine). Relative levels are expressed as the mean-centered ratio of the normalized signal intensity in the infected erythrocyte extract at each time point (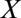 N/
N/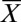 ) from two independent replicates.
) from two independent replicates.
Metabolic disruption also affected certain metabolic pathways in a complex manner that was not confined to decreased intermediates under isoleucine starvation. For example, the tricarboxylic acid (TCA) metabolism intermediates succinate and citrate were elevated under isoleucine-starved conditions toward the end of the time course (Fig. S4). This elevation was concomitant with decreased levels of fumarate and, to a lesser extent, malate under isoleucine-starved conditions. Moreover, the glycolytic intermediates phosphoenolpyruvate and 3-phosphoglycerate were elevated under isoleucine-starved conditions, whereas other intermediates of the same pathway were suppressed.
PfeIF2α Responds Specifically to Isoleucine Levels.
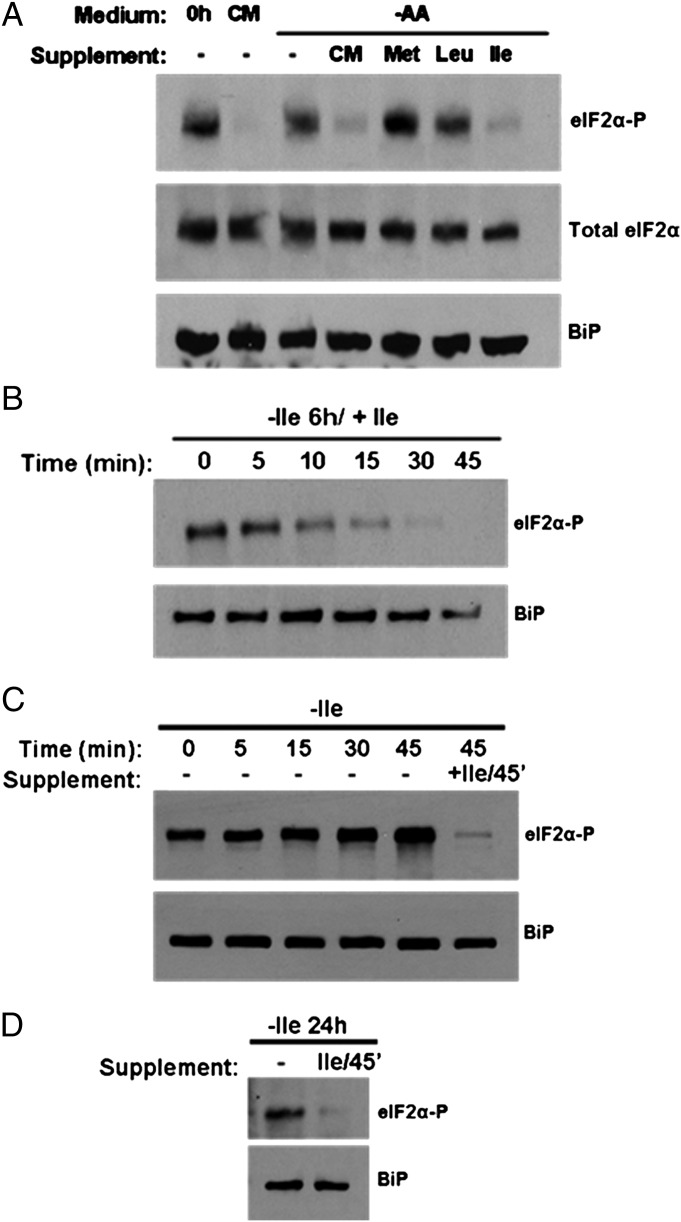
In P. falciparum, exposure to amino acid-free RPMI results in the phosphorylation of parasite eIF2α (PfeIF2α) (15). Considering that isoleucine is the only amino acid for which the parasite can truly be starved, we examined the specificity of this response. We incubated parasites in amino acid-free RPMI, followed by supplementation with CM containing all 20 amino acids or with the single amino acids isoleucine, methionine, or leucine, which represent amino acids that respectively are absent, in low abundance, or abundantly present in human hemoglobin (6). As seen previously (15), phosphorylation of PfeIF2α in amino acid-free conditions was observed readily by Western blot analysis using antibodies specific for the phosphorylated motif (Fig. 6A). Addition of CM resulted in the dephosphorylation of PfeIF2α, but single amino acid supplementation with methionine or leucine did not elicit this response. Only the addition of isoleucine to the amino acid-starved cultures resulted in dephosphorylation of PfeIF2α similar to that achieved with CM supplementation. Furthermore, we observed that when cultures maintained in isoleucine-free RPMI were supplemented with isoleucine, PfeIF2α dephosphorylation could be detected within 10 min, with complete loss of phosphorylation within 45 min of incubation (Fig. 6B). Remarkably, phosphorylation of eIF2α could be detected within minutes after isoleucine withdrawal (Fig. 6C). Even the short processing period for the zero time sample was enough to allow phosphorylation, suggesting the absence of a significant intracellular isoleucine store. Collectively, these data indicate that the eIF2α-mediated starvation response of P. falciparum is exquisitely sensitive to isoleucine availability.
Fig. 6.
Parasite eIF2α phosphorylation status depends on the isoleucine environment. (A) Resupplementation of starved parasites. Synchronous parasites cultured for 6 h in RPMI lacking all amino acids were resupplemented with CM or the indicated single amino acids (at the concentration found in complete RPMI) for 45 min. Parasite lysates were prepared for SDS/PAGE followed by immunoblotting with antibodies against phosphorylated eIF2α (eIF2α-P), total eIF2α, and the endoplasmic reticulum marker BiP as a loading control. (B) Time course of resupplementation. Synchronous parasites were starved for 6 h and then were resupplemented with isoleucine for the indicated times. Samples were processed for analysis as in A. (C) Time course of starvation. Synchronous parasites were washed in isoleucine-free medium, centrifuged briefly, and replated in isoleucine-free medium. Samples were taken at the indicated times and processed for analysis as in A. (D) Synchronous parasites were maintained in isoleucine-free RPMI medium for 24 h and then were resupplemented with isoleucine for 45 min. Samples were processed for analysis as in A.
Recovery from Starvation Is Independent of PfeIK1 Signaling.
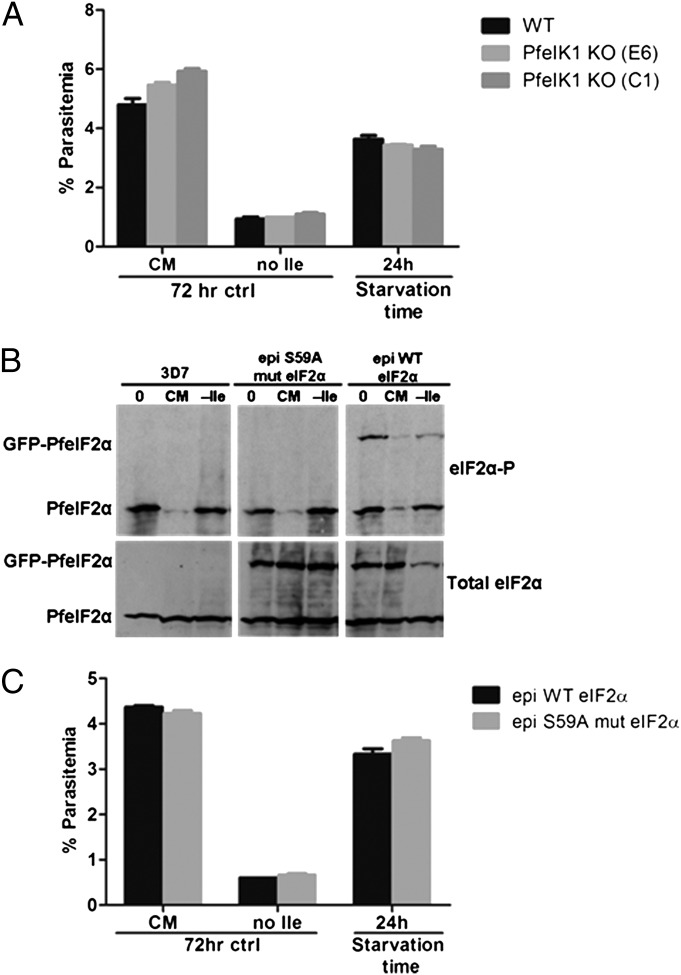
The amino acid-sensing eIF2α kinase of P. falciparum was identified recently as PfeIK1, an ortholog of yeast GCN2. Unlike the parental strain, parasite mutants lacking PfeIK1 do not phosphorylate PfeIF2α in response to amino acid-limiting conditions in vitro (15). Because long-term starved wild-type parasites maintain PfeIF2α phosphorylation (Fig. 6D) and are able to recover growth after resupplementation (Fig. 1C), we investigated whether pfeik1− mutants lose viability after prolonged starvation. We incubated a clonal line of pfeik1− parasites (15) in isoleucine-free RPMI for 24 h and then supplemented the culture with isoleucine and measured outgrowth by flow cytometry. The pfeik1− parasites starved for 24 h recovered growth similar to the wild-type parental strain (Fig. 7A), suggesting that parasite viability during starvation does not depend on PfeIK1 signaling. The pfeik1− parasites exhibited a reduction in the metabolic incorporation of [35S] methionine and cysteine during amino acid starvation (Fig. S5), comparable to that seen in wild-type parasites in Fig. 1D, but phosphorylation of PfeIF2α remained undetectable in the mutant parasites (Fig. S6 and ref. 15).
Fig. 7.
PfeIK1 activity is not required to maintain viability during hibernation. (A) Viability of pfeIk1-knockout parasites after isoleucine starvation. Synchronous 3D7 parasites were incubated for 24 h in isoleucine-free medium (no Ile). Isoleucine was added back, and parasites were allowed to recover in CM for 72 h. Parental strain, black bars; pfeik1-knockout clones, light (E6) and dark (C1) gray bars. Control parasites were fed (CM) or starved for isoleucine without refeeding (no Ile) for 72 h. Parasitemia of all other cultures was measured by flow cytometry after 72 h of recovery. Data shown represent the mean parasitemia ± SEM; n = 3. (B) Response of wild-type PfeIF2α and the PfeIF2α S59A phosphorylation mutant to starvation. Synchronous parasites expressing an episomal GFP-tagged copy of wild-type (epi WT) or mutant (epi S59A mut) PfeIF2α and a parental line of 3D7 were incubated in CM or isoleucine-free RPMI (−Ile) for 5 h. Parasite lysates were prepared for SDS/PAGE followed by immunoblotting with antibodies against phosphorylated eIF2α (eIF2α-P) and total eIF2α. (C) Viability of the PfeIF2α S59A phosphorylation mutant after isoleucine starvation. Black bars represent parasites expressing an episomal GFP-tagged wild-type copy of PfeIF2α (epi WT PfeIF2α); gray bars represent parasites expressing an episomal GFP-tagged mutant copy of PfeIF2α (epi S59A mut eIF2α). A growth-recovery assay was performed as in A. Data shown represent the mean parasitemia ± SEM, n = 3.
To assess further whether PfeIF2α phosphorylation plays a role in regulating the starvation stress response of P. falciparum, we generated parasites episomally expressing a nonphosphorylatable form of PfeIF2α. When exposed to isoleucine-free conditions, the mutant PfeIF2α-S59A was not phosphorylated, although both a wild-type episomal control and the endogenous PfeIF2α were robustly phosphorylated (Fig. 7B). In other organisms, the expression of a phosphorylation-insensitive eIF2α typically dampens the adaptive response to stress, resulting in reduced fitness (10, 29); in contrast, parasites expressing mutant PfeIF2α-S59A grew well and recovered as well as the wild-type–expressing control after extended isoleucine starvation and resupplementation (Fig. 7C). These data indicate that although PfeIK1 phosphorylation of PfeIF2α is induced by isoleucine starvation in a wild-type background, the regulation of parasite growth during starvation and concomitant entry into and exit from the hibernating state are independent of both PfeIK1 activity and PfeIF2α phosphorylation.
Discussion
In this study, we have shown that starvation for the single amino acid isoleucine elicits a metabolic response in P. falciparum that results in slowed parasite growth. This starvation-induced stasis is reversed upon isoleucine resupplementation, demonstrating the remarkable resilience of Plasmodium. We liken this phenomenon to hibernation, in which an organism is able to decrease its metabolic rate dramatically to conserve energy and resources, ultimately leading to increased survival once growth-permissive conditions are restored. Although asexual growth is recoverable poststarvation, our data suggest that limitations exist regarding the duration of starvation tolerable by the parasite. By 72 h, viability starts to tail off. Initially we suspected an increase in gametocyte conversion with extended starvation, considering that reductions in asexual parasitemia often correlate with induction of gametocytogenesis (30) and that certain starvation responses in yeast result in induction of the sexual/mating type differentiation program (31). However, gametocytemia was virtually absent in the recovered cultures, suggesting that amino acid stress does not necessarily skew parasite commitment toward sexual differentiation and that starvation-induced hibernation can protect the parasite for only a limited time before viability is compromised. Optimal growth of in vitro-cultured P. falciparum requires an isoleucine concentration above 20 μM; however, slow, continuous growth is still observed in cultures maintained in lower isoleucine concentrations (4). In natural infections of malnourished children, where blood isoleucine levels can fall to single-digit micromolar levels (7), we propose that P. falciparum adjusts its metabolic growth accordingly, allowing it to survive and persist. This response appears to be specific to isoleucine, because glucose starvation led to rapid death; presumably, blood glucose concentrations rarely fall to levels that threaten parasite survival even in malnourished patients.
Notably, entry into the hibernating state did not confer protection against treatment with the antimalarial artemisinin, which has been reported to induce a putative quiescent state in P. falciparum (24). However, artemisinin tolerance and quiescence have been reported only for ring forms (23, 24), and isoleucine-starved parasites gradually progress past this stage. We conclude that the biological mechanisms for drug-associated dormancy and starvation-induced hibernation may differ, at least in the control of cell-cycle progression. Nevertheless, it would be of great interest to investigate PfeIF2α phosphorylation status in artemisinin-tolerant ring stages.
Proteolysis plays a role in maintaining the parasite in a growth-competent state, because inhibition of this activity adversely affected recovery after isoleucine resupplementation. As starved parasites slowly progress through the trophozoite stage, peak proteolytic activity appears to coincide with gradual maturation. Moreover, this activity becomes particularly crucial during long-term starvation. Hemoglobin, which lacks isoleucine, makes up 95% of the soluble host-cell protein (20), leaving the other 5% to supply a limited pool of isoleucine to sustain the parasite during extended hibernation. Alternatively, autophagy of parasite proteins could provide a source of isoleucine in such conditions. Given the involvement of DV proteases in starvation survival, the autophagy route would have to involve the DV. Autophagy pathways in intraerythrocytic P. falciparum are largely uncharacterized. Plasmodium encodes nine putative autophagy-related genes (ATG), representing less than 30% of the complement of ATG genes in other eukaryotes (14); interestingly, none of these genes appeared to be specifically induced during starvation (Dataset S3). Also, there does not appear to be a significant isoleucine store, because parasites preconditioned in subsistence levels of isoleucine (20 μM) survived starvation as well as those conditioned in high (200 μM) isoleucine and because parasites detect isoleucine withdrawal almost immediately, as evidenced by rapid phosphorylation of PfeIF2α.
Isoleucine starvation did not activate an alternative transcriptional program, the hallmark of a conventional starvation response, in the parasite (12, 13). This observation further illustrates the parasite’s astonishingly limited capacity for transcriptional regulation (32–34) and is consistent with the lack of homologs for starvation-response regulators such as GCN4 and ATF4. Additionally, homologs of the prokaryotic transcription effector RelA, which regulates the amino acid starvation response in bacteria (35), could not be identified in the parasite. Although infected RBCs reportedly take up amino acids, including isoleucine, an order of magnitude more efficiently than uninfected RBCs (36), expression of putative amino acid transporters (e.g., PFL1515c, PF11_0334, and PFL0420w) was not up-regulated during starvation (Dataset S3). Field isolates of P. falciparum exhibit differential expression profiles, including one described as having starvation-response characteristics (37). This phenomenon was not observed in our conditions; in fact, isoleucine-starved parasites continued to express genes normally associated with active growth but with significantly delayed kinetics in comparison with a fed control. Indeed, the most remarkable feature of the transcriptome from starved parasites in this report is the apparent delay in both the decay and accumulation of stage-related transcripts, constituting a 60% decrease in the rate of developmental progression.
LC/MS analysis of P. falciparum-infected erythrocytes revealed that, under isoleucine starvation, metabolic pathways associated with central-carbon metabolism were disrupted. Of the total metabolome detected (87 metabolites), the most notable changes included reduced levels of glycolytic and pentose phosphate pathway intermediates under starved conditions. The reduction in metabolite pools during the time course was not a delayed response, as observed in the transcriptional analysis, but was a consistent suppression that became more apparent at later stages of the time course, presumably because these intermediates are accumulating in the maturing parasite (28). Glycolysis and the pentose phosphate pathway are important for progression through the IDC, because the parasite’s energy and carbon requirements increase dramatically during trophozoite maturation and schizogony (38–40). Interestingly, pyrimidine biosynthesis, methyl-group transfers, and the TCA pathway also are disrupted under isoleucine starvation and likewise are critical for parasite maturation. The suppression of these pathways in isoleucine-starved conditions is consistent with the morphological disruption observed, the failure to reach cell division.
We note that measuring the steady-state metabolite pools of the infected erythrocyte during isoleucine starvation can reveal which pathways are perturbed without a priori knowledge of the mechanism involved. However, this steady-state approach cannot explain how the flux through each pathway causes the changes in steady-state metabolite pools observed. Stable-isotope labeling, although a major endeavor, would overcome this limitation and determine how the flux through these central-carbon metabolic pathways during isoleucine starvation causes the elevation and suppression of intermediates of the same pathway.
The disruption of one or more enzymatic steps could inhibit flux through a pathway, causing the accumulation of upstream intermediates and the decrease of downstream intermediates. Under isoleucine starvation, the glycolytic intermediates phosphoenolpyruvate and 3-phosphoglycerate were elevated, and the downstream product acetyl-CoA was reduced. This result may be explained by inhibition of either pyruvate kinase or pyruvate dehydrogenase. Likewise, inhibition of succinate dehydrogenase could explain the elevated level of succinate and the concomitant suppression of fumarate and malate pools when isoleucine was absent. However, this explanation remains incomplete, because the differential changes in metabolite levels within each pathway do not overlap completely at each time point. Further stable-isotope studies are required to ascertain the mechanisms involved.
Given that isoleucine participates directly in very few metabolic processes (other than protein synthesis) and is disconnected from central-carbon metabolism, disruption of central-carbon metabolism is unlikely to be a direct result of decreased substrate availability. Therefore, it remains unclear how central-carbon metabolism is altered. The parasite has a clear transcriptional response to isoleucine removal, but this response is a generalized slowing of developmental progression and not a specific amino acid-starvation response. Determining if central-carbon metabolism is disrupted because of reduced metabolic enzyme translation or because of a more dynamic response that may involve posttranslational modification of metabolic enzymes is of ongoing interest. This question is particularly important in regards to the observation that PfeIF2α is dephosphorylated within 10 min upon isoleucine supplementation. This finding indicates that the parasite has some dynamic response to isoleucine supplementation, but whether any other posttranslational modifications occur in response to changes in isoleucine availability remains uncertain.
In other eukaryotes, such as yeast, plants, and mammals, the amino acid-starvation response has been studied extensively, and many conserved biological features [e.g., TOR signaling (9), up-regulation of amino acid biosynthetic enzymes (12), and induction of autophagy (41)] are inherently translatable to other model systems. However, components of this response that are found in other organisms appear to be missing in Plasmodium (3, 14). Even the one component involved in the canonical starvation response that is conserved, the GCN2 ortholog PfeIK1, is seemingly dispensable, because its absence does not compromise the viability of parasites under isoleucine-limiting conditions, despite a well-documented ability to phosphorylate PfeIF2α during starvation. In prototrophic yeast, lack of GCN2 does not affect logarithmic growth in most single amino acid-dropout medium conditions because of compensatory crosstalk between other amino acid-regulatory pathways that function independently of GCN2-mediated signaling (42). However, there is no evidence to suggest that such metabolic complexity exists in P. falciparum, especially considering that the parasite is deficient in amino acid biosynthesis and is absolutely dependent on exogenous isoleucine. The Plasmodium genome encodes another blood stage-expressed eIF2α kinase, PfPK4, which has been shown to be essential for completion of the IDC (18), but we suspect that functional redundancy is not at play during isoleucine starvation, because PfeIF2α remains unphosphorylated in pfeik1− parasites. Indeed, we have shown that amino acid starvation of wild-type P. falciparum results in the phosphorylation of PfeIF2α, with remarkable selective specificity for isoleucine. Although generally such a response is coupled to growth arrest, that does not appear to be the case for blood-stage P. falciparum. These observations raise questions about the role played by PfeIK1: Why did malaria parasites maintain this enzyme? Could it have other functions independent of PfeIF2α phosphorylation? The surprisingly fast response in PfeIF2α dephosphorylation upon isoleucine repletion implies an efficient signaling system starting with an isoleucine sensor and feeding into an effector phosphatase. It would be of great interest to elucidate this pathway.
Presumably as an adaptation to erythrocyte parasitism, the malaria organism has evolved a stripped-down starvation-response pathway. In light of our findings, we propose that growth regulation in P. falciparum operates predominantly by reaction rates. Low isoleucine concentrations slow the rate-limiting steps of translational processivity, thereby restricting growth. This primitive response to amino acid limitation is enough to allow the parasite to survive in its host for several days, waiting for nutrient repletion.
Materials and Methods
Parasite Culturing.
Plasmodium falciparum strain 3D7 and derived knockout clones were cultured (43) in human O+ erythrocytes in complete RPMI 1640, containing all 20 amino acids, supplemented with 27 mM NaHCO3, 22 mM glucose, 0.37 mM hypoxanthine, 10 μg/mL gentamicin, and 5 g/L Albumax (Invitrogen). Homemade complete, glucose-free, isoleucine-free, and amino acid-free RPMI were prepared according to the RPMI 1640 recipe provided by Invitrogen and supplemented with RPMI 1640 vitamins (Sigma), the appropriate respective amino acids (Sigma) at the concentrations found in RPMI 1640, and the appropriate additional supplements mentioned above. A clone of P. falciparum strain 3D7 (IG06) that has a 38-h cycle was used for most analyses. This fast-growing strain allowed us to perform more extensive time courses. Both 38-h and conventional 48-h clones were able to recover from starvation, and correlation of gene-expression profiles for corresponding stages was consistent.
Generation of Episomal PfeIF2α (WT and S59A)-GFP–Expressing Parasites.
Full-length PfeIF2α (PF07_0117) (omitting the stop codon) was PCR amplified from 3D7 genomic DNA using primers 5′-AATTCTCGAGATGACTGAAATGCGAGTAAAAGCAGATTTG-3′ (XhoI site underlined) and 5′-AATTCCTAGGATCTTCCTCCTCCTCGTCTTCACTAGTATT-3′ (AvrII site underlined), digested with XhoI and AvrII, and ligated into the same sites of the pIRCTGFP vector (44), containing the promoter region for PfHsp86, a C-terminal GFP tag, and a human dihydrofolate reductase (hDHFR) drug-selection cassette. A point mutation was introduced to change Ser59 to Ala59 in PfeIF2α using the QuikChange XL mutagenesis kit (Stratagene) and the primers 5′-GGAAGGTATGATTTTAATGTCCGAACTAGCCAAAAGAAGATTCAGAAG-3′ and 5′-CTTCTGAATCTTCTTTTGCGTAGTTCGGACATTAAAATCATACCTTCC-3′. All cloning steps were confirmed by sequencing.
Ring-stage 3D7 parasites were transfected by electroporation (45) with 100 μg of purified vector DNA. Parasites carrying the plasmid were selected by adding 10 nM WR99210 to the culture medium. Sixty percent of transfected parasites were green by fluorescence microscopy, a typical plasmid maintenance result for this organism.
Flow Cytometry.
Parasite samples were fixed in 4% paraformaldehyde/0.015% glutaraldehyde in PBS and were stored at 4 °C. For analysis, the cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature. One-half of the sample was treated with 100 μg/mL RNase A (Qiagen) in PBS for 20 min at 37 °C. All samples were stained with 0.5 μg/mL Acridine Orange (Molecular Probes) in PBS, and 3 × 104 to 1 × 105 cells were counted on a BD Biosciences FACS Canto flow cytometer. Total cell number was measured on the forward- and side-scattering channels. Fluorescence was detected on both the FITC-H and the PerCP-Cy5-H channels, and parasitemia gates were defined by fluorescence intensity, with highly fluorescent infected RBCs distinctly separated from low-fluorescence uninfected RBCs. Alternatively, live cells were stained directly with 0.5 μg/mL Acridine Orange and were counted immediately on the FACS Canto using the parasitemia-gating parameters described above. Data were analyzed using FlowJo software (TreeStar Inc.).
Isoleucine Titration Assay.
P. falciparum 3D7 parasites were adapted to complete RPMI medium containing 20 µM isoleucine for approximately 1 wk under normal culturing conditions. Early ring-stage parasites were sorbitol synchronized (46), subcultured to 5% parasitemia, washed twice in PBS, equally partitioned, and replated in isoleucine-free RPMI. Isoleucine was added at various concentrations, and the cultures were incubated at 37 °C with 5% CO2 for 41 h. Live parasite samples were stained with 0.5 µg/mL Acridine Orange (Molecular Probes) in PBS and analyzed by flow cytometry as described above.
Growth Recovery Assay.
P. falciparum 3D7 parasites and clonal knockout lines were sorbitol synchronized (46) to the late ring stage, cultured in complete RPMI at 2% hematocrit, and subcultured to ∼1% parasitemia. The cultures were washed twice in PBS, partitioned, and washed in complete RPMI, glucose-free RPMI, or isoleucine-free RPMI; then the parasites were replated in triplicate in their respective medium and were incubated at 37 °C with 5% CO2. Control fed, glucose-starved, or isoleucine-starved parasites were grown for 72 h and prepared for flow cytometry to assess parasitemia. Remaining starved cultures were supplemented with isoleucine (382 μM final) or glucose (22 mM final) after starvation for various periods of time and were allowed to recover for an additional 72 h. During extended isoleucine starvation, isoleucine-free culture medium was refreshed every other day. Parasites were prepared for flow cytometry after recovery.
In the experiments in which drug was added, 10 μM transepoxysuccinyl-l-leucylamino (4-guanidine)-butane (E-64d) (Sigma), 5 μM pepstatin A (Sigma), or 50 nM artemisinin (Sigma) was added to fed or starved cultures for a 24-h period at various times during the incubation. After the 24-h exposure, cultures were washed twice in PBS and replated in CM for recovery or isoleucine-free medium for extended starvation followed by isoleucine supplementation for recovery. After the 72-h recovery, parasites were prepared for flow cytometry.
Microarray Analysis.
A large-scale sorbitol-synchronized (46) P. falciparum 3D7 culture at 8–10% parasitemia was washed twice in PBS, equally partitioned, and washed in complete or isoleucine-free RPMI; then the parasites were replated in their respective medium and were incubated at 37 °C with 5% CO2. Samples were harvested initially and at 3- or 6-h intervals over a 48-h period. Culture medium was changed every 12 h, and parasites incubated in CM were subcultured just before schizont rupture to maintain parasitemia between 8–10% after reinvasion. The infected RBC pellet was washed with PBS and resuspended in TRIzol reagent (Invitrogen). Chloroform was added, followed by centrifugation at 7,500 × g for 1 h at 4 °C. Isopropanol was added to the aqueous phase to precipitate the RNA. After centrifugation, the isolated RNA pellet was washed with 70% (vol/vol) ethanol, dried, and dissolved in diethylpyrocarbonate-treated water.
Amino-allyl cDNA was synthesized from isolated RNA and labeled with Cy5 for hybridization against a Cy3-labeled reference pool as described previously (27). DNA microarray hybridizations were performed on a new P. falciparum Agilent platform DNA microarray (47). The Cy5/Cy3 ratio, representing relative expression levels, was calculated for each sample and log2 transformed for statistical analysis. The resulting data were analyzed by hierarchical clustering using the Princeton Microarray database (PUMAdb) and visualized with Java TreeView (48). R2 correlation values were calculated in Excel (Microsoft) by comparing the transformed relative expression data for each respective sample. Pearson coefficients were calculated by comparing the transformed relative expression data from this dataset with the corresponding values generated for the high-resolution IDC transcriptome reported in ref. 27.
Metabolite Extraction and Analysis.
Metabolic profiling was performed using methods previously described (28), with minor modifications. Forty-eight–hour metabolite assays were carried out with highly synchronized P. falciparum-infected erythrocyte cultures (10% parasitemia) at 2% hematocrit. Cell counts were performed with an Improved Neubauer Hemacytometer, and cultures were adjusted accordingly. Time courses were initiated by resuspending cultures with fresh, prewarmed (37 °C) RPMI 1640 medium (either isoleucine supplemented or isoleucine free). Every 6 h two biological replicates were collected, and the medium was replaced on each remaining sample. For each assay, an uninfected erythrocyte culture at the same hematocrit was incubated for 24 h before the experiment and then was treated identically to the P. falciparum-infected erythrocyte time course (incubated with control or isoleucine-free RPMI).
At each time point, infected or uninfected erythrocytes were cooled rapidly to 4 °C, washed once in ice-cold PBS, centrifuged for 1 min at 15,000 × g, and extracted with 20 volumes of 90% (vol/vol) methanol at 4 °C. Cell debris was cleared by centrifugation, and the supernatant was collected, dried under nitrogen, and stored at −80 °C until analysis.
Metabolite extracts were analyzed by reversed-phase ion-pairing LC coupled by electrospray ionization (ESI) (negative mode) to a high-resolution, high-accuracy mass spectrometer (Exactive; Thermo Fisher) operated in full-scan mode (49). This analysis was complemented with LC coupled by ESI (negative mode) to a Thermo Scientific TSQ Quantum triple-quadrupole mass spectrometer operating in single-reaction monitoring mode (50). Amino acids were derivatized with benzyl chloroformate before their quantitation by negative-mode LC-ESI-MS/MS (51).
Data analysis and metabolite identification were performed using the Metabolomic Analysis and Visualization Engine (MAVEN) software program (52). Metabolites were excluded from analysis if they failed to give a signal greater than 1,000 counts in any sample analyzed. Each metabolite signal was normalized to the signal of the internal standard U-13C-aspartate in the same sample to control for variation between samples. Normalized data were expressed as fold change relative to an uninfected erythrocyte sample from the equivalent time point. Final data represented as heatmaps are presented as mean-centered ratios and are row clustered.
Starvation Assay.
P. falciparum 3D7 parasites and clonal lines of pfeik1− parasites (15) were prepared and assayed as described previously (15). Briefly, parasites were sorbitol synchronized (46) to the late ring stage, cultured in complete RPMI at 2% hematocrit, and grown to ∼8–10% parasitemia. The parasites were washed twice in PBS, equally partitioned, and washed in complete, isoleucine-free, or amino acid-free RPMI and then were replated in their respective medium. Cultures were incubated at 37 °C with 5% CO2 for various increments of time. After incubation, parasites were harvested immediately or were supplemented with complete RPMI or with single amino acids (isoleucine, methionine, leucine) (Sigma) at concentrations found in complete RPMI and were reincubated at 37 °C for up to 45 min. After harvesting, infected RBCs were lysed with 100 hemolytic units (HU) of tetanolysin (List Biological), washed with PBS buffer containing Complete protease inhibitor mixture (Roche), 2 mM NaF, and 2 mM Na3VO4. Samples were resuspended in SDS-Laemmli buffer. Parasite proteins were resolved by SDS/PAGE and were transferred to nitrocellulose for immunoblotting (53).
Antibodies and Immunoblotting.
Rabbit anti-phosphorylated eIF2α (Ser51) and mouse anti-eIF2α were purchased from Cell Signaling Technology. Rabbit and rat anti-BiP were acquired from the Malaria Research and Reference Reagent Resource Center (ATCC). Secondary antibodies conjugated with HRP were from GE Healthcare Life Sciences. For immunoblotting, nitrocellulose membranes were blocked with 5% (wt/vol) BSA in Tris-buffered saline-0.1% Tween 20 (TBST) for 1 h at room temperature. Rabbit anti-phosphorylated eIF2α (Ser51) was diluted 1:1,000 in TBST. Mouse anti-eIF2α was diluted 1:1,000 in TBST. Rabbit or rat anti-BiP was diluted 1:10,000 in TBST. Respective secondary antibodies were diluted 1:20,000. Bound antibodies were detected with Western Lightning Chemiluminescence reagent (Perkin-Elmer).
Metabolic Labeling and TCA Precipitation.
P. falciparum 3D7 parasites and clonal lines of pfeik1− parasites were sorbitol synchronized (46) to the late ring stage, cultured in complete RPMI at 2% hematocrit, and grown to ∼8–10% parasitemia. The parasites were washed twice in PBS, equally partitioned, and washed in complete or isoleucine-free labeling RPMI that did not contain methionine or cysteine. The parasites then were replated in their respective medium in the presence or absence of 10 μg/mL cycloheximide and were incubated at 37 °C with 5% CO2 for 6 h. During the last hour of incubation, 0.1 mCi (1175 Ci/mmol) [35S] Express protein labeling mix (Perkin-Elmer) was added to each culture. After harvesting, labeled cultures were washed with PBS buffer containing Complete protease inhibitor mixture (Roche) and were lysed with 100 HU of tetanolysin (List Biological). Portions of the samples were resuspended in SDS-Laemmli buffer, followed by SDS/PAGE, Coomassie staining, and autoradiography. Remaining samples were TCA-precipitated by adding one-fourth volume of 100% (wt/vol) TCA to the parasite pellet resuspended in 200 μL PBS. Samples were incubated on ice for 10 min and centrifuged. The precipitated protein pellet was washed with ice-cold acetone, dried, resuspended in water, and pipetted onto FilterMat (Skatron Instruments). After the filters dried, they were placed in vials with Ultima Gold scintillation fluid (Perkin-Elmer) and were counted on a Beckman LS6000 scintillation counter.
Supplementary Material
Acknowledgments
We thank Mark Drew and Paul Sigala for helpful suggestions, Anna Oksman for technical assistance, Jacobus Pharmaceuticals for WR99210, and MR4/John Adams for antisera. M.L. received support from the Burroughs Wellcome Fund for Investigators in Pathogenesis of Infectious Disease, National Institutes of Health Grant1DP2OD001315, and Center for Quantitative Biology Grant P50 GM071508.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data are provided in spreadsheet format in Datasets S1, S2, and S3.
See Author Summary on page 19061 (volume 109, number 47).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209823109/-/DCSupplemental.
References
- 1.Sherman IW. Transport of amino acids and nucleic acid precursors in malarial parasites. Bull World Health Organ. 1977;55(2-3):211–225. [PMC free article] [PubMed] [Google Scholar]
- 2.Sims PF, Hyde JE. Proteomics of the human malaria parasite Plasmodium falciparum. Expert Rev Proteomics. 2006;3(1):87–95. doi: 10.1586/14789450.3.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Payne SH, Loomis WF. Retention and loss of amino acid biosynthetic pathways based on analysis of whole-genome sequences. Eukaryot Cell. 2006;5(2):272–276. doi: 10.1128/EC.5.2.272-276.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Istvan ES, Gluzman IY, Gross J, Goldberg DE. Plasmodium falciparum ensures its amino acid supply with multiple acquisition pathways and redundant proteolytic enzyme systems. Proc Natl Acad Sci USA. 2006;103(23):8840–8845. doi: 10.1073/pnas.0601876103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Divo AA, Geary TG, Davis NL, Jensen JB. Nutritional requirements of Plasmodium falciparum in culture. I. Exogenously supplied dialyzable components necessary for continuous growth. J Protozool. 1985;32(1):59–64. doi: 10.1111/j.1550-7408.1985.tb03013.x. [DOI] [PubMed] [Google Scholar]
- 6.Soeters PB, van de Poll MC, van Gemert WG, Dejong CH. Amino acid adequacy in pathophysiological states. J Nutr. 2004;134(6) Suppl:1575S–1582S. doi: 10.1093/jn/134.6.1575s. [DOI] [PubMed] [Google Scholar]
- 7.Baertl JM, Placko RP, Graham GG. Serum proteins and plasma free amino acids in severe malnutrition. Am J Clin Nutr. 1974;27(7):733–742. doi: 10.1093/ajcn/27.7.733. [DOI] [PubMed] [Google Scholar]
- 8.Shankar AH. Nutritional modulation of malaria morbidity and mortality. J Infect Dis. 2000;182(Suppl 1):S37–S53. doi: 10.1086/315906. [DOI] [PubMed] [Google Scholar]
- 9.Kim J, Guan KL. Amino acid signaling in TOR activation. Annu Rev Biochem. 2011;80:1001–1032. doi: 10.1146/annurev-biochem-062209-094414. [DOI] [PubMed] [Google Scholar]
- 10.Dever TE, et al. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68(3):585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- 11.Hinnebusch AG. Gene-specific translational control of the yeast GCN4 gene by phosphorylation of eukaryotic initiation factor 2. Mol Microbiol. 1993;10(2):215–223. doi: 10.1111/j.1365-2958.1993.tb01947.x. [DOI] [PubMed] [Google Scholar]
- 12.Hinnebusch AG, Natarajan K. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot Cell. 2002;1(1):22–32. doi: 10.1128/EC.01.1.22-32.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab. 2009;20(9):436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennand A, et al. Autophagy in parasitic protists: Unique features and drug targets. Mol Biochem Parasitol. 2011;177(2):83–99. doi: 10.1016/j.molbiopara.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Fennell C, et al. PfeIK1, a eukaryotic initiation factor 2alpha kinase of the human malaria parasite Plasmodium falciparum, regulates stress-response to amino-acid starvation. Malar J. 2009;8:99. doi: 10.1186/1475-2875-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Möhrle JJ, Zhao Y, Wernli B, Franklin RM, Kappes B. Molecular cloning, characterization and localization of PfPK4, an eIF-2alpha kinase-related enzyme from the malarial parasite Plasmodium falciparum. Biochem J. 1997;328(Pt 2):677–687. doi: 10.1042/bj3280677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M, et al. The Plasmodium eukaryotic initiation factor-2alpha kinase IK2 controls the latency of sporozoites in the mosquito salivary glands. J Exp Med. 2010;207(7):1465–1474. doi: 10.1084/jem.20091975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang M, et al. PK4, a eukaryotic initiation factor 2α(eIF2α) kinase, is essential for the development of the erythrocytic cycle of Plasmodium. Proc Natl Acad Sci USA. 2012;109(10):3956–3961. doi: 10.1073/pnas.1121567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konrad C, Wek RC, Sullivan WJ., Jr A GCN2-like eukaryotic initiation factor 2 kinase increases the viability of extracellular Toxoplasma gondii parasites. Eukaryot Cell. 2011;10(11):1403–1412. doi: 10.1128/EC.05117-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francis SE, Sullivan DJ, Jr, Goldberg DE. Hemoglobin metabolism in the malaria parasite Plasmodium falciparum. Annu Rev Microbiol. 1997;51:97–123. doi: 10.1146/annurev.micro.51.1.97. [DOI] [PubMed] [Google Scholar]
- 21.Sijwali PS, Rosenthal PJ. Gene disruption confirms a critical role for the cysteine protease falcipain-2 in hemoglobin hydrolysis by Plasmodium falciparum. Proc Natl Acad Sci USA. 2004;101(13):4384–4389. doi: 10.1073/pnas.0307720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Codd A, Teuscher F, Kyle DE, Cheng Q, Gatton ML. Artemisinin-induced parasite dormancy: A plausible mechanism for treatment failure. Malar J. 2011;10:56. doi: 10.1186/1475-2875-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witkowski B, et al. Increased tolerance to artemisinin in Plasmodium falciparum is mediated by a quiescence mechanism. Antimicrob Agents Chemother. 2010;54(5):1872–1877. doi: 10.1128/AAC.01636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teuscher F, et al. Artemisinin‐induced dormancy in plasmodium falciparum: Duration, recovery rates, and implications in treatment failure. J Infect Dis. 2010;202(9):1362–1368. doi: 10.1086/656476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bozdech Z, et al. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1(1):E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Roch KG, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301(5639):1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 27.Llinás M, Bozdech Z, Wong ED, Adai AT, DeRisi JL. Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res. 2006;34(4):1166–1173. doi: 10.1093/nar/gkj517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olszewski KL, et al. Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell Host Microbe. 2009;5(2):191–199. doi: 10.1016/j.chom.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joyce BR, Queener SF, Wek RC, Sullivan WJ., Jr Phosphorylation of eukaryotic initiation factor-2alpha promotes the extracellular survival of obligate intracellular parasite Toxoplasma gondii. Proc Natl Acad Sci USA. 2010;107(40):17200–17205. doi: 10.1073/pnas.1007610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dyer M, Day KP. Commitment to gametocytogenesis in Plasmodium falciparum. Parasitol Today. 2000;16(3):102–107. doi: 10.1016/s0169-4758(99)01608-7. [DOI] [PubMed] [Google Scholar]
- 31.van Werven FJ, Amon A. Regulation of entry into gametogenesis. Philos Trans R Soc Lond B Biol Sci. 2011;366(1584):3521–3531. doi: 10.1098/rstb.2011.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deitsch K, et al. Mechanisms of gene regulation in Plasmodium. Am J Trop Med Hyg. 2007;77(2):201–208. [PubMed] [Google Scholar]
- 33.Callebaut I, Prat K, Meurice E, Mornon JP, Tomavo S. Prediction of the general transcription factors associated with RNA polymerase II in Plasmodium falciparum: Conserved features and differences relative to other eukaryotes. BMC Genomics. 2005;6:100. doi: 10.1186/1471-2164-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coulson RM, Hall N, Ouzounis CA. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum. Genome Res. 2004;14(8):1548–1554. doi: 10.1101/gr.2218604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma UK, Chatterji D. Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity. FEMS Microbiol Rev. 2010;34(5):646–657. doi: 10.1111/j.1574-6976.2010.00223.x. [DOI] [PubMed] [Google Scholar]
- 36.Martin RE, Kirk K. Transport of the essential nutrient isoleucine in human erythrocytes infected with the malaria parasite Plasmodium falciparum. Blood. 2007;109(5):2217–2224. doi: 10.1182/blood-2005-11-026963. [DOI] [PubMed] [Google Scholar]
- 37.Daily JP, et al. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature. 2007;450(7172):1091–1095. doi: 10.1038/nature06311. [DOI] [PubMed] [Google Scholar]
- 38.Pfaller MA, Krogstad DJ, Parquette AR, Nguyen-Dinh P. Plasmodium falciparum: Stage-specific lactate production in synchronized cultures. Exp Parasitol. 1982;54(3):391–396. doi: 10.1016/0014-4894(82)90048-0. [DOI] [PubMed] [Google Scholar]
- 39.Roth EF, Jr, Raventos-Suarez C, Perkins M, Nagel RL. Glutathione stability and oxidative stress in P. falciparum infection in vitro: Responses of normal and G6PD deficient cells. Biochem Biophys Res Commun. 1982;109(2):355–362. doi: 10.1016/0006-291x(82)91728-4. [DOI] [PubMed] [Google Scholar]
- 40.Zolg JW, Macleod AJ, Scaife JG, Beaudoin RL. The accumulation of lactic acid and its influence on the growth of Plasmodium falciparum in synchronized cultures. In Vitro. 1984;20(3 Pt 1):205–215. doi: 10.1007/BF02618189. [DOI] [PubMed] [Google Scholar]
- 41.Kourtis N, Tavernarakis N. Autophagy and cell death in model organisms. Cell Death Differ. 2009;16(1):21–30. doi: 10.1038/cdd.2008.120. [DOI] [PubMed] [Google Scholar]
- 42.Zaborske JM, Wu X, Wek RC, Pan T. Selective control of amino acid metabolism by the GCN2 eIF2 kinase pathway in Saccharomyces cerevisiae. BMC Biochem. 2010;11:29. doi: 10.1186/1471-2091-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193(4254):673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 44.Russo I, Oksman A, Goldberg DE. Fatty acid acylation regulates trafficking of the unusual Plasmodium falciparum calpain to the nucleolus. Mol Microbiol. 2009;72(1):229–245. doi: 10.1111/j.1365-2958.2009.06639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fidock DA, Wellems TE. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci USA. 1997;94(20):10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65(3):418–420. [PubMed] [Google Scholar]
- 47.Kafsack BF, Painter HJ, Llinás M. New Agilent platform DNA microarrays for transcriptome analysis of Plasmodium falciparum and Plasmodium berghei for the malaria research community. Malar J. 2012;11:187. doi: 10.1186/1475-2875-11-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 49.Lu W, et al. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem. 2010;82(8):3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu W, Bennett BD, Rabinowitz JD. Analytical strategies for LC-MS-based targeted metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871(2):236–242. doi: 10.1016/j.jchromb.2008.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kraml CM, Zhou D, Byrne N, McConnell O. Enhanced chromatographic resolution of amine enantiomers as carbobenzyloxy derivatives in high-performance liquid chromatography and supercritical fluid chromatography. J Chromatogr A. 2005;1100(1):108–115. doi: 10.1016/j.chroma.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 52.Clasquin MF, Melamud E, Rabinowitz JD. 2012. LC-MS data processing with MAVEN: A metabolomic analysis and visualization engine. Current Protocols in Bioinformatics Chapter 14:Unit14.11. [DOI] [PMC free article] [PubMed]
- 53.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA. 1979;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]