Abstract
Rift Valley fever and Toscana viruses are human pathogens for which no effective therapeutics exist. These and other phleboviruses have segmented negative-sense RNA genomes that are sequestered by a nucleocapsid protein (N) to form ribonucleoprotein (RNP) complexes of irregular, asymmetric structure, previously uncharacterized at high resolution. N binds nonspecifically to single-stranded RNA with nanomolar affinity. Crystal structures of Rift Valley fever virus N-RNA complexes reconstituted with defined RNAs of different length capture tetrameric, pentameric and hexameric N-RNA multimers. All N-N subunit contacts are mediated by a highly flexible α-helical arm. Arm movement gives rise to the three multimers in the crystal structures and also explains the asymmetric architecture of the RNP. Despite the flexible association of subunits, the crystal structures reveal an invariant, monomeric RNP building block, consisting of the core of one N subunit, the arm of a neighboring N, and four RNA nucleotides with the flanking phosphates. Up to three additional RNA nucleotides bind between subunits. The monomeric building block is matched in size to the repeating unit in viral RNP, as visualized by electron microscopy. N sequesters four RNA bases in a narrow hydrophobic binding slot and has polar contacts only with the sugar-phosphate backbone, which faces the solvent. All RNA bases, whether in the binding slot or in the subunit interface, face the protein in a manner that is incompatible with base pairing or with “reading” by the viral polymerase.
Keywords: RNA binding, RNA packaging, viral protein, segmented genome, negative-sense RNA virus
Viruses of the Phlebovirus genus (Bunyaviridae family) are transmitted by arthropod vectors and cause a variety of severe diseases worldwide. The Rift Valley fever virus is a highly infectious, mosquito-borne pathogen endemic to sub-Saharan Africa. RVFV infects livestock and humans and generally causes a flu-like illness; however, 1% of cases result in hemorrhagic fever disease, which has a 50% case-fatality rate (1). The closely related Toscana virus (TOSV) is endemic to the Mediterranean basin, is transmitted by infected phlebotomine sandflies and causes neurological dysfunction in humans (2). The membrane envelope of bunyaviruses encloses a three-segment, negative-sense RNA genome that is encapsidated by a nucleocapsid protein (N), forming the ribonucleoprotein (RNP) (3). However, the size and sequence of N vary extensively among the five genera of the Bunyaviridae family. Phlebovirus N are highly similar, but they appear unrelated to N of the other four bunyavirus genera.
We showed previously that phleboviruses have a unique genome-packaging strategy and an RNP that lacks the helical symmetry observed in some other negative-sense RNA viruses (NSVs) (4, 5). However, the detailed interactions between phlebovirus N and the viral RNA genome are unknown. The crystal structure of a RVFV N monomer revealed a compact helical fold with two lobes (5). The structure of an RVFV N hexamer demonstrated conformational flexibility in N and showed a putative RNA binding site on the inner surface of the hexameric ring (6). An α-helical arm, which is sequestered within the subunit in the N monomer structure, extends from the monomer to mediate subunit contacts in the hexamer. However, both structures lack RNA and a detailed explanation for the nonhelical structure of the N-RNA polymer has not been provided.
EM visualization of authentic RNPs from phlebovirus-infected cells revealed an extended, open RNP that lacks higher-order structure or symmetry (5, 7). Nucleocapsid protein–RNA (N-RNA) complexes extracted from viral RNPs by extensive ribonuclease treatment or expressed recombinantly have asymmetric ring-like structures of variable size (5, 6). Single-particle EM analysis suggested a heterogeneous population of multimers, each with three to seven N subunits. The heterogeneous, recombinant N-RNA multimers did not crystallize even after extensive ribonuclease digestion and purification.
In this study, we used fluorescence polarization to investigate the N–genome interaction using RNA-free N and defined RNA and DNA oligomers. We used EM to optimize nucleic acid lengths for crystallization trials of reconstituted N-RNA and N-DNA. Homogeneous N multimer preparations led to crystal structures of three different reconstituted N-RNA complexes and one N-DNA complex. The crystal structures show the tremendous flexibility of the α-helical arm, which allows phlebovirus N to form several distinct multimers. The N-RNA structures reveal a hydrophobic binding slot, where RNA bases are sequestered from solvent, and a single-subunit RNP building block. The structures provide exquisite detail about N organization and RNA binding and explain the observed asymmetry of phlebovirus RNP.
Results
RVFV and TOSV N Bind RNA and DNA Nonspecifically.
RNA-free N bound with high affinity to single-stranded nucleic acid, based on measurement of binding affinities by fluorescence polarization using labeled single-stranded oligomers of RNA or DNA (Table S1 and Fig. S1 A and B). The RNA-free N from two phleboviruses, RVFV and Toscana virus (TOSV), had similar affinities (6–26 nM) for RNA and DNA oligomers over a broad size range (10–35 nt). In this direct test of affinity for nucleic acid, RNA-free N displayed no sequence specificity. The high affinity for DNA is likely irrelevant to biological function, because N is not thought to enter the nucleus. Phleboviruses replicate in the host cytoplasm and, thus, have no need to discriminate RNA from DNA. The only experimental factor that affected N affinity for RNA was the degree of nucleic acid contamination of the N preparation, as judged by the ratio of absorbances at 260 and 280 nm (Table S1 and Fig. S1C).
Reconstitution and Crystallization of N–Nucleic Acid Complexes.
The equal-affinity binding of N to nucleic acid oligomers of different lengths is consistent with the appearance of N-RNA or N-DNA in negative-stain EM visualization experiments (Fig. S2 A and B) (5, 6). The reconstituted N–nucleic acid complexes appeared identical to both authentic RNP from virus-infected cells following extensive ribonuclease treatment and recombinant N-RNA that had not been stripped of RNA (5). The mixed population of multimers formed predominantly closed ring structures, but some appeared as incomplete or open rings (Fig. S2C). Therefore, to select a nucleic acid target for crystallization, we used negative-stain EM to screen oligomers for the length that generated the most homogeneous multimer population. Previous studies showed that N pentamers and hexamers were the most prevalent species of digested authentic viral RNP and of recombinant N-RNA (5, 6). Additionally, the electrophoretic mobility of RNA extracted from recombinant N-RNA had a broad size distribution centered between 30 and 40 nt (Fig. S1D), consistent with the length of a positively charged cleft on the inner surface of hexameric N in a crystal structure (6). We screened complexes reconstituted with DNA oligomers of length 25–45 nt. The N-DNA35 complex yielded the most homogeneous multimer population, with ring-like structures of predominantly five or six N subunits (Fig. S2 C and D).
Reconstituted RVFV N-DNA and N-RNA complexes were purified by gel filtration and subjected to crystallization screening, along with RNA-free N from RVFV and TOSV. Crystals were obtained for RVFV N-RNA, RVFV N-DNA, RVFV RNA-free N, and TOSV N (49% identical to RVFV N). Initial N-DNA35 crystals diffracted to 3.6 Å, but further optimization of the crystallization conditions and DNA length and purity resulted in N-DNA30 crystals with a diffraction limit of 2.7 Å (Table S2). Two crystal forms of N-RNA35 and one of N-RNA28 diffracted to 3.9, 3.4, and 2.15 Å, respectively.
N Multimer Formation.
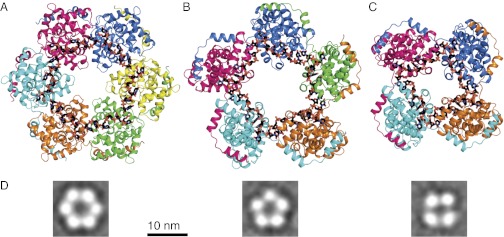
All crystals, with or without nucleic acid, contained N multimers (Fig. 1 and Fig. S3). RVFV RNA-free N, N-DNA30, and TOSV N crystallized as hexamers (N6 or N6-DNA30). N-RNA35 crystallized in identical conditions as either a hexamer (N6-RNA35) or a pentamer (N5-RNA35), whereas N-RNA28 crystallized as a tetramer (N4-RNA28). In all cases, electron density for the nucleic acid oligomers was continuous and lacked clear ends, indicating that ends of the nucleic acid oligomers were distributed randomly within the crystallized multimers (Fig. S4). The 65 independent views of the N subunit from the six crystal structures (Table S2) and 7 views from two published structures (5, 6) are identical in the subunit core, but they differ in the position of the helical N-terminal arm (amino acids 1–27) that extends from the core of each subunit to contact the core of a neighbor (Figs. 1 and 2). The arm-to-core interactions are identical in all subunits of all crystal structures (Fig. S5) and account for nearly all protein–protein contacts in the N oligomers. Remarkably, the crystallized multimers deviate from perfect four-, five-, or sixfold symmetry.
Fig. 1.
Structural plasticity of phlebovirus N-RNA. (A) N6-RNA35. (B) N5-RNA35. (C) N4-RNA28. N subunits are rendered in ribbon form with contrasting colors. The helical arm of each subunit wraps around the neighboring subunit on the outside of the multimer. Single-stranded RNA, rendered in stick form with black carbon atoms, binds to the inner surface of the multimer with all bases pointing into the protein. (D) EM visualization of reconstituted hexamer, pentamer, and tetramer multimers. The class averages, from Fig. S2C, are representative of the multimers released from viral RNP by ribonuclease digestion (5).
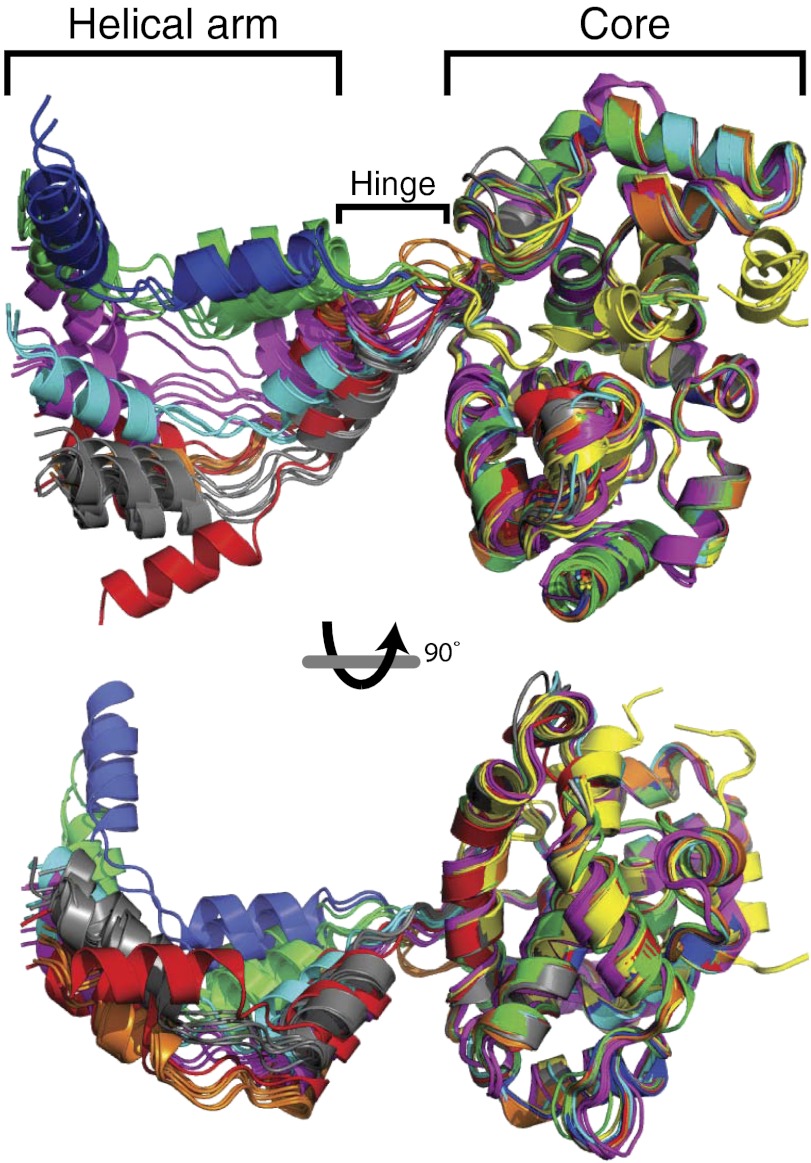
Fig. 2.
Flexible hinge between the helical arm and N core. The cores of all N subunits (residues 40–245) are superimposed. N subunits are rendered in ribbon form and colored by structure. Orange, RVFV N6-RNA35; green, RVFV N5-RNA35; blue, RVFV N4-RNA28; gray, RVFV N6; cyan, RVFV N6 [PDB ID code 3OV9 (6)]; yellow, RVFV N monomer [PDB ID code 3LYF (5)]; magenta, TOSV N6; and red, RVFV N6-DNA30.
N-RNA Binding.
RVFV N binds single-stranded RNA using an unusual base sequestration mechanism. Each N subunit possesses a deep, narrow RNA-binding slot, which becomes a continuous groove on the inner surface of the ring of four to six N subunits (Figs. 3A and 4). The inner surface of the groove is lined with conserved hydrophobic amino acids, whereas the rim has several conserved positively charged residues (Fig. S6). Nucleic acid binds with the bases inserted into the slot and the sugar-phosphate backbone oriented toward the center of the N multimer (Fig. 3A and Fig. S6). The high affinity of N for single-stranded nucleic acid is explained by extensive hydrophobic contacts of bases with amino acids in the RNA-binding slot and by base stacking. Although we crystallized N with oligomers of pyrimidine nucleotides, the RNA-binding slot is deep enough to accommodate purines (Fig. S6B).
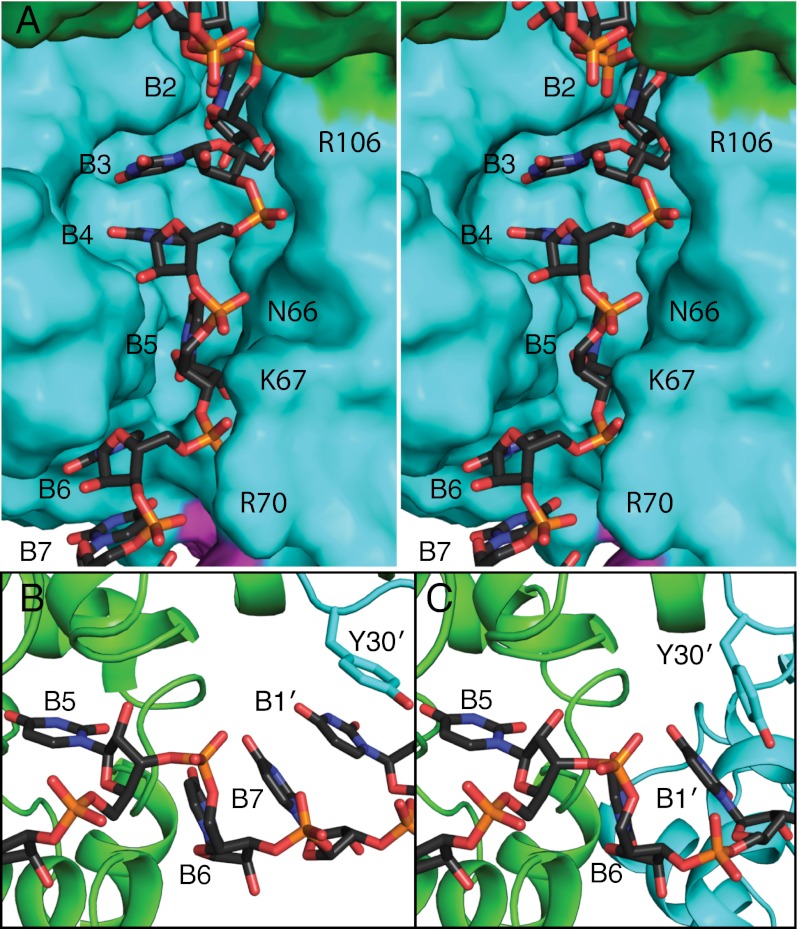
Fig. 3.
RNA sequestered in the RNA-binding slot. (A) Stereo image of RNA binding in the RNA-binding slot of one N subunit. RNA is drawn in stick form with black C atoms, and the surfaces of adjacent N subunits are colored green, cyan, and magenta. (B) RNA binding between subunits in N4-RNA28. Three RNA bases (B6, B7, B1') stack with Tyr30 at the interface of N subunits in N4-RNA28. B5, B6, and B7 are more closely associated with the green subunit and B1' with the cyan subunit. (C) RNA binding between N subunits of N6-RNA35, where only two RNA bases stack with Tyr30. In the hexamer (C), the cyan N subunit is closer to the green subunit than in the tetramer in B. RNA bases between subunits (B6, B7, B1′) point toward the protein interface and are inaccessible to other proteins or RNA.
Fig. 4.
RNA–Ncore–arm building block of phlebovirus RNP. The core of an N subunit (cyan surface) binds four RNA nucleotides (sticks) and the helical arm (green surface) of an adjacent N. These interactions on opposite sides of the N core create the compact RNA–Ncore–arm building block.
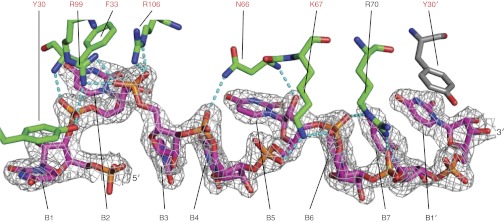
Each N subunit binds four RNA nucleotides in the RNA-binding slot and 2–3 additional nucleotides in the subunit interface. RNA is secured into the binding slot by contacts with 18 conserved amino acids (Fig. 5 and Figs. S6C and S7). In each N subunit, the 5′-most base (B1) stacks with Tyr30 in the hinge region between the helical arm and the N core. B2 stacks with Phe33 in a “back pocket” of the RNA-binding slot (Fig. S6A). The B3 and B4 bases are stacked in the central compartment of the RNA-binding slot, which is lined with the side chains of Ala109, Ala110, Pro147, Ile180, Pro199, and Ala202. B5 occupies a narrow pocket lined with the side chains of Gly65, Leu126, Pro127, and Phe176 (Fig. S6C). B6 and B7 stack with B1′ at the downstream N-N′ interface in the N5-RNA35 pentamer and in the N4-RNA28 tetramer (Fig. 3B). The N6-RNA35 hexamer lacks a B7, and B6 is stacked with B1′ (Fig. 3C). At the N-N interface, the positions of B1 and B2 create a sharp bend in the RNA backbone, which is more pronounced in the tetramer structure (∼80°) than in the pentamer (∼100°) and hexamer (∼105°) structures (Fig. 1 and Fig. S4). In addition to the hydrophobic and base-stacking interactions, N forms a network of polar contacts with the RNA 5′ phosphates (P1 to P7) of all nucleotides except P1: the Phe33 amide and Tyr30 hydroxy with P2; Arg99 with P2 and P3; Arg106 with P3; Asn66 with P4 and the 2′-hydroxy of R5; Lys67 with P5 and P6; and Arg70 with P6 and P7 (Fig. 5 and Fig. S7). Consistent with sequence-independent RNA binding, the N subunit forms no H bonds with RNA bases.
Fig. 5.
RVFV N RNA-binding interactions. Amino acids and RNA nucleotides are rendered in stick form. The carbon atoms in amino acids from adjacent subunits are colored green and gray. The 2.15-Å electron density (2Fo–Fc, contoured at 1σ, unbiased density in Fig. S7) shows RNA bound to an N subunit in RVFV N4-RNA28. Amino acids labeled in red are invariant in phlebovirus N proteins. RNA bases 1–7 are labeled B1 to B7 and B1′ is B1 of the adjacent N subunit. Dashed lines represent hydrogen bonds.
Discussion
The crystal structures of reconstituted RVFV N-RNA complexes offer a high-resolution view of genome packaging in segmented negative-sense RNA viruses. The unusual RNA-binding mechanism and limited N-N interactions in the RNP represent an assembly paradigm not previously observed. This work established three important properties of phlebovirus N: the mechanism of flexible subunit contacts in RNP, the nonspecific sequestration of nucleotide bases in the RNA-binding slot, and the variable base stacking between N subunits.
Strikingly, a four-nucleotide RNA core binds all N subunits identically, regardless of multimer size. The RNA core includes nucleotides 2–5 and the flanking phosphates (Figs. 3 and 4 and Fig. S6). Similarly, contacts of the helical arm of N with the neighboring subunit are identical in all subunits of all multimers (Fig. S5). These invariant RNA–protein and protein–protein contacts, which are located on opposite faces of the N core domain, define a compact unit of identical structure, the “RNA–Ncore–arm” unit (Fig. 4), in all RNA-N multimers.
Despite the identical RNA–protein and protein–protein contacts, the structures vary considerably among and within multimers. The variability derives from differences in the number of stacked bases between N subunits, in the hinge between the N core and its helical arm, and in the sharp bend between nucleotides 1 and 2. These structural features are located at the N-N subunit interface, where a lack of contacts between N cores also facilitates variability (Fig. 3). The arm hinge (amino acids 28–35, including Tyr30 stacked on RNA B1) varies among the multimers over a range of at least 55° (Fig. 2). Hinge flexibility was evident in the earlier structure of an N hexamer (6), but the six structures described here reveal a far greater range of arm motion (Fig. 2).
The crystal structures of N multimers show the RNA–protein interaction in exquisite detail, but the multimers are not building blocks for RNP (Fig. 6) (5, 7, 8). The width of the RNP matches the size of the invariant RNA–Ncore–arm monomer unit found in all three N-RNA crystal structures. The EM images of viral RNP show that phlebovirus RNP is a flexible chain of monomer-sized building blocks, and the crystal structures show that the building block is the invariant RNA–Ncore–arm unit (Figs. 4 and 6, Inset). Multimers (tetramer, pentamer, hexamer) result from N binding to an RNA oligomer (Fig. 1) or to a sheared RNA polymer (5). The irregular, asymmetric structure of phlebovirus RNP is fully explained by flexibly linked RNA–Ncore–arm building blocks that can move relative to one another over the range captured in the crystal structures (∼55°). The deep RNA-binding slot in each subunit encapsidates the RNA fully, including the nucleotides between subunits (Fig. 3). This rather primitive system for genome encapsidation requires few protein–protein contacts and sequesters all RNA bases away from access to other proteins. It is utterly incompatible with base pairing, even for the nucleotides bound between N subunits. The deep RNA sequestration explains the RNP insensitivity to ribonuclease, and the hydrophobic base-binding slot explains the resistance to high-salt treatment (5). N displayed a lack of RNA sequence specificity in protein contacts (Fig. 5 and Fig. S7) and in binding affinity (Table S1), consistent with its primary biological function to encapsidate a single-stranded RNA genome. Neither the crystal structures nor the binding data explain the slight sequence preferences reported for RNA aptamers developed to RVFV N (9). Rather N protects the entire viral genome within the RNP, which serves as the substrate for transcription and replication by the RNA-dependent RNA polymerase (RdRp) (10).
Fig. 6.
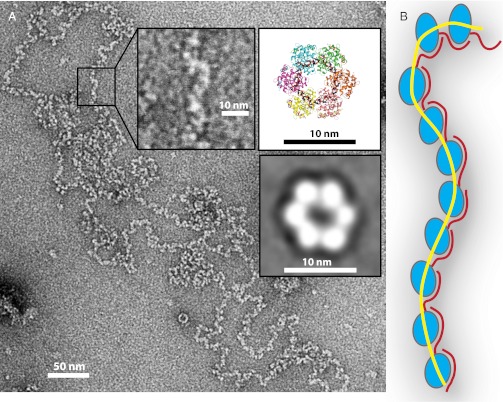
RVFV RNP: a flexible chain of monomer building blocks. (A) Negative-stain EM visualization of RVFV RNP isolated from infected BSR-T7/5 cells (5). The insets show an enlarged section of the viral RNP, the structure of the N6-RNA35 hexamer (Fig. 1A), and a predominant EM class average of N6-DNA35 (Fig. S2C). The scale bar shows that the hexamer is too large to be the repeating unit in RVFV RNP. (B) Cartoon representation of RNP constructed from RNA–Ncore–arm building blocks. The orientation of the helical arm (red) relative to the N core (blue) reflects the range of motion visualized in the crystal structures. RNA is represented as a yellow line.
The crystal structures and EM visualization demonstrate the ability of phlebovirus N to self-assemble in a simple linear or circular form (Figs. 1 and 6 and Figs. S2–S4). Circular RNPs have been visualized by EM for several segmented negative-sense viruses, including RVFV, influenza virus, the arenavirus Pichinde, the orthobunyavirus La Crosse, and the phlebovirus Uukuniemi (4, 5, 7, 11–13). Short complementary sequences (14–16 nt in RVFV) at the 5′ and 3′ ends of each genomic segment of these viruses create a base-paired “panhandle” structure. In influenza virus, the panhandle serves as a platform for RdRp attachment and as a promoter (14). Phlebovirus RNP may form circles in two ways, one by RNA panhandles and the other by N-N and N-RNA contacts. A duplex structure for the panhandle appears to be important, because increasing the number of potential base pairs increased promoter activity and transcription in Uukuniemi virus and the orthobunyavirus Bunyamwera (15, 16). Preparations of authentic RVFV RNP contain both N and RdRp (5), so the base-paired panhandle could function as an RdRp attachment site for each genomic segment of phleboviruses, similar to the influenza panhandle. The exclusively single-stranded, high-affinity N-RNA interaction observed in the crystal structures implies that N would bind tightly to any RNA strand released by duplex breathing. Thus, the RdRp may protect the panhandle duplex, perhaps by binding the 5′ and 3′ ends throughout its catalytic cycles. Bunyavirus transcription and replication require both N and the panhandle sequences (15–19). The N-RNA structures do not illuminate the essential function of N in replication and transcription, but they show that N must be stripped from the RNA for RdRp “reading” during transcription and replication. The hinge between the N core and helical arm is sufficiently flexible for the RdRp to create a moving bubble in the RNP by inserting itself between RNA and N during transcription or replication.
The phlebovirus RNPs have some of the smallest N proteins and represent the extreme of asymmetry among a range of architectures displayed by RNPs of negative-sense viruses. The arenaviruses also have an asymmetric RNP (4, 12), but the N protein is unrelated to the phlebovirus N and has an additional exoribonuclease domain (20, 21). The RNP architecture is understood in greatest detail for those viruses with RNP of highest symmetry. Respiratory syncytial virus (RSV), vesicular stomatitis virus (VSV), and rabies virus have highly symmetric, helical RNPs, the structures of which determine the morphology of the rod- or bullet-shaped virus particles (22–25). In contrast, spherical phlebovirus particles have T = 12 icosahedral symmetry that does not include the RNP (26). N-RNA multimers from RSV, VSV, and rabies virus have a strict 10- or 11-fold circular symmetry that is clearly related to the helical symmetry of their respective RNPs (22, 24, 25). The homologous N proteins from these viruses and from Borna disease virus (27) have protruding chain termini that mediate subunit contacts, but, unlike the phlebovirus N, the protrusions are in fixed positions and there are also substantial Ncore-Ncore contacts. The segmented RNP of influenza virus has an intermediate degree of symmetry. Like the phlebovirus N, the influenza N protein can occur in multiple associated states (4, 14, 28, 29), but its RNP is helical (4, 14, 30). Thus, the N proteins of negative-sense viruses with segmented genomes (RVFV, arenavirus, influenza) have flexible conformations and association modes unlike the N of nonsegmented viruses (RSV, VSV, rabies, Borna disease).
Phleboviruses use a base-in binding mode with a deep hydrophobic slot that buries four nucleotide bases in each N subunit and sequesters two to three more bases between subunits (Fig. 3). No base is available for “reading” by proteins or nucleic acids. In contrast, N proteins of other negative-sense viruses achieve sequence-independent RNA binding in a manner that sequesters the RNA bases only partially. The Lassa virus N protein binds eight RNA nucleotides with the sugar-phosphate backbone directed into an RNA-binding pocket. Some bases are directed into a deep pocket, similar to the phleboviruses, and others are solvent-exposed but with protein contacts (20). RSV, VSV, and rabies virus also have a mixed binding mode, where three stacked bases point into an RNA-binding pocket and another four or six bases face the solvent (22, 24, 25). In these viruses with partial or complete base-out RNA binding, the RdRp may be able to access the genome without complete disassembly of the helical RNP (24, 25).
The structure of N bound to RNA enables calculation of the RNP volume and, thus, the quantity of encapsidated genetic material that can pack in a virus particle. The inner volume of the RVFV virus particle (26) could accommodate a predicted average 1:4:4 ratio of the large:medium:small genomic segments (19). However, this would require packing at the maximum density possible, at least threefold greater density than in other negative-sense viruses (23, 25, 30).
The crystal structures of RVFV N bound to RNA and EM visualization of viral and reconstituted RNPs provide a detailed understanding of how phleboviruses encapsidate their RNA genomes. The virus uses a primitive but effective system using limited protein–protein interactions and a deep RNA-binding slot where all RNA bases are inaccessible to other proteins or nucleic acids. The resulting nuclease-resistant RNP lacks symmetry and becomes the template for genome replication and transcription by the RdRp. Shielding of the viral RNA bases in the RNA-binding slot prevents base pairing and protects the RNA from the cellular antiviral response; however, it requires that the RdRp must strip the genome of N to access the genomic information during replication and transcription.
Materials and Methods
Detailed methods are given in the SI Text.
Recombinant RVFV and TOSV N proteins were purified under native or denaturing conditions from an Escherichia coli expression system. Fluorescence polarization experiments were performed using natively purified or refolded, RNA-free N proteins and PAGE-purified, fluorescently labeled, single-stranded RNA or DNA oligomers. N–nucleic acid complexes were reconstituted by incubating RNA-free N with nucleic acid at a molar ratio of 1:8 and purified by gel-filtration chromatography before crystallization. Diffraction data were collected at General Medicine/Cancer (GM/CA) beamlines 23ID-B and 23ID-D at the Advanced Photon Source (Argonne National Laboratory), and structures were solved by molecular replacement. Data collection and refinement statistics are presented in Table S2, and representative electron density for nucleic acids are shown in Fig. 5 and Figs. S4 and S7. For EM, samples were prepared onto carbon-coated grids using conventional negative-staining protocols and imaged at room temperature.
Supplementary Material
Acknowledgments
We thank the staff of the GM/CA beamlines [supported by the National Institutes of Health (NIH) Institute of General Medical Sciences and National Cancer Institute] at the Advanced Photon Source (supported by the United States Department of Energy).This work was supported by NIH Grant P01-AI055672 (to J.L.S.), by NIH Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research Program Award U54-AI057153 (to S.R.G.), and by the Rackham Graduate School at the University of Michigan (S.R.G.). D.D.R. was supported by NIH Molecular Biophysics Training Grant T32-GM008270. M.E.P. was supported by NIH Cell and Molecular Biology Training Grant T32-GM007315.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID codes 4H6F and 4H6G (RVFV N6-RNA35), 4H5O (RVFV N5-RNA35), 4H5P (RVFV N4-RNA28), 4H5Q (RVFV N6-DNA30), 4H5M (RVFV N6), and 4H5L (TOSV N6)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213553109/-/DCSupplemental.
References
- 1.Pepin M, Bouloy M, Bird BH, Kemp A, Paweska J. Rift Valley fever virus(Bunyaviridae: Phlebovirus): An update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet Res. 2010;41(6):61. doi: 10.1051/vetres/2010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valassina M, Cusi MG, Valensin PE. A Mediterranean arbovirus: The Toscana virus. J Neurovirol. 2003;9(6):577–583. doi: 10.1080/13550280390247678. [DOI] [PubMed] [Google Scholar]
- 3.Schmaljohn CS, Nichol ST. In: Fields Virology. Knipe DM, et al., editors. Philadelphia, PA: Lippincott, Williams & Wilkins; 2007. pp. 1741–1789. [Google Scholar]
- 4.Ruigrok RW, Crépin T, Kolakofsky D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr Opin Microbiol. 2011;14(4):504–510. doi: 10.1016/j.mib.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 5.Raymond DD, Piper ME, Gerrard SR, Smith JL. Structure of the Rift Valley fever virus nucleocapsid protein reveals another architecture for RNA encapsidation. Proc Natl Acad Sci USA. 2010;107(26):11769–11774. doi: 10.1073/pnas.1001760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferron F, et al. The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes. PLoS Pathog. 2011;7(5):e1002030. doi: 10.1371/journal.ppat.1002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pettersson RF, von Bonsdorff CH. Ribonucleoproteins of Uukuniemi virus are circular. J Virol. 1975;15(2):386–392. doi: 10.1128/jvi.15.2.386-392.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hewlett MJ, Pettersson RF, Baltimore D. Circular forms of Uukuniemi virion RNA: An electron microscopic study. J Virol. 1977;21(3):1085–1093. doi: 10.1128/jvi.21.3.1085-1093.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellenbecker M, Sears L, Li P, Lanchy JM, Lodmell JS. Characterization of RNA aptamers directed against the nucleocapsid protein of Rift Valley fever virus. Antiviral Res. 2012;93(3):330–339. doi: 10.1016/j.antiviral.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piper ME, Sorenson DR, Gerrard SR. Efficient cellular release of Rift Valley fever virus requires genomic RNA. PLoS ONE. 2011;6(3):e18070. doi: 10.1371/journal.pone.0018070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruigrok RW, Baudin F. Structure of influenza virus ribonucleoprotein particles. II. Purified RNA-free influenza virus ribonucleoprotein forms structures that are indistinguishable from the intact influenza virus ribonucleoprotein particles. J Gen Virol. 1995;76(Pt 4):1009–1014. doi: 10.1099/0022-1317-76-4-1009. [DOI] [PubMed] [Google Scholar]
- 12.Young PR, Howard CR. Fine structure analysis of Pichinde virus nucleocapsids. J Gen Virol. 1983;64(Pt 4):833–842. doi: 10.1099/0022-1317-64-4-833. [DOI] [PubMed] [Google Scholar]
- 13.Obijeski JF, Bishop DH, Palmer EL, Murphy FA. Segmented genome and nucleocapsid of La Crosse virus. J Virol. 1976;20(3):664–675. doi: 10.1128/jvi.20.3.664-675.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klumpp K, Ruigrok RW, Baudin F. Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J. 1997;16(6):1248–1257. doi: 10.1093/emboj/16.6.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flick K, et al. Functional analysis of the noncoding regions of the Uukuniemi virus (Bunyaviridae) RNA segments. J Virol. 2004;78(21):11726–11738. doi: 10.1128/JVI.78.21.11726-11738.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barr JN, Wertz GW. Bunyamwera bunyavirus RNA synthesis requires cooperation of 3′- and 5′-terminal sequences. J Virol. 2004;78(3):1129–1138. doi: 10.1128/JVI.78.3.1129-1138.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mir MA, Brown B, Hjelle B, Duran WA, Panganiban AT. Hantavirus N protein exhibits genus-specific recognition of the viral RNA panhandle. J Virol. 2006;80(22):11283–11292. doi: 10.1128/JVI.00820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohl A, Dunn EF, Lowen AC, Elliott RM. Complementarity, sequence and structural elements within the 3′ and 5′ non-coding regions of the Bunyamwera orthobunyavirus S segment determine promoter strength. J Gen Virol. 2004;85(Pt 11):3269–3278. doi: 10.1099/vir.0.80407-0. [DOI] [PubMed] [Google Scholar]
- 19.Gauliard N, Billecocq A, Flick R, Bouloy M. Rift Valley fever virus noncoding regions of L, M and S segments regulate RNA synthesis. Virology. 2006;351(1):170–179. doi: 10.1016/j.virol.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 20.Hastie KM, et al. Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding. Proc Natl Acad Sci USA. 2011;108(48):19365–19370. doi: 10.1073/pnas.1108515108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi X, et al. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature. 2010;468(7325):779–783. doi: 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albertini AA, et al. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313(5785):360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- 23.Ge P, et al. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science. 2010;327(5966):689–693. doi: 10.1126/science.1181766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green TJ, Zhang X, Wertz GW, Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006;313(5785):357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- 25.Tawar RG, et al. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science. 2009;326(5957):1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- 26.Freiberg AN, Sherman MB, Morais MC, Holbrook MR, Watowich SJ. Three-dimensional organization of Rift Valley fever virus revealed by cryoelectron tomography. J Virol. 2008;82(21):10341–10348. doi: 10.1128/JVI.01191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rudolph MG, et al. Crystal structure of the borna disease virus nucleoprotein. Structure. 2003;11(10):1219–1226. doi: 10.1016/j.str.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 28.Coloma R, et al. The structure of a biologically active influenza virus ribonucleoprotein complex. PLoS Pathog. 2009;5(6):e1000491. doi: 10.1371/journal.ppat.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye Q, Krug RM, Tao YJ. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature. 2006;444(7122):1078–1082. doi: 10.1038/nature05379. [DOI] [PubMed] [Google Scholar]
- 30.Noda T, Kawaoka Y. Structure of influenza virus ribonucleoprotein complexes and their packaging into virions. Rev Med Virol. 2010;20(6):380–391. doi: 10.1002/rmv.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.