Abstract
Neuroblastoma is a pediatric tumor of the sympathetic nervous system. MYCN (V-myc myelocytomatosis viral-related oncogene, neuroblastoma derived [avian]) is amplified in 20% of neuroblastomas, and these tumors carry a poor prognosis. However, tumors without MYCN amplification also may have a poor outcome. Here, we identified downstream targets of MYCN by shRNA-mediated silencing MYCN in neuroblastoma cells. From these targets, 157 genes showed an expression profile correlating with MYCN mRNA levels in NB88, a series of 88 neuroblastoma tumors, and therefore represent in vivo relevant MYCN pathway genes. This 157-gene signature identified very poor prognosis tumors in NB88 and independent neuroblastoma cohorts and was more powerful than MYCN amplification or MYCN expression alone. Remarkably, this signature also identified poor outcome of a group of tumors without MYCN amplification. Most of these tumors have low MYCN mRNA levels but high nuclear MYCN protein levels, suggesting stabilization of MYCN at the protein level. One tumor has an MYC amplification and high MYC expression. Chip-on-chip analyses showed that most genes in this signature are directly regulated by MYCN. MYCN induces genes functioning in cell cycle and DNA repair while repressing neuronal differentiation genes. The functional MYCN-157 signature recognizes classical neuroblastoma with MYCN amplification, as well as a newly identified group marked by MYCN protein stabilization.
Neuroblastoma is a pediatric solid tumor derived from the sympathetic nervous system. The prognosis is highly variable and is associated with parameters such as age at diagnosis, dissemination at time of diagnosis, tumor stage, and grade of differentiation of the primary tumor (1). Neuroblastoma stages 1 and 2 have a very good prognosis, but survival in stage 4 neuroblastoma is below 30%. Amplification of MYCN (V-myc myelocytomatosis viral-related oncogene, neuroblastoma derived [avian]) is associated with poor outcome. It occurs in about 20% of the tumors but is confined to high-stage neuroblastoma.
Together with MYC and MYCL, MYCN belongs to the MYC transcription factor family, whose role in cancer has been studied extensively. A series of investigators have manipulated MYCN expression in neuroblastoma cell lines by overexpression or silencing. High expression of MYCN is associated with fast proliferation and induction of cell cycle genes [(2–5) and reviewed by Bell (5)]. Although cell cycle genes were identified, the actual number of MYC-regulated genes is a small minority compared with all other genes claimed to be regulated in such experiments. In fact, thousands of candidate target genes of MYC and/or MYCN currently are known, which has complicated pinpointing of the relevant set of genes regulated by MYC genes and responsible for the aggressive phenotype.
In a very early study, MYCN protein expression was found to be a poor prognostic factor (6). Recent experiments in cell lines showed that MYCN protein stability is decreased after phosphorylation by glycogen synthase kinase-3β (GSK3β), which itself is inactivated by AKT. Accordingly, activation of the PI3K/AKT pathway in neuroblastoma cell lines resulted in stabilization of MYCN protein (7, 8). Inactivation of the pathway using a PI3K inhibitor resulted in reduced levels of MYCN (9). At another level, FBXW7 (F-box/WD repeat-containing protein 7) and AURKA (aurora kinase A) are involved in MYCN protein stability (10). However, the relevance of MYCN protein stability and its consequence on MYCN target gene expression are ill defined in neuroblastoma.
In this study, we integrated in vitro regulation by MYCN and in vivo correlation to identify relevant genes in neuroblastoma. The unique MYCN-157 gene signature predicts outcome in neuroblastoma. The signature recognizes all tumors with MYCN amplification, but surprisingly also an equally large group of tumors without MYCN amplification. These tumors have low MYCN mRNA levels but high nuclear MYCN protein levels, suggesting stabilization of MYCN at the protein level.
Results
Gene Regulation by MYCN in MYCN-Amplified Neuroblastoma: MYCN-157 Signature.
Many cell line experiments have suggested that MYC oncogenes control expression of thousands of genes functioning in multiple pathways. We attempted to identify MYCN-regulated genes relevant for in vivo functioning of MYCN in neuroblastoma. We began by establishing downstream genes of the MYCN pathway in MYCN-amplified neuroblastoma cell line IMR32, which expressed high levels of MYCN. MYCN was silenced in IMR32 by the use of a lentiviral shRNA construct. A time series experiment was performed in duplo using shMYCN and a control shRNA virus without a target. RNAs isolated from t = 0 to t = 72 h were analyzed on Affymetrix HG U133 Plus 2.0 arrays.
For the analysis of such time series data and integration with expression profiles of tumor series, we developed the bioinformatics program R2 (http://r2.amc.nl). Fig. 1A summarizes the steps in R2 that we used in this study to analyze MYCN-regulated genes in neuroblastoma. The “Time Series” tool was used to select the genes regulated after silencing of MYCN. A total of 905 genes were regulated with 2logfold >1 and a minimal expression of 50 units. This group consisted of 442 up- and 463 down-regulated genes.
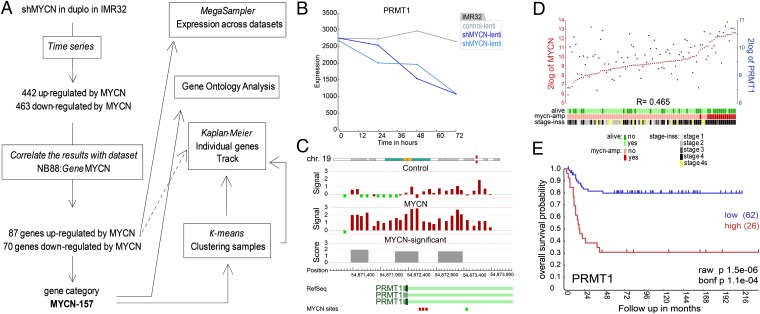
Fig. 1.
Gene regulation by MYCN. (A) Overview of R2 bioinformatics tools used to identify MYCN-regulated genes relevant to neuroblastoma. In the Time Series module, genes are selected for regulation by MYCN. Correlation with MYCN in neuroblastoma tumors (NB88) reduces the group to 157 regulated genes relevant to tumors. These genes are analyzed individually or as a group (gene category: MYCN-157) with the different tools indicated on the right. R2 programs are indicated with boxes, and tool names are in italics. (B–E) PRMT1 is regulated by MYCN. (B) IMR32 was transduced with shMYCN (blue lines) or control virus (gray line). The expression graph shows the PRMT1 mRNA expression of the Affymetrix profiling. (C) ChIP-on-chip analysis in IMR32 with anti-MYCN or control antibody. Signal: raw data of binding ratios (2logfold) in red bars. Score: the significant binding region (2logfold) (FDR: < 0.05). The three reference PRMT1 transcripts (exon dark, intron light green) are aligned to their chromosomal position, and MYCN consensus sites are indicated below (CACGTG in red and the alternative site in green; Dataset S1). (D) YY plot showing the correlation of MYCN (red) and PRMT1 (blue) in neuroblastoma tumor series NB88. Below the graph tracks for survival, MYCN amplification and International Neuroblastoma Staging System stages are shown. (E) Overall survival for PRMT1 expression in the NB88 set.
To select genes relevant in neuroblastoma tumors, we combined the cell line experiment with gene expression profiles of neuroblastoma. The NB88 tumor series consists of 88 primary neuroblastoma tumors of all stages that we have profiled on the Affymetrix U133 Plus 2.0 microarrays. We analyzed which of the 905 genes regulated by MYCN in the cell line experiment had an expression pattern that correlated with MYCN (P < 0.01) expression in the tumor series. There were 87 genes up-regulated by MYCN in vitro that showed a positive correlation with MYCN in the NB88 series (Dataset S1). Of the in vitro down-regulated genes, 70 genes correlated negatively with MYCN expression in the NB88 series (Dataset S1). For only 157 of the 905 genes (17%) regulated by MYCN in vitro, we have evidence for regulation by MYCN in vivo.
We validated the regulation of three genes in detail. The mRNA expression of PRMT1 decreased after shRNA-mediated silencing of MYCN in cell line IMR32 (Fig. 1B) and correlated positive (R = 0.465) with MYCN in tumor series NB88 (Fig. 1D). Protein analysis confirmed the up-regulation of PRMT1 by MYCN in neuroblastoma cell lines IMR32 and SKNBE (Fig. S1A). Similar results were obtained for MYCN–up-regulated gene MTAP (Fig. S2 A and C). CLU is an example of a gene down-regulated by MYCN in vitro and negatively correlating with MYCN in the NB88 series (Fig. S2 A and C). These results also were confirmed at the protein level (Fig. S1A). The R2 program also can generate Kaplan-Meier curves to establish prognostic value of gene expression levels in tumor series. High expression of PRMT1 and MTAP and low expression of CLU are significantly associated with a poor prognosis, which is in line with their relationship to MYCN (Fig. 1E and Fig. S2D). Data on all 157 genes are summarized in Dataset S1 and correlation and Kaplan-Meier plots themselves can be generated at http://r2.amc.nl. The combination of in vitro regulation by MYCN and in vivo correlation in a neuroblastoma tumor series identified a potentially interesting dataset of 157 genes (MYCN-157).
Prognostic Value of MYCN-157 Gene Signature.
We postulated that the MYCN-157 gene set forms a signature reflecting the activity of the MYCN pathway in neuroblastoma. As a first analysis of this signature, we investigated the prognostic value of the MYCN-157 gene set. We used K-means clustering to subdivide the NB88 series in two groups with high or low activity of the MYCN-157 signature (Fig. S3). One cluster of 33 tumors included all the MYCN-amplified samples, showed high expression of up-regulated genes and low expression of down-regulated genes, and therefore was assigned MYCN-157-POS. A cluster of 55 neuroblastomas with low expression of up-regulated genes and high expression of down-regulated genes was designated MYCN-157-NEG. Kaplan-Meier analysis of the two clusters showed that tumors with a MYCN-157-POS signature conferred an extremely poor prognosis (log-rank test P = 2.0 × 10−11, Fig. 2A). Surprisingly, this cluster also included 17 MYCN single-copy tumors. When the tumors with MYCN amplification were left out of the Kaplan-Meier analysis, the poor prognosis of the remaining tumors with a MYCN-157-POS signature was still highly significant (P = 1.6 × 10−7, Fig. 2B). Amplification of MYCN is an established marker for poor prognosis, but it was less significant than the MYCN-157 signature (P = 2.0 × 10−8, Fig. S4A). Also, high MYCN mRNA expression is associated with poor prognosis but is less significant (P = 7.7 × 10−7, Fig. S4B). In contrast to the MYCN-157 signature, MYCN mRNA expression is no longer significant when tumors with MYCN amplification are excluded (Fig. S4C). The prognostic value of the MYCN-157 signature therefore is superior to MYCN amplification or expression. This was confirmed in a regression analysis showing a hazard ratio for survival of 14.87 for the MYCN-157 signature versus 6.25 for MYCN amplification (Table S1). In addition, using clinical markers as stratus, the MYCN-157 signature adds to the predictive power of MYCN amplification, LOH1p, and clinical stages (Table S1).
Fig. 2.
The expression of MYCN-157 signature predicts clinical outcome of neuroblastoma. Overall survival analysis was performed in tumor series NB88 (A and B) and NB251 (C and D). The gene expression of MYCN-157 in neuroblastoma tumors was clustered with K-means, resulting in two groups: MYCN-157-POS and MYCN-157-NEG (see Fig. S3 for NB88). Overall survival is shown for the clustering according to MYCN-157 with (A and C) and without (B and D) MYCN-amplified (MNA) tumors.
We could validate these observations in an independent dataset of 251 neuroblastoma tumors (11), the expression profile of which was determined on custom-made two-color arrays, which included probes for 79 of the MYCN-157 genes. Expression data of these genes were used for K-means clustering in two groups. The MYCN-157-POS cluster consisted of 77 patients, including 30 patients with a MYCN amplification. The MYCN-157-NEG cluster consisted of 174 patients, including one MYCN-amplified tumor. The MYCN-157-POS cluster had an extremely poor prognosis (P = 1.6 × 10−20, Fig. 2C). Importantly, after removal of the tumors with MYCN amplification in this series, Kaplan-Meier analysis of the remaining tumors still showed a poor prognosis for tumors classified with a MYCN-157-POS signature (P = 5.5 × 10−14, Fig. 2D). Also in this independent series of 251 tumors, amplification or expression of MYCN was less significant than MYCN-157 (Fig. S4 D–F). Interestingly, the single tumor with a MYCN amplification in the MYCN-157-NEG cluster was from a patient who survived.
We conclude that the MYCN-157 signature predicts clinical outcome in two independent neuroblastoma series. Surprisingly, although the signature is based on regulation by MYCN, it also predicts outcome in neuroblastoma without MYCN amplification.
Neuroblastomas Without MYCN Amplification but With a MYCN-157-POS Signature Have High MYCN Protein Expression.
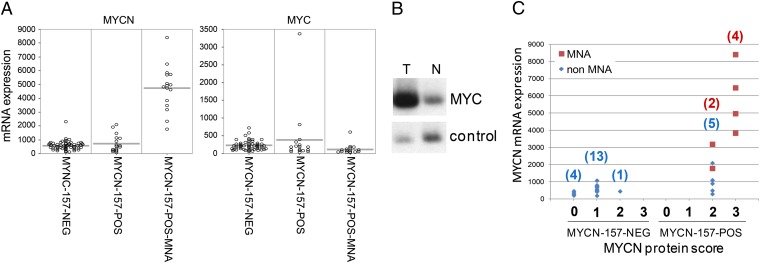
The MYCN-157-POS group identified in the K-means clustering of the NB88 series included 16 tumors with MYCN amplification, but also 17 tumors without MYCN amplification. We investigated why tumors without MYCN amplification can have a MYCN-157-POS signature. One tumor showed very high MYC expression, as well as amplification of the MYC gene (Fig. 3 A and B). This suggests that the positive MYCN signature in this tumor is the result of MYC amplification. The remaining 16 tumors had no elevated MYCN or MYC mRNA expression, compared with the MYCN-157-NEG group in the K-means clustering.
Fig. 3.
MYC amplification and high MYCN protein expression in neuroblastoma tumors clustering together with MYCN-amplified tumors. (A) MYCN and MYC mRNA expression detected by Affymetrix profiling in MYCN-157-NEG and MYCN-157-POS tumors. The MYCN-157-POS group was divided by absence or presence of MYCN amplification (MNA). (B) Southern blot of tumor (T) and normal (N) DNA of patient itcc0184 showing MYC amplification. The control probe is L.2.3 (29). (C) MYCN protein expression analysis on tissue array compared with MYCN mRNA expression. Tumors were scored for expression of nuclear MYCN according to the following categories of expression: none (0), weak (1), moderate (2), or strong (3). The y axis shows the expression of MYCN mRNA. MYCN-amplified tumors are indicated in red and nonamplified tumors in blue.
We therefore asked whether tumors with low MYCN mRNA levels might nevertheless have high MYCN protein levels, resulting in a positive MYCN signature. Destabilization of MYCN protein levels by inactivation of PI3K has been observed in neuroblastoma cell lines (8, 9). However, to our knowledge this has not been investigated in human neuroblastoma series. We analyzed the levels of MYCN protein expression by immunohistochemical analysis of a tissue array with 29 neuroblastoma tumors from the NB88 series (Fig. S5). Staining intensities were scored as absent (0), low (1), moderate (2), or strong (3). Of the 18 tumors with a MYCN-157-NEG signature, 4 had no MYCN staining, 13 had low staining, and only 1 had moderate staining (Fig. 3C). Of the 11 tumors with a MYCN-157-POS signature, 6 had MYCN amplification, 4 of which scored as strong and 2 as moderate on MYCN protein staining. Interestingly, all five tumors with a MYCN-157-POS signature but without MYCN amplification also showed moderate staining (Fig. 3C). Their MYCN mRNA level was similar to that of tumors of the MYCN-157-NEG group, but they nevertheless had much higher MYCN protein staining and a MYCN-157-POS signature. This analysis shows that a group of neuroblastoma tumors exists without MYCN amplification but with a high MYCN protein expression, which is not explained by an increased MYCN mRNA level. This suggests that these tumors have an increased MYCN protein expression due to increased protein stabilization. In the K-means clustering, they have a MYCN-157-POS signature and a poor prognosis.
MYCN Binds Preferentially to the Promoters of Induced Genes.
As the MYCN-157 signature appeared to be a clinically more relevant parameter for outcome than MYCN amplification status or MYCN mRNA expression, we further analyzed the function and regulation of these 157 genes by MYCN. We first searched for the genes regulated by the MYCN transcription factor. We performed ChIP-on-chip assays in IMR32. MYCN-bound DNA was isolated, amplified, and hybridized to NimbleGen promoter arrays, covering the −5 to +3 kb genomic regions relative to the transcription start site (TSS) of genes. As a control, we used a FLAG antibody, which has no target in these cells. Examples of the binding data are shown for the promoters of PRMT1, MTAP, and CLU (Fig. 1C and Fig. S2B). The MYCN binding extends over a large region both up- and downstream of the TSS, but is scored significantly stronger than the control values only in the regions marked red (PRMT1 and MTAP, high confidence binding) and orange (CLU, indicative of binding).
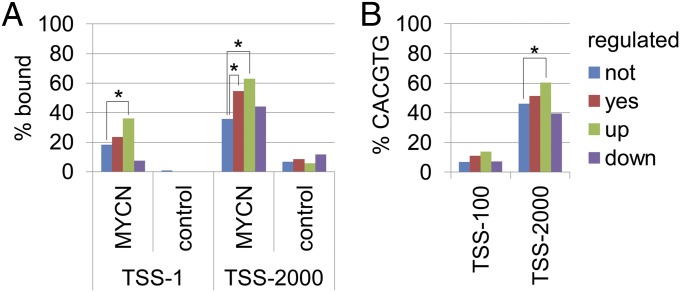
The areas with the highest confidence of binding (false discovery rate [FDR] ≤ 0.05) were used for genome-wide analyses. Of the MYCN-157 signature, 154 genes were represented on the promoter array. MYCN showed significant binding around the TSS of all genes in the genome, MYCN-157 signature genes, and up-regulated genes of the signature and scattered over a larger region for the down-regulated genes (Fig. S6). Because the most prominent binding appears around the TSS, we analyzed binding at the TSS (TSS1, Fig. 4A) and the presence of MYCN binding sites in the −100 to +100 bp range (Fig. 4B; TSS-100, a region covering two NimbleGen probes, determines binding at TSS1). At the TSS, 18% of all genes bind MYCN (Fig. 4A). This increased significantly to 36% in the up-regulated genes. Although 36% of up-regulated genes bind at the TSS, only 14% have a canonical (CACGTG) consensus site within 100 bp (TSS-100, Fig. 4B). For overall binding, we analyzed the 2-kb region up- or downsteam of the TSS. Fifty-five percent of MYCN-157 genes bind MYCN to the promoter (TSS-2000, Fig. 4A). This percentage increases to 63% when only up-regulated genes are considered. Canonical sites are present in the TSS-2000 region of 60% of up-regulated genes, explaining most up-regulated and MYCN-bound genes (Fig. 4B and Dataset S1). We also scanned the regions for alternative MYCN consensus sequences. Alternative sites are present in promoter regions of genes such as E2F5 and MCM3 but are absent in others, such as MCM5 and ATAD2 (Dataset S1). No significant enrichment was observed among down-regulated genes; thus, down-regulation of MYCN-157 genes occurs mainly via alternative ways. Several mechanisms have been described, including interference with transcription factors MIZ1 and Sp1 (12, 13) and induction of microRNA expression (14, 15).
Fig. 4.
MYCN binds preferentially to up-regulated genes. (A) ChIP-on-chip analysis of MYCN and control in IMR32. The percentage of significant (FDR < 0.05) bound promoters at the TSS (TSS-1) and ± 2,000 bp (TSS-2000) is shown. (B) The percentage of genes with a CACGTG ± 100 bp (TSS-100) or ± 2,000 bp (TSS-2000) is shown. The analysis was performed for MYCN-157 (regulated: yes), the up-and down-regulated subsets, and all other genes (regulated: no). *Significant association (P < 0.01) between bound/unbound genes or presence/absence of CACGTG and regulation in the MYCN-157 sets (two-sided Fisher’s exact test).
Overall, 84 of the 154 genes analyzed for MYCN binding showed significant binding. K-means clustering with only MYCN-bound genes and with only unbound genes resulted in strong prognostic clustering in both groups (Fig. S4 G and H). We conclude that genes indirectly regulated by MYCN also form an important part of the MYCN-157 signature.
Biological Function of MYCN in Neuroblastoma Tumors.
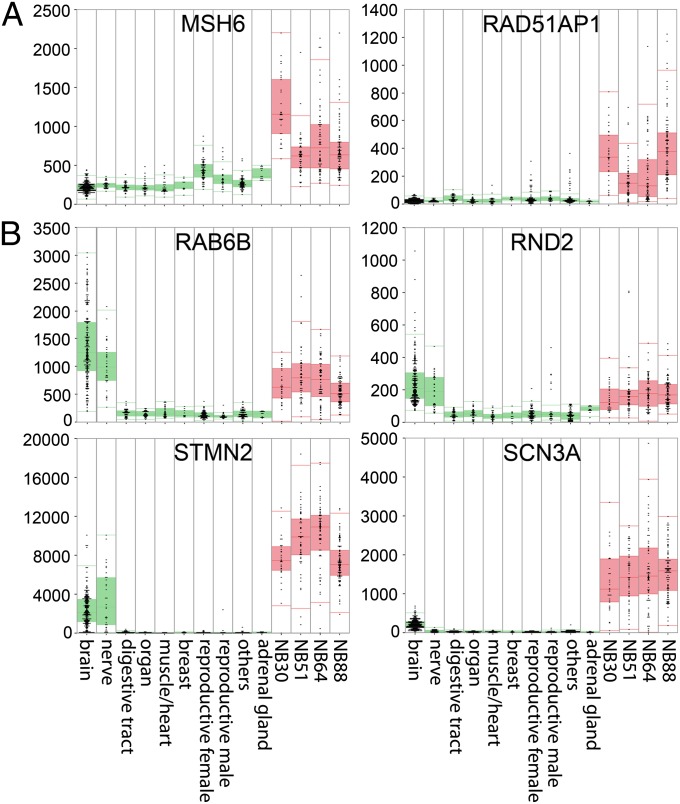
The functional role of the MYCN-157 signature was investigated by Gene Ontology (GO) analyses. The most significant overrepresentation was observed for the GO categories “cell cycle” and “DNA repair,” and these genes are up-regulated by MYCN (Table 1). This confirms previous reports; genes such as MCM3 and MCM5 were directly bound and induced by MYCN (16). A remarkable class of induced genes is “DNA repair” (Table 1). We used the MegaSampler tool of R2 (http://r2.amc.nl) to compare the expression in four independent neuroblastoma series and normal tissues. MSH6 and RAD51AP1 are strongly overexpressed in four independent neuroblastoma tumor series compared with any normal tissue (Fig. 5A), with RAD51AP1 almost completely absent in normal tissues. The GO analysis also identified genes of the category “small GTPase mediated signal transduction” (Table 1), which typically are down-regulated. RAB6B and RND2 are highly expressed in all four neuroblastoma series, but also in normal neuronal tissues (assigned brain and nerve [Fig. 5B]). This suggests that MYCN might down-regulate genes with a neuronal tissue-specific expression.
Table 1.
Gene ontology classes enriched in MYCN-157
| GO description | Total no. genes in set | No. genes regulated | P value | Gene symbols* |
| Cell cycle | 807 | 20 | 1.50E-05 | OIP5, E2F5, SKA3, NCAPH, POLA2, PHGDH, LIG3, MCM3, MCM5, POLD2, POLE2, TIPIN, HJURP, PKIA, RAD51, RRM2, ANAPC1, E2F8, RAD54L, KIF23 |
| DNA replication | 144 | 7 | 1.20E-05 | POLA2, LIG3, MCM3, MCM5, POLD2, POLE2, RRM2 |
| Mitotic cell cycle | 299 | 10 | 8.00E-05 | E2F5, NCAPH, POLA2, MCM3, MCM5, POLD2, POLE2, RRM2, ANAPC1, KIF23 |
| S phase of mitotic cell cycle | 105 | 5 | 4.90E-04 | POLA2, MCM3, MCM5, POLD2, POLE2 |
| G1/S transition of mitotic cell cycle | 129 | 5 | 3.30E-03 | POLA2, MCM3, MCM5, POLE2, RRM2 |
| DNA repair | 250 | 7 | 7.80E-03 | RAD51AP1, MSH6, LIG3, POLD2, POLE2, RAD51, RAD54L |
| Small GTPase-mediated signal transduction | 277 | 7 | 0.02 | RUNDC3A, ARL4D, RHOC, ARHGEF3, RAB6B, RND2, DIRAS3 |
*Underlined symbols are down-regulated by MYCN; the others are up-regulated by MYCN.
Fig. 5.
DNA repair and neuronal differentiation genes are highly expressed in neuroblastoma tumors. MegaSampler analysis in normal and tumor tissues. The box plots show the expression of DNA repair genes (A) and neuronal differentiation genes (B) in neuroblastoma and normal tissue. Normal tissues [green (30)] are divided in the 10 groups indicated below. Four independent neuroblastoma series [red: NB30 (31), NB51, NB64 (32), and NB88] are named after the sample number in the group.
We investigated this idea further by comparing the expression of all genes of the signature. Based on high expression in neuronal tissues, 30% of the genes (21 of 70) down-regulated by MYCN are neuronal tissue-derived mRNAs (Fig. 5B and Fig. S7A), including STMN2 and SNC3A. Of the genes up-regulated by MYCN, only 2 of 87 (2%) were highly expressed in brain tissue (Fig. S7B). These data suggest that MYCN suppresses genes associated with neuronal differentiation. This is in line with the phenotypes observed after shMYCN-mediated MYCN silencing in neuroblastoma cell lines IMR32 and SK-N-BE, which displayed neuronal differentiation 72 h after MYCN silencing (Fig. S1B). This was even more evident by siRNA-mediated MYCN silencing in IMR32 (Fig. S1C).
The genes in the MYCN-157 signature show that activation of cell cycle and DNA repair genes and silencing of neuronal differentiation genes are essential for MYCN function in neuroblastoma.
Discussion
In this study, we identified a MYCN-157 gene signature by integration of in vitro regulation by MYCN in MYCN-amplified neuroblastoma cells and in vivo correlation.
Clustering of tumors based on this signature was highly predictive for prognosis, even in a large neuroblastoma series analyzed on a different platform (11) (Fig. 2). The signature identified a poor-prognosis group including MYCN-amplified tumors as well as MYCN single-copy tumors. The latter group was shown to have low MYCN mRNA expression but increased MYCN protein expression. The MYCN-157 signature therefore recognizes a new type of neuroblastoma marked by stabilization of MYCN at the protein level.
Two early studies reported that high MYCN protein levels carry a poor prognostic value, but MYCN mRNA levels or signatures were not evaluated in these studies (6, 17). Much later, MYCN protein stabilization was studied in neuroblastoma cell lines. The MYCN protein was found to be destabilized by phosphorylation by GSK3β (7, 8). As GSK3β in turn is inactivated by AKT, activation of the PI3K/AKT pathway resulted in stabilization of MYCN in cell lines. Also, FBXW7 and AURKA have been shown to regulate MYCN protein stability (10). Our present study connects these cell line data with neuroblastoma tumor data and shows that a class of neuroblastoma exists with low MYCN mRNA levels but stabilization of MYCN at the protein level and active MYCN signaling, as suggested by a positive MYCN-157 signature. These tumors have a poor prognosis.
Several gene signatures predict outcome in neuroblastoma. Large tumor series were profiled, resulting in prognostic signatures (11, 18–20). Selection based on comparison of favorable and unfavorable tumors also has resulted in different sets of prognostic signatures (19–21). A biological profile was derived from the MYC target database (22). These different neuroblastoma profiling platforms resulted in mainly nonoverlapping but prognostic profiles. Usually, MYCN gene regulation and binding to promoter regions are performed in the SHEP-21N cell line with ectopic expression of MYCN (2, 16, 23). Signatures derived from SHEP-21N are prognostic and can account for survival differences among stages (22–24). Overlap between different profiles is limited. Also, MYCN-157 derived from MYCN silencing in IMR32 resulted in a completely different set of genes. DKC1 is the only gene in multiple prognostic signatures and directly regulated by MYC(N) in cell lines (Dataset S1 and refs. 22 and 23).
Although the overlap between different signatures is very limited, the signatures consistently identify the cell cycle as induced by MYCN and correlated with a poor prognosis. The MYCN-157 signature confirms that MYCN induces cell cycle genes. Recent silencing of MYCN with siRNA (25) or inducible shRNA (4) in MYCN-amplified neuroblastoma cell lines shows that MYCN regulates the cycle via induction of known cell cycle genes. Here we show that, in addition, DNA repair genes are induced by MYCN and overexpressed in neuroblastoma tumors with the MYCN-157-POS signature. The induction of DNA repair genes might add to the aggressive nature of MYCN-amplified tumors and lack of response to therapy. PRMT1 recently was shown to direct the recruitment of RAD51 to DNA damage sites (26). PRMT1-deficient cells were hypersensitive to the DNA-damaging agent etoposide. DNA repair genes might be interesting drug targets in poor-prognosis tumors from the MYCN-157-POS cluster. As pharmacological targeting of MYC and MYCN thus far has been unsuccessful (27), targeting of MYC(N) downstream pathway genes might provide an alternative option. Interestingly, we recently showed that CDK2 is synthetically lethal to MYCN, as silencing of CDK2 was lethal only in neuroblastoma cells with MYCN overexpression (28).
A low differentiation grade is seen as a histological mark for aggressive neuroblastoma (1). Phenotypic silencing of MYCN induces neuronal differentiation [Fig. S1 and (4)]. In addition to known differentiation markers, we identified 21 genes that were highly expressed in normal neural tissues (Fig. 5 and Fig. S7) and down-regulated by MYCN. A biological profile derived from the literature that predicts outcome has been described before (22). Two separate profiles were identified, one based on high MYC transcription targets, the other on low expression of nine differentiation genes. We show that both these processes are under control of MYCN.
In conclusion, we identified a MYCN-157 signature that predicts an extremely poor clinical outcome for a group of neuroblastoma tumors, only half of which have MYCN amplification. Tumors clustering with MYCN-amplified tumors, but without MYCN amplification, have increased nuclear MYCN protein expression due to MYCN protein stabilization.
Materials and Methods
Patient Samples.
The NB88 set contains samples from patients with neuroblastoma of all stages (stage 1, n = 8; stage 2, n = 15; stage 3, n = 13, with 1 MYCN amplification; stage 4, n = 40, with 15 MYCN amplifications; and stage 4s, n = 12). All neuroblastoma samples were derived from primary tumors of untreated patients. Samples with over 80% tumor content were used for profiling (28). mRNA was isolated and analyzed using Affymetrix Human Genome U133 Plus 2.0 arrays and were normalized using MAS5.0 (accession no. GSE16476). Written informed consent was obtained from patients' parents in accordance with review board policies and procedures for research dealing with tumor specimen and clinical information. The medical-ethics committee of the Academic Medical Center (AMC) in Amsterdam approved the study.
Cell Lines and Transduction.
Neuroblastoma cell lines were cultured in DMEM, supplemented with 10% (vol/vol) FCS, 1× nonessential amino acids, 2 mM l-glutamine, 10 units/mL penicillin, and 10 units/mL streptomycin.
Lentiviral particles were made by transfecting HEK293T cells with the lentiviral packaging vectors pMD2.G, pMDLg/pRRE, pRSV-Rev (Addgene), and either the MYCN shRNA vector (TRCN0000020696, Sigma) or control shRNA vector (SHC002, Sigma) with Lipofectamin 2000 (Invitrogen). Lentivirus particles were collected on days 2 and 3 after transfection and filtered through a 0.45-μm filter (Millipore). The viral particles were purified and concentrated by ultracentrifugation. The viral titer was calculated using the p24 ELISA (Cambridge Biosciences), with viral aliquots stored at −80 °C. Cells were transduced with a multiplicity of infection ratio of 3. After 24 h, the medium was replaced for longer time points. The mRNA expression was profiled on U133 Plus 2.0 arrays and analyzed in R2 (accession no. GSE39218).
R2 Description.
A complete description of the bioinformatics tool R2 may be found at http://r2.amc.nl. Datasets used in this study are described in SI Materials and Methods.
Western Blotting.
Primary antibodies used were MYCN (clone B8.4.B, BD Pharmingen), CLU (Santa Cruz Biotechnology), MTAP, PRMT1, and ACTB (Abcam). Western blotting was done according to the manufacturer’s protocol with secondary antibodies linked to IRDyes (Odyssey, Li-Cor Biosciences).
Southern Blot.
Tumor and normal, leukocyte DNA was analyzed by Southern blot as previously described (29). The filters were hybridized with MYC and control probe L.2.3 (29).
ChIP-on-Chip Analysis.
ChIP was performed with MYCN antibody; DNA was purified and amplified (SI Materials and Methods). The samples were labeled, hybridized to the 2.1M Deluxe Promoter Array, and analyzed (NimbleGen). The data were visualized in the R2 program.
Tissue Microarray.
Nuclear MYCN expression was scored on tissue microarray (SI Materials and Methods) by two investigators independently.
Supplementary Material
Acknowledgments
This research was supported by the Tom Voûte Fund and Kinderen Kankervrij (KIKA) Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. W.A.W. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE7307, GSE13136, GSE16237, GSE12460, GSE16476, and GSE39218).
See Commentary on page 19041.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208215109/-/DCSupplemental.
References
- 1.Park JR, Eggert A, Caron H. Neuroblastoma: Biology, prognosis, and treatment. Hematol Oncol Clin North Am. 2010;24(1):65–86. doi: 10.1016/j.hoc.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Lutz W, et al. Conditional expression of N-myc in human neuroblastoma cells increases expression of alpha-prothymosin and ornithine decarboxylase and accelerates progression into S-phase early after mitogenic stimulation of quiescent cells. Oncogene. 1996;13(4):803–812. [PubMed] [Google Scholar]
- 3.Jiang R, Xue S, Jin Z. Stable knockdown of MYCN by lentivirus-based RNAi inhibits human neuroblastoma cells growth in vitro and in vivo. Biochem Biophys Res Commun. 2011;410(2):364–370. doi: 10.1016/j.bbrc.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 4.Henriksen JR, et al. Conditional expression of retrovirally delivered anti-MYCN shRNA as an in vitro model system to study neuronal differentiation in MYCN-amplified neuroblastoma. BMC Dev Biol. 2011;11:1. doi: 10.1186/1471-213X-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell E, et al. MYCN oncoprotein targets and their therapeutic potential. Cancer Lett. 2010;293(2):144–157. doi: 10.1016/j.canlet.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 6.Chan HS, et al. MYCN protein expression as a predictor of neuroblastoma prognosis. Clin Cancer Res. 1997;3(10):1699–1706. [PubMed] [Google Scholar]
- 7.Kenney AM, Widlund HR, Rowitch DH. Hedgehog and PI-3 kinase signaling converge on Nmyc1 to promote cell cycle progression in cerebellar neuronal precursors. Development. 2004;131(1):217–228. doi: 10.1242/dev.00891. [DOI] [PubMed] [Google Scholar]
- 8.Chesler L, et al. Inhibition of phosphatidylinositol 3-kinase destabilizes Mycn protein and blocks malignant progression in neuroblastoma. Cancer Res. 2006;66(16):8139–8146. doi: 10.1158/0008-5472.CAN-05-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segerström L, et al. Effects of small molecule inhibitors of PI3K/Akt/mTOR signaling on neuroblastoma growth in vitro and in vivo. Int J Cancer. 2011;129(12):2958–2965. doi: 10.1002/ijc.26268. [DOI] [PubMed] [Google Scholar]
- 10.Otto T, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15(1):67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Oberthuer A, et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;24(31):5070–5078. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
- 12.Peukert K, et al. An alternative pathway for gene regulation by Myc. EMBO J. 1997;16(18):5672–5686. doi: 10.1093/emboj/16.18.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iraci N, et al. A SP1/MIZ1/MYCN repression complex recruits HDAC1 at the TRKA and p75NTR promoters and affects neuroblastoma malignancy by inhibiting the cell response to NGF. Cancer Res. 2011;71(2):404–412. doi: 10.1158/0008-5472.CAN-10-2627. [DOI] [PubMed] [Google Scholar]
- 14.Schulte JH, et al. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int J Cancer. 2008;122(3):699–704. doi: 10.1002/ijc.23153. [DOI] [PubMed] [Google Scholar]
- 15.Lovén J, et al. MYCN-regulated microRNAs repress estrogen receptor-alpha (ESR1) expression and neuronal differentiation in human neuroblastoma. Proc Natl Acad Sci USA. 2010;107(4):1553–1558. doi: 10.1073/pnas.0913517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koppen A, et al. Direct regulation of the minichromosome maintenance complex by MYCN in neuroblastoma. Eur J Cancer. 2007;43(16):2413–2422. doi: 10.1016/j.ejca.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Hiyama E, Hiyama K, Yokoyama T, Ishii T. Immunohistochemical analysis of N-myc protein expression in neuroblastoma: Correlation with prognosis of patients. J Pediatr Surg. 1991;26(7):838–843. doi: 10.1016/0022-3468(91)90151-i. [DOI] [PubMed] [Google Scholar]
- 18.Schramm A, et al. Prediction of clinical outcome and biological characterization of neuroblastoma by expression profiling. Oncogene. 2005;24(53):7902–7912. doi: 10.1038/sj.onc.1208936. [DOI] [PubMed] [Google Scholar]
- 19.Ohira M, et al. Expression profiling using a tumor-specific cDNA microarray predicts the prognosis of intermediate risk neuroblastomas. Cancer Cell. 2005;7(4):337–350. doi: 10.1016/j.ccr.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 20.Warnat P, et al. Cross-study analysis of gene expression data for intermediate neuroblastoma identifies two biological subtypes. BMC Cancer. 2007;7:89. doi: 10.1186/1471-2407-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberthuer A, et al. Classification of neuroblastoma patients by published gene-expression markers reveals a low sensitivity for unfavorable courses of MYCN non-amplified disease. Cancer Lett. 2007;250(2):250–267. doi: 10.1016/j.canlet.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 22.Fredlund E, Ringnér M, Maris JM, Påhlman S. High Myc pathway activity and low stage of neuronal differentiation associate with poor outcome in neuroblastoma. Proc Natl Acad Sci USA. 2008;105(37):14094–14099. doi: 10.1073/pnas.0804455105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westermann F, et al. Distinct transcriptional MYCN/c-MYC activities are associated with spontaneous regression or malignant progression in neuroblastomas. Genome Biol. 2008;9(10):R150. doi: 10.1186/gb-2008-9-10-r150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vermeulen J, et al. Predicting outcomes for children with neuroblastoma using a multigene-expression signature: a retrospective SIOPEN/COG/GPOH study. Lancet Oncol. 2009;10(7):663–671. doi: 10.1016/S1470-2045(09)70154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo CW, et al. Use of RNA interference to elucidate the effect of MYCN on cell cycle in neuroblastoma. Pediatr Blood Cancer. 2008;50(2):208–212. doi: 10.1002/pbc.21195. [DOI] [PubMed] [Google Scholar]
- 26.Yu Z, Chen T, Hébert J, Li E, Richard S. A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation. Mol Cell Biol. 2009;29(11):2982–2996. doi: 10.1128/MCB.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clausen DM, et al. In vitro cytotoxicity and in vivo efficacy, pharmacokinetics and metabolism of 10074-G5, a novel small-molecule inhibitor of c-Myc/Max dimerization. J Pharmacol Exp Ther. 2010;335(3):715–727. doi: 10.1124/jpet.110.170555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molenaar JJ, et al. Inactivation of CDK2 is synthetically lethal to MYCN over-expressing cancer cells. Proc Natl Acad Sci USA. 2009;106(31):12968–12973. doi: 10.1073/pnas.0901418106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caron H, et al. Allelic loss of chromosome 1p36 in neuroblastoma is of preferential maternal origin and correlates with N-myc amplification. Nat Genet. 1993;4(2):187–190. doi: 10.1038/ng0693-187. [DOI] [PubMed] [Google Scholar]
- 30.Roth RB, et al. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics. 2006;7(2):67–80. doi: 10.1007/s10048-006-0032-6. [DOI] [PubMed] [Google Scholar]
- 31.Łastowska M, et al. Identification of candidate genes involved in neuroblastoma progression by combining genomic and expression microarrays with survival data. Oncogene. 2007;26(53):7432–7444. doi: 10.1038/sj.onc.1210552. [DOI] [PubMed] [Google Scholar]
- 32.Fix A, et al. Characterization of amplicons in neuroblastoma: High-resolution mapping using DNA microarrays, relationship with outcome, and identification of overexpressed genes. Genes Chromosomes Cancer. 2008;47(10):819–834. doi: 10.1002/gcc.20583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.