Abstract
Marine viruses are major evolutionary and biogeochemical drivers in marine microbial foodwebs. However, an in-depth understanding of the cellular mechanisms and the signal transduction pathways mediating host–virus interactions during natural bloom dynamics has remained elusive. We used field-based mesocosms to examine the “arms race” between natural populations of the coccolithophore Emiliania huxleyi and its double-stranded DNA-containing coccolithoviruses (EhVs). Specifically, we examined the dynamics of EhV infection and its regulation of cell fate over the course of bloom development and demise using a diverse suite of molecular tools and in situ fluorescent staining to target different levels of subcellular resolution. We demonstrate the concomitant induction of reactive oxygen species, caspase-specific activity, metacaspase expression, and programmed cell death in response to the accumulation of virus-derived glycosphingolipids upon infection of natural E. huxleyi populations. These subcellular responses to viral infection simultaneously resulted in the enhanced production of transparent exopolymer particles, which can facilitate aggregation and stimulate carbon flux. Our results not only corroborate the critical role for glycosphingolipids and programmed cell death in regulating E. huxleyi–EhV interactions, but also elucidate promising molecular biomarkers and lipid-based proxies for phytoplankton host–virus interactions in natural systems.
Phytoplankton are the basis of marine foodwebs and are responsible for nearly half the global primary production (1). Although they grow rapidly and form massive blooms in ocean surface waters, phytoplankton cell fate is regulated by a suite of abiotic (e.g., nutrients and light availability) and biotic (e.g., grazers and viruses) interactions. The coccolithophore Emiliania huxleyi (Prymnesiophyceae, haptophyte) is a cosmopolitan unicellular photoautotroph that plays a prominent role in the marine carbon cycle. Its intricate calcite coccoliths account for ∼1/3 of the total marine CaCO3 production (2), and it is a key producer of dimethylsulfide, a bioactive gas that plays a significant role in climate regulation by enhancing cloud formation (3).
E. huxleyi forms massive annual spring blooms in the North Atlantic that have been shown to be routinely terminated by lytic, giant, double-stranded, DNA-containing (dsDNA) coccolithoviruses (EhVs) (4, 5). Viruses are the most abundant biological entities in aquatic environments and turn over more than a quarter of the photosynthetically fixed carbon, thereby fueling microbial foodwebs and short-circuiting carbon transfer to higher trophic levels and export to the deep sea (6, 7). However, very little is known about the molecular mechanisms mediating phytoplankton host–virus interactions and the associated regulation of cell fate. Consequently, we lack cellular biomarkers to constrain and quantify active viral infection, and this hinders our understanding of the role of viruses and virus-mediated processes in the oceans.
Coccolithoviruses use a sophisticated coevolutionary “arms race,” centered on control of the programmed cell death (PCD) machinery, to mediate host–virus interactions. Lytic viral infection of E. huxleyi not only activates the PCD biochemical machinery, including metacaspase expression and caspase catalytic activity, but also actively recruits and requires them for successful viral replication (8). It is reminiscent of the “Red Queen” dynamic (9) that drives plant–pathogen and animal–pathogen interactions (10, 11) and is facilitated by lateral gene transfer between host and virus (12, 13). Genome analysis of E. huxleyi virus 86 (EhV86), the type strain for the Coccolithoviruses, revealed an unexpected suite of putative sphingolipid biosynthetic genes (14), a pathway never before described in a viral genome and one that ostensibly derived from the host’s sphingolipid biosynthetic pathway (13). The EhV86 genome encodes an active serine palmitoyltransferase (SPT) (15), which initiates and rate-limits de novo sphingolipid biosynthesis (16), ultimately leading to ceramide production, a potent inducer of PCD in animals and plants (17, 18). In turn, these ceramides are glycosylated, via yet-unknown mechanisms that are presumably derived from the host (19), to yield viral glycosphingolipids (vGSLs). Importantly, vGSLs critically regulate host–virus interactions by inducing host PCD in noninfected cells and facilitating viral production (20), potentially serving as a promising diagnostic biomarker for viral infection.
The recent discoveries of virus-induced PCD (8) and its lipid regulation (20) have yielded unprecedented mechanistic insight into this coevolutionary “arms race” between cultured strains of E. huxleyi and EhVs, but it is currently unknown whether these pathways regulate natural host–virus interactions among diverse E. huxleyi-EhV genotypes in the modern ocean. Likewise, although physiological and biochemical experiments and genome-wide in silico analyses have collectively shown that an operational PCD molecular machinery was established early in the evolution of unicellular eukaryotic phytoplankton (21), the incidence and ecological relevance of PCD remain unknown in natural populations. Here, we interrogate host–virus interactions during a bloom and subsequent demise of natural coccolithophore and coccolithovirus assemblages using a specific suite of molecular, biochemical, and physiological stress and PCD biomarkers. Our analysis reveals a functional conservation of subcellular and molecular responses and modes of mortality in the face of considerable host–virus microdiversity, which regulate cell fate and can influence carbon export. We present an interrogation of natural phytoplankton host–virus interactions at subcellular levels of resolution and demonstrate PCD activation in natural populations. Our work provides a foundation for using diverse and diagnostic cellular markers to quantify and constrain the influence of viral infection in the sea.
Results and Discussion
We used mesocosm enclosures, deployed in Norwegian coastal waters, as an experimental platform to test our recent laboratory-based findings of a lipid-based and PCD-mediated arms race (22) on natural populations of E. huxleyi and EhVs. We focused specifically on the subcellular dynamics of E. huxleyi-EhV over the course of a bloom using a diverse suite of molecular tools that target different cellular processes. Mesocosms are an effective way to address fundamental questions around in situ host–virus dynamics of E. huxleyi and its associated dsDNA viruses (4, 23, 24) because they mimic the conditions of natural bloom succession with native host and viral genotypes. Annual spring E. huxleyi blooms occur in these Norwegian coastal waters (4) so the enclosures contained the representative pool of genotypes and biotic interactions.
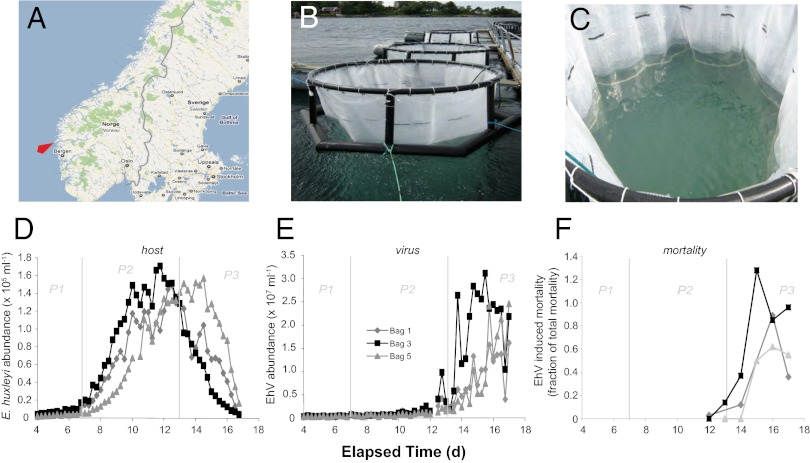
E. huxleyi bloom dynamics consisted of three distinct phases: phase 1 (days 2–7), characterized by a lagged phase of low E. huxleyi abundance; phase 2 (days 7–13), characterized by exponential growth of the E. huxleyi population (peaking at 1.7 × 105 cells/mL in bag 3); and phase 3 (days 13–17), representing the concomitant demise of E. huxleyi host cells and rise in EhV populations. The pattern of host–virus dynamics was generally similar in the three triplicate mesocosm enclosures, with notable differences in the timing of bloom initiation and infection (Fig. 1 D–F), perhaps indicative of subtle differences in the inoculum populations. Rapid cell loss was observed in all three enclosures from days 13–15 (phase 3) along with a concurrent exponential increase in EhV concentrations, indicative of virus-induced bloom termination. On the basis of flow cytometry data, the numerical contribution of other phytoplankton, such as nanoeukaryotes and Synechococcus, was relatively minor. E. huxleyi numerically dominated the total phytoplankton population up to the end of the mesocosm experiment, comprising an average of 74% and 66% across all three mesocosms during bloom (phase 2; days 7–13) and demise (phase 3; days 13–17) respectively. Signs of community succession started to appear at the end of the experiment, but, even on day 16, E. huxleyi still accounted for 48% of the total population, with the next largest contribution coming from Synechococcus at 21%.
Fig. 1.
Location, setup, and coccolithophore/coccolithovirus bloom dynamics for the 2008 mesocosm field experiment in Norway. (A) Map showing the location of the Espeland Marine Biological Station (University of Bergen, Espelandsveien 232 NO-5258 Blomsterdalen, Norway: designated by red arrow). (B and C) Photos of the mesocosm setup whereby replicate (×3) bags were filled with surrounding fjord water and used as community inoculate for daily spikes of nitrate and phosphate at Redfield ratios to induce a bloom. (D) Dynamics of host E. huxleyi and (E) EhV abundance in the three replicate bags over an ∼2-wk time course. (F) EhV-induced mortality, taken as the number of E. huxleyi cells killed by EhV, was calculated using EhV concentration, EhV production, and published viral decay rates and burst sizes based on equations from ref. 4. Note that the largest viral impact was observed for bag 3.
To quantitatively attribute the observed host cell losses to EhV infection, we estimated the fraction of virus-induced mortality (VIM) using EhV concentration and EhV production as input data, along with published viral decay rates (0.4 d−1) and viral burst sizes (500) from similar E. huxleyi blooms (4, 25). VIM reached maxima values on days 15–16 in all bags ranging between 62–128% (Fig. 1F). VIM in bag 5 was notably lower and more stable (50–62%) over this period, with delayed host demise and viral production, perhaps due to different biological interactions affecting mortality, such as release of antiviral infochemicals (26), selective grazing on viral-infected cells (27), or the development of a resistant haploid subpopulation (28). Conversely, VIM in bags 1 and 3 showed a dramatic increase on days 15–16, with the latter slightly exceeding 100% total net mortality (128%), perhaps due to underestimates of cell mortality in the face of simultaneous E. huxleyi growth (4). Given that maximum EhV abundance in these bags occurred on day 15 (Fig. 1E), followed by a slow decline in EhV particles, we posit that our VIM calculations accurately reflect the infection dynamics. The consistent pattern of tight regulation of E. huxleyi bloom dynamics by EhV motivated us to “zoom” into the molecular nature of these host–virus interactions. Due to the notably different timing and scale of host–virus dynamics between the replicate mesocosms, we treated them independently for subsequent analyses.
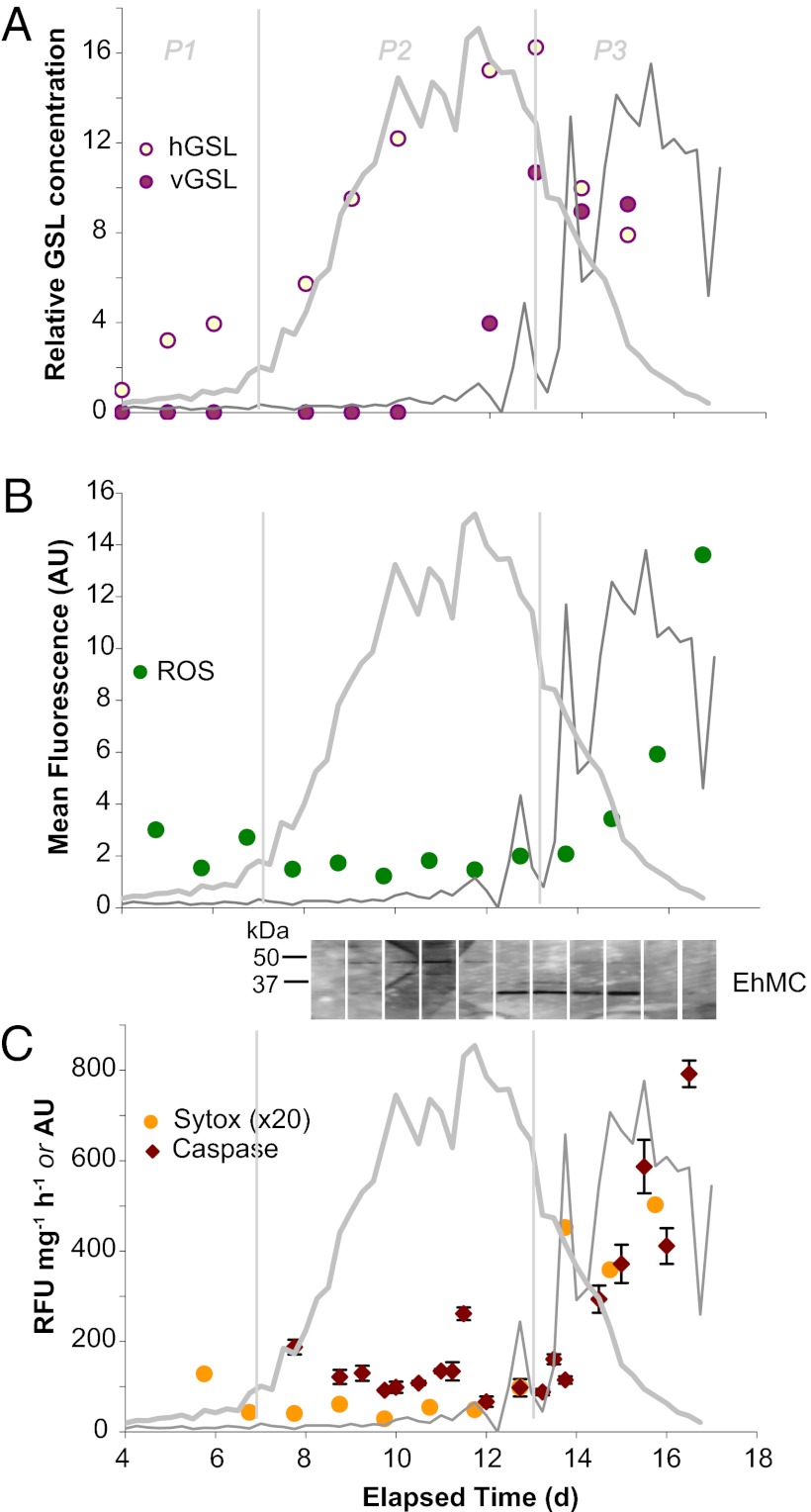
A unique class of polar lipid molecules, glycosphingolipids, are central to the interactions between E. huxleyi and its EhV viruses in culture (20, 22) and may critically “lubricate” viral infection in natural systems. Indeed, host-specific glycosphingolipids (hGSLs), consisting of palmitoyl-based glycosphingolipids (29), and viral glycosphingolipids (vGSLs), consisting of myristoyl-based glycosphingolipids, play important functional roles in the infection of E. huxleyi (20). Using high-performance liquid chromatography/electrospray ionization mass spectrometry (HPLC/ESI-MS) (30), we examined the abundances and chemical compositions of both hGSLs and vGSLs associated with resident E. huxleyi and EhV populations over the course of the mesocosm bloom and demise. The ability to sample at high time resolution and correlate our molecular data with a diverse array of biological and ecological parameters allowed us to substantiate our laboratory-based findings with an extensive dataset and put these lipids into a unique ecological context. In our mesocosms, we detected hGSLs, similar to those seen in healthy, uninfected E. huxleyi cultures and in the North Atlantic (20), in all three triplicate enclosures throughout the bloom, with the relative hGSL content clearly tracing E. huxleyi cell abundances (Fig. 2A; Figs. S1 and S2) and, thereby, serving as a specific proxy for healthy coccolithophore populations. The dominant hGSL species consisted of a C22:2 hydroxy fatty acid and a peculiar d19:3 long-chain base (Fig. S3), which was also observed in our laboratory-based culture experiments. The occurrence of such an unusual sphingoid base has also been described in sphingomyelin from squid nerve and starfish spermatozoa (31, 32), suggesting that these conjugated polyunsaturated, branched sphingoid bases play an important role in glycosphingolipid signaling in the marine environment.
Fig. 2.
Cellular response of natural E. huxleyi populations to EhV infection for replicate mesocosm bag 3. The traces of host and viral abundances are provided as thick gray and thin gray lines, respectively. Time courses of the following are given: (A) hGSLs and vGSLs, provided as the concentration relative to hGSL at 4 d; the analytical error of vGSL measurements is ∼10% (20), and thus the observed increases in glycosphingolipids during infection are distinct from background levels. (B) Levels of cellular ROS, detected by in vivo fluorescent staining with CM-H2DCFDA and flow cytometry. (C) Caspase-specific activity (RFU mg−1⋅h−1) and cell death (SYTOX staining; arbitrary units). (Inset) Metacaspase protein expression via immunoreactivity to polyclonal EhMC antisera; all lanes were run on the same gel, had identical exposure times, and are positioned to correspond with elapsed time on the x-axis. Bloom phases are indicated by thin vertical gray lines (P1, prebloom; P2, bloom; P3, crash/demise). RFU, relative fluorescence units.
The unique vGSL chemical signatures were also consistently detected within the triplicate mesocosms (Fig. 2A; Figs. S1 and S2). Remarkably, as with the hGSLs, the distribution of vGSLs from these native populations of E. huxleyi and EhV genotypes in the mesocosms mirrored the major vGSLs detected in laboratory-based cultures (20). Initial levels of vGSL were very low with induction on day 12 corresponding with the onset of E. huxleyi cell demise and tracing increases in both EhV abundance and VIM. Given the functional role of vGSLs in regulating host–virus interactions through pronounced increases in host E. huxelyi cells during the onset of lytic phase (20), the observed vGSL induction can be attributed to the accumulation of infected cells before bloom demise. Furthermore, given that vGSLs are an integral component of virion membranes (20), our dataset substantiates the possibility that vGSLs can serve as a lipid-based marker to trace EhVs and their subsequent fate during natural bloom succession in contemporary oceans, as well as in the distant past through preservation in ancient marine sediments (22, 33).
Collectively, our detection of naturally occurring hGSL and vGSL chemical signatures corroborates the expression analysis of the two host and viral genes involved in the de novo sphingolipid biosynthetic pathway in the very same E. huxleyi mesocosm populations (34). SPT, the first gene in the pathway that rate-limits de novo sphingolipid production, and dihydroceramide desaturase (DCD), the last gene in the pathway that produces ceramide, are present in both host and viral genomes (13, 14). Viral SPT and DCD genes were strongly up-regulated (by one to two orders of magnitude) during the virus-induced bloom demise (phase 3), whereas the corresponding host gene homologs were suppressed (34).
Elevated production of intra- and extracellular reactive oxygen species (ROS) during late lytic phase of infection of E. huxleyi cultures (35) suggests that virus-induced cell death is mediated by vGSL-induced intracellular ROS production (22). ROS generation is a widely known cellular response to abiotic and biotic stress, which can induce PCD (36). We used the fluorescent probe 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate (CM-H2DCFDA) coupled with analytical flow cytometry to monitor ROS generation associated with host–virus interactions over the course of “bloom and bust” cycles in natural E. huxleyi communities. ROS clearly accumulated in E. huxleyi cells during bloom demise and concomitant increases in EhV abundance (phase 3; from day 14 onward); the mean fluorescence per E. huxleyi cell increased by a factor of 5–6 in enclosure 3 over this time period (Fig. 2B), with similar increases observed in bags 1 and 5 (Figs. S1B and S2B). Notably, ROS staining was associated with the accumulation of EhVs, which is consistent with late-lytic-phase ROS production rather than with the early stages of infection. Similar dynamics were reported for infected E. huxleyi cultures, whereby ROS production was attributed to compromised photophysiology (35).
ROS production may be required for viral replication in E. huxleyi by triggering metacaspase expression and caspase activity. Bidle and coworkers (8) demonstrated late-phase induction and active recruitment of host caspase activity by coccolithoviruses as part of their replication strategy; a subset of virally encoded proteins possessed internal caspase cleavage recognition sequences. We examined caspase-specific activity in cell extracts of natural mesocosm populations through the in vitro cleavage of the canonical fluorogenic tetrapeptide substrate (IETD-AFC) (Fig. 2C). Induction of caspase-specific activity increased significantly with the onset of phase 3 (from day 14 onward), tracking viral productivity and host cell death, as indicated by the increased staining with SYTOX Green, a membrane integrity DNA-binding fluorescent stain (Fig. 2C). Given that measurements of in vitro activity are derived from protein extracts encompassing all captured cells in the mixed microbial population, in addition to healthy and dying E. huxleyi microdiversity, the true catalytic activity of these enzymes during viral demise is likely underestimated.
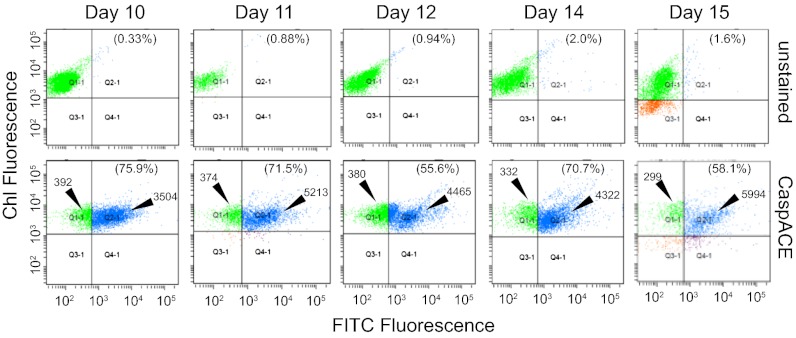
Cellular caspase activation within the E. huxleyi population was independently assessed by direct staining with a fluorescein isothiocyanate (FITC) conjugate of the broad-spectrum caspase inhibitor, z-Val-Ala-Asp-fluoromethyl-ketone (z-VAD-FMK), and the flow cytometry VAD-FMK-FITC (CaspACE) freely diffuses into cells and irreversibly binds to activated caspases, thus serving as an in vivo marker. Plots of chlorophyll fluorescence versus FITC fluorescence (Fig. 3) confirmed high percentages of CaspACE-positive E. huxleyi subpopulations during the viral-induced demise phase (days 10–15): 55.6–75.9% of E. huxleyi cells were positively stained (Fig. 3; quadrant Q2-1) with 9- to 20-fold higher FITC fluorescence per cell compared with unstained cells (Fig. 3; quadrant Q1-1). This corresponded remarkably well with ∼63% CaspACE-positive cells observed during late-phase EhV infection of E. huxleyi cultures compared with ∼3% at the time of infection (8). Notably, vGSL production corresponded with induction of caspase activity, suggesting that it triggered the PCD biochemical cascade. These observations are consistent with findings that exogenous application of purified vGSL to uninfected E. huxleyi cells in a dose-dependent manner triggered cellular caspase activity and PCD, biomimicking viral infection (20).
Fig. 3.
Cellular caspase activation in natural E. huxleyi populations of bag 3 during EhV1 infection in phase 3 of bloom demise as determined by CaspACE (FITC-VAD-FMK) staining and flow cytometry analysis. Plots of chlorophyll fluorescence and FITC fluorescence distribution for unstained and CaspACE-stained E. huxleyi cell populations are given for each designated sampling day (5,000 cells counted). Quadrant plots Q1-1 and Q2-1 correspond to negatively and positively stained E. huxleyi cells, respectively. The percentage of positively stained cells (Q2-1 quadrant) is provided in parentheses. The mean FITC fluorescence value is also indicated for negatively and positively stained CaspACE-stained cells.
We also assessed metacaspase expression with Western blot analysis using a polyclonal antibody raised against a purified E. huxleyi metacaspase (protein ID 440238). We detected immunoreactive proteins (Fig. 2C, Inset) of ∼36 and ∼42 kDa in natural E. huxleyi populations of all three enclosures during the course of bloom development, which showed remarkable similarities to EhV infection of E. huxleyi cultures (8). These protein sizes correspond with the predicted molecular weights of several putatively annotated metacaspase proteins (36.8, 40.6, 41.1, and 45.2 kDa) in the Ehux1516 genome (http://genome.jgi-psf.org/Emihu1/Emihu1.home.html), including the putative size for the aforementioned protein ID 440238 (36.8 kDa). In bags 3 and 5, we observed strong up-regulation of the 36-kDa metacaspase protein at the onset of bloom demise and EhV production (day 12). Similar dominance of these two metacaspase proteins was observed in cell extracts from bag 1 (Fig. S1), but here the induced expression of the 42-KDa protein best correlated with the onset of viral infection. Importantly, the timing and shift in metacaspase expression during host–virus dynamics corresponded with the induction of other stress and PCD cellular markers, supporting previous findings that metacaspase expression is linked to host-cell turnover and viral replication. Metacaspase expression has been shown to be diagnostic of successful viral infection of cultured sensitive and resistant E. huxleyi strains (8, 37), although they are likely not responsible for the observed caspase activities, given their altered substrate requirements (38). Our research demonstrates in vivo caspase activation and induced metacaspase expression during natural phytoplankton bloom dynamics. We propose that they can be used as diagnostic biochemical markers to assess in situ physiological stress and PCD in field populations.
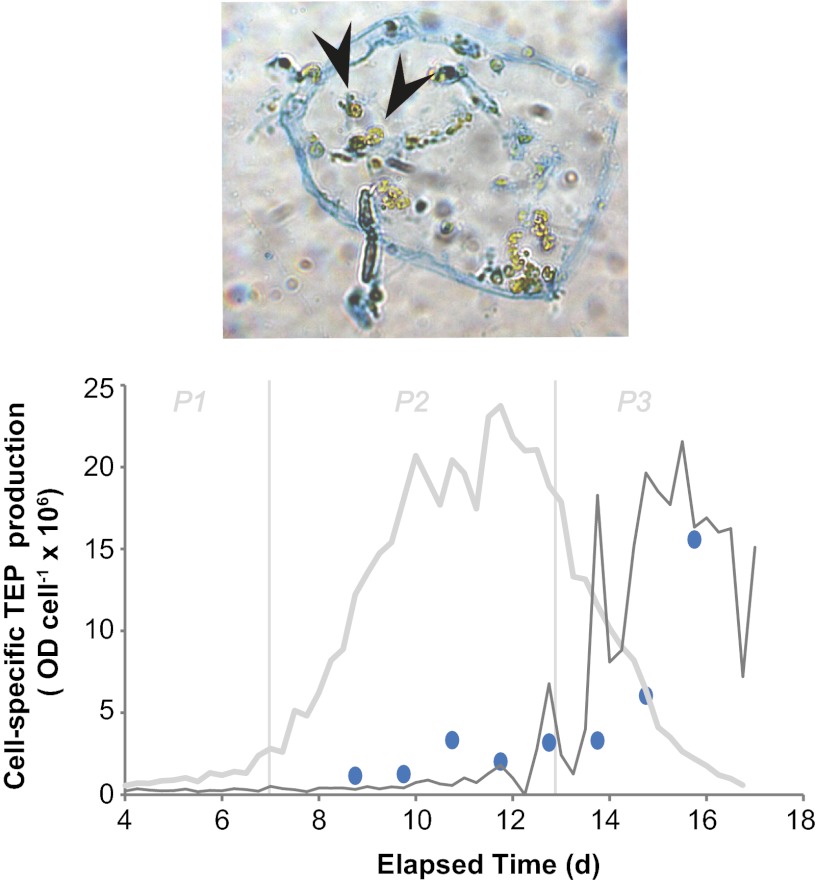
Under physiological stress, algal cells secrete dissolved polysaccharides (39), which can accumulate and catalyze the formation of transparent exopolymer particles (TEP). Due to its surface-reactive nature, TEP production, in turn, facilitates the formation of large sinking aggregates (i.e., marine snow) and thereby serves as a mechanism to accelerate carbon export to deeper water (40). Our observations of virus-induced subcellular stress and PCD markers in natural E. huxleyi blooms motivated us to explore its mechanistic link to TEP production in diatoms and cyanobacteria. There is a documented mechanistic coupling between impaired photosynthetic efficiency, oxidative stress, and caspase activation in phytoplankton with TEP production and associated cell–cell adhesion and vertical transport (41, 42). Using the acidic polysaccharide-specific stain alcian blue coupled with microscopy and spectrophotometry, we visualized and quantified TEP production over the course of the E. huxleyi bloom. Cell-specific TEP production increased in bag 3 during virus-induced demise (Fig. 4), closely corresponding with the induction of stress and PCD markers (i.e., caspase activity, ROS formation, and compromised membrane integrity) (Fig. 2). Similar trends were generally observed for bags 1 and 5, although not to the same degree; relative cell-specific TEP production was higher at earlier time points in these bags (Figs. S1 and S2). Microscopic examination of samples exhibited E. huxleyi cells that were encased within larger sheath-like TEP particles (Fig. 4, Upper). Our findings linking virus-induced mortality and TEP production during E. huxleyi bloom termination provide experimental support for previous modeling efforts (43) and may have important implications for the fate of both particulate organic carbon (POC) and particulate inorganic carbon (PIC). The contribution of viral infection to POC/PIC sinking flux is currently unknown, but the use of vGSLs as diagnostic markers of EhV infection now provides a framework to quantitatively address this issue.
Fig. 4.
Time course of cell-specific TEP production during E. huxleyi bloom and demise via EhV infection in mesocosm bag 3. The traces of host and viral abundances are provided as thick gray and thin gray lines, respectively. Bloom phases are indicated by thin vertical gray lines (P1, prebloom; P2, bloom; P3, crash/demise). (Upper) Visualization of TEP by alcian blue staining and light microscopy. Note how E. huxleyi cells can become entangled in TEP particles. Cells can be differentiated by their green chlorophyll (indicated by arrowheads).
Virus-induced TEP production may represent another intriguing “layer” in the chemical arms race between E. huxleyi and coccolithoviruses (22). First, the accumulation of TEP and the associated changes in cell-surface stickiness and aggregation might provide E. huxleyi host cells with a protective mucilage layer against viral absorption and/or viral release. Indeed, ecosystem modeling of Phaeocystis globosa-PgV (another Prymnesiaophyte-Phycodnaviridae host–virus system) interactions during mesocosm experiments demonstrated that viral absorption to TEP, which was derived from disintegration of host colonies, significantly reduced the fraction of infectious viruses and subsequent host infection (44). Second, the enhanced cell densities in aggregates may provide microhabitats favorable to cell–cell chemical signaling whereby effective infochemical threshold concentrations may be reached (45). Purified vGSLs trigger a dose-dependent PCD response in uninfected E. huxleyi cells (20) and function as a bloom termination signal, perhaps as a viral exclusion strategy. Third, entrainment of EhVs in TEP may serve to effectively eliminate them from surface waters. A significant fraction of viral abundance has been observed with TEP, especially in water masses characterized by low renewal rates (46). These ideas need to be substantiated with further research.
The transfer of a vGSL biosynthetic pathway into coccolithoviruses and its role in regulating induction and recruitment of host PCD machinery points to an elegant coevolution of host–virus interactions in the marine environment. The Red Queen hypothesis governing the evolution of host–parasite relationships (9) argues for an intense arms race between host and viral strains, whereby the former evade viral infection through mutations and development of resistance and the latter seek to maximize infectivity (47). At the same time, viral infection of E. huxleyi induces transition to resistant haploid sexual stage cells in a “Cheshire cat” evasion strategy (48). These intense evolutionary selection pressures serve to stimulate genetic diversity and coexistence of host and virus under diverse conditions (49). An important outcome of this study is that the E. huxleyi subcellular response to viral infection (vGSL production, ROS accumulation, caspase activation, TEP production) is collectively conserved in the face of both community-wide genetic diversity and documented transition to haploid phases. Genetic variability was observed both in the calcium-binding protein (GPA) of E. huxleyi hosts and the major capsid protein of infectious EhVs during early and prebloom stages in the present mesocosm experiment (50). Similar diversity has also been documented in previous mesocosm experiments (51) and in a coccolithophore bloom in the North Sea (52). Furthermore, Combined CaCO3 Optical Detection- Fluorescent in situ Hybridization (COD-FISH) along with haploid-specific transcript analysis documented the presence and dynamics of noncalcified haploid E. huxleyi cells in the very same mesocosm enclosures (28). Haploid cell markers peaked during the phase 2 to phase 3 bloom transition, when host cell numbers declined due to viral demise. Haploid-specific gene expression was not detected after day 14, suggestive of syngamy into new diploid genotypes or transit abundance of haploid cells. Nonetheless, the dynamics of the diverse suite of lipid, subcellular stress, and PCD markers that we observed in this study were remarkably similar to those observed in EhV-infected cultures of individual sensitive strains (8, 20). Our findings indicate that these cellular responses are conserved across the natural genetic diversity and ecological backdrop of E. huxleyi and EhV populations and serve to control bloom fate.
Our findings demonstrate that PCD mechanistically mediates algal bloom termination in response to virus-induced infochemicals (e.g., vGSLs), consistent with viral exclusion as an evolutionary driver in unicellular organisms (21) and supportive of an ecological role in aquatic environments. Our results also demonstrate that the respective hGSL and vGSL productions not only critically regulate host–virus interactions, but also can function as unique lipid-based proxies to ecologically diagnose host–virus bloom dynamics and to assess the ecological and biogeochemical roles of coccolithoviruses in the oceans.
Materials and Methods
Mesocosm Setup and Sampling.
Mesocosms were carried out for 17 d (June 5–21, 2008) in Raunefjorden at the University of Bergen’s Marine Biological Station, Norway (60.38° N; 5.28° E). Triplicate experimental enclosures consisted of transparent polyethylene bags (11 m3; 4 m deep and 2 m wide; 90% photosynthetically active radiation) mounted on floating frames and moored to a raft (Fig. 1 B and C) (for details see ref. 53). The source water used for the mesocosm was obtained from the surrounding fjord. Nutrients were added daily to triplicate enclosures at a nitrogen:phosphorous ratio of 15:1 (1.5 µM NaNO3; 0.1 µM KH2PO4), as previously described (25). Mixing was ensured by continuously pumping water from the bottom of each bag to the surface. Surface-water samples were collected daily at 6-h intervals (0600, 1200, 1800, and 2400 h) using 20-L carboys and immediately processed in the laboratory.
Cells were collected by gentle vacuum filtration onto 47-mm diameter, 0.8-µm pore-size, mixed cellulose ester filters (AAWP; Millipore) after prescreening with 100-µm pore-size Nitex mesh to remove microzooplankton grazers. Biomass collected on filters was scraped off with a clean razor blade into a sterile petri dish, resuspended in 0.2 µm of filtered seawater, and transferred to a sterile 1.5-mL microfuge tube. Cells were pelleted via centrifugation (2 min, maximum speed, room temperature), frozen immediately in liquid N2, and stored at –80 °C until processing.
Virus-Induced Mortality.
Viral-induced lysis was quantified by comparing the estimated number of host cells killed by viral lysis to the overall total net cell mortality, as previously described (4). The number of E. huxleyi cells killed by viral lysis is estimated from the number of viruses produced over time (change in EhV abundance mL−1), the burst size (number of EhV released per lysed cell), and the estimated loss of EhV due to viral decay. Assuming no growth of E. huxleyi, the net total mortality of E. huxleyi cells during a defined time period can be estimated as the observed decrease in E. huxleyi abundance (cells per mL). Virus-induced mortality rates of E. huxleyi were estimated for the days of the experiment where a decrease in E. huxleyi cell abundance coincided with an increase in the abundance of EhV.
Measurement of Caspase Activity and Metacaspase Expression.
Frozen cell pellets were resuspended in caspase activity buffer and sonicated on ice, and cellular debris was pelleted by centrifugation as previously described (8). Cell extracts were incubated in triplicate with the canonical caspase-8 fluorogenic peptide substrate (Ile-Glu-Trp-Asp-7-amido-4-trifluoromethylcoumarin and IETD-AFC) (Calbiochem) at a final concentration of 50 µM. Kinetic analysis of substrate cleavage was performed over a 4-h period at 25 °C with measurements taken at 10-min intervals using a Spectra Max Gemini XS plate reader (excitation 400 nm, emission 505 nm; Molecular Devices) and the SoftMax Pro-3.1.1 analysis program. Cleavage rates were normalized to protein, as determined by the BCA Protein Assay Kit (Pierce).
In vivo caspase activity was assessed in E. huxleyi cells by direct staining with VAD-FMK-FITC (CaspACE; Promega). Cells were pelleted by centrifugation, washed once with PBS (pH 7.5), and resuspended in PBS before the addition of VAD-FMK-FITC (final concentration, 20 μM). Cells were stained for 20 min at 18 °C in the dark, after which they were pelleted by centrifugation, washed once with PBS, fixed with 2% (vol/vol) formalin/PBS, and stored at 4 °C until analyzed. Five thousand E. huxleyi cells were analyzed for the percentage of positively stained cells and the level of intensity of the staining at 520 nm using a FACSAria flow cytometer (BD Biosciences) equipped with an Octagon (488-nm) blue laser with standard filter set up. E. huxleyi cells were discriminated from other phytoplankton species on the basis of dot plots of side-scatter signal versus red (chlorophyll a) fluorescence signal (695/40).
Western Blot Analysis.
Frozen pellets were resuspended in PBS containing protease inhibitor mixture (Sigma P2714), sonicated (as described above), and centrifuged (14,000 × g for 5 min at 4 °C). Supernatants containing equal amounts of protein were loaded onto 12% (wt/vol) Criterion Tris⋅HCl polyacrylamide gels (Bio-Rad), subjected to SDS/PAGE (200 V; 1.5 h) and transferred onto PVDF membranes (100 V; 1 h). Membranes were probed using a polyclonal antisera raised against a purified, recombinant E. huxleyi metacaspase (EhMC) protein (titer = 1:500) as previously described (8), followed by polyclonal goat anti-rabbit IgG-HRP (Pierce; titer = 1:10,000). A horseradish peroxidase chemiluminescence system was used for detection (SuperSignal; Pierce) by exposing BioMax XAR film (Kodak) for 10–30 s.
Cell Death and ROS Production.
Cell death was also determined by assaying the cell membrane integrity with SYTOX Green (Invitrogen). After 15 min of incubation with 1 µM SYTOX in the dark, cells were quantified by analysis of fresh samples on a FacScan flow cytometer (Becton Dickinson) equipped with a 15-mW laser exciting at 488 nm. Samples were analyzed at a high flow rate (∼70 μL min–1) with 500–8,000 E. huxleyi cells being counted for the analysis depending on the stage of the bloom. Intracellular ROS was measured by the fluorescent probe 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate (CM-H2DCFDA; Invitrogen; Ex 488 nm, Em 522 nm). The CM-H2DCFDA probe was stored in 50-μg aliquots at −20 °C until use. Fresh 1-mM stock solutions were made up before use by dilution in ethanol, and a final CM-H2DCFDA concentration of 5 μM was used (35). The average per cell green fluorescence of the E. huxleyi population was used as a relative measure of ROS compared with the control population.
Lipid Analysis.
Five-liter water samples were collected and filtered on precombusted GF/F filters, which were snap-frozen in liquid nitrogen and stored at −80 °C until processed. Lipids were extracted using a modified Bligh–Dyer method, as previously described (20). Cellular polar membrane lipids were analyzed by HPLC/ESI-MS as described (54) using an Agilent 1100 HPLC and Thermo Finnigan LCQ Deca XP ion-trap mass spectrometer. Authentic glycosphingolipid standards (brain cerebrosides; Avanti Polar Lipids) were used for initial identification of retention times and to identify characteristic MS2 and MS3 fragmentation. These authentic standards were also used to construct standard curves for quantification. A subset of the samples was analyzed using identical HPLC and ESI conditions on a Thermo FTQ high-resolution Fourier-transform ion cyclotron resonance mass spectrometer for confirmation of elemental formulas in glycosphingolipid molecular ions and MS2 fragment ions.
TEP Measurements
Relative changes in TEPs were determined by using a spectrophotometric alcian blue staining technique (55). Seawater samples were filtered on 0.4-µm polycarbonate filters, stained with a 0.02% (wt/vol) alcian blue solution, rinsed with distilled water, and immediately frozen. Triplicate filters were processed for each water sample. Filters were soaked in 80% (vol/vol) H2SO4 for 2 h before measuring the absorbance at 787 nm. The filters were also prepared for light microscopy using a transfer-freeze technique whereby freshly stained filters were applied to a frozen microscope slide and peeled off. Intact TEP particles remaining on the slide were promptly fixed with gelatin. Slides were stored at 4 °C.
Supplementary Material
Acknowledgments
We thank the mesocosm organizers W. Wilson (Bigelow Laboratory for Ocean Sciences, East Boothbay, ME), M. Allen (Plymouth Marine Laboratory, Plymouth, UK), and A. Aadnesen (Marine Biological Station, Blomsterdalen, Norway), along with other international participants for assistance in sampling and fruitful discussions This research was supported by National Science Foundation Grants IOS-0717494 and OCE-1061883 (to K.D.B.) and OCE-1061883 and OCE-1031143 (to B.A.S.V.M.); by Office of Naval Research Grants N000140910091 (to B.A.S.V.M.); by grants from the Legacy Heritage Fund (Israel Science Foundation) and Edith and Nathan Goldenberg Career Development Chair to A.V.; by a Norwegian Research Council Grant [“Viral lysis and programmed cell death in marine phytoplankton” (VIPMAP)] 186142 (to A.L.); and by resources from the Natural Environment Research Council to organizers W. Wilson and M. Allen.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208895109/-/DCSupplemental.
References
- 1.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science. 1998;281(5374):237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 2.Iglesias-Rodriguez MD, et al. Representing key phytoplankton functional groups in ocean carbon cycle models: Coccolithophorids. Global Biogeochem Cycles. 2002;16(4):1100–1120. [Google Scholar]
- 3.Charlson RJ, Lovelock JE, Andreae MO, Warren SG. Oceanic phytoplankton, atmospheric sulfur, cloud albedo and climate. Nature. 1987;326:655–661. [Google Scholar]
- 4.Bratbak G, Egge JK, Heldal M. Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar Ecol Prog Ser. 1993;93:39–48. [Google Scholar]
- 5.Schroeder DC, Oke J, Malin G, Wilson WH. Coccolithovirus (Phycodnaviridae): Characterisation of a new large dsDNA algal virus that infects Emiliana huxleyi. Arch Virol. 2002;147(9):1685–1698. doi: 10.1007/s00705-002-0841-3. [DOI] [PubMed] [Google Scholar]
- 6.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399(6736):541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 7.Suttle CA. Marine viruses: Major players in the global ecosystem. Nat Rev Microbiol. 2007;5(10):801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 8.Bidle KD, Haramaty L, Barcelos E, Ramos J, Falkowski PG. Viral activation and recruitment of metacaspases in the unicellular coccolithophore, Emiliania huxleyi. Proc Natl Acad Sci USA. 2007;104(14):6049–6054. doi: 10.1073/pnas.0701240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.VanValen L. A new evolutionary law. Evol Theory. 1973;1:1–30. [Google Scholar]
- 10.Bergelson J, Kreitman M, Stahl EA, Tian D. Evolutionary dynamics of plant R-genes. Science. 2001;292(5525):2281–2285. doi: 10.1126/science.1061337. [DOI] [PubMed] [Google Scholar]
- 11.Staskawicz BJ, Mudgett MB, Dangl JL, Galan JE. Common and contrasting themes of plant and animal diseases. Science. 2001;292(5525):2285–2289. doi: 10.1126/science.1062013. [DOI] [PubMed] [Google Scholar]
- 12.Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. 2005;438(7064):86–89. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]
- 13.Monier A, et al. Horizontal gene transfer of an entire metabolic pathway between a eukaryotic alga and its DNA virus. Genome Res. 2009;19(8):1441–1449. doi: 10.1101/gr.091686.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson WH, et al. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science. 2005;309(5737):1090–1092. doi: 10.1126/science.1113109. [DOI] [PubMed] [Google Scholar]
- 15.Han G, et al. Expression of a novel marine viral single-chain serine palmitoyltransferase and construction of yeast and mammalian single-chain chimera. J Biol Chem. 2006;281(52):39935–39942. doi: 10.1074/jbc.M609365200. [DOI] [PubMed] [Google Scholar]
- 16.Hanada K. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim Biophys Acta. 2003;1632(1–3):16–30. doi: 10.1016/s1388-1981(03)00059-3. [DOI] [PubMed] [Google Scholar]
- 17.Hannun YA, Obeid LM. Ceramide: An intracellular signal for apoptosis. Trends Biochem Sci. 1995;20(2):73–77. doi: 10.1016/s0968-0004(00)88961-6. [DOI] [PubMed] [Google Scholar]
- 18.Liang H, et al. Ceramides modulate programmed cell death in plants. Genes Dev. 2003;17(21):2636–2641. doi: 10.1101/gad.1140503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michaelson LV, Dunn TM, Napier JA. Viral trans-dominant manipulation of algal sphingolipids. Trends Plant Sci. 2010;15(12):651–655. doi: 10.1016/j.tplants.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Vardi A, et al. Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton. Science. 2009;326(5954):861–865. doi: 10.1126/science.1177322. [DOI] [PubMed] [Google Scholar]
- 21.Bidle KD, Falkowski PG. Cell death in planktonic, photosynthetic microorganisms. Nat Rev Microbiol. 2004;2(8):643–655. doi: 10.1038/nrmicro956. [DOI] [PubMed] [Google Scholar]
- 22.Bidle KD, Vardi A. A chemical arms race at sea mediates algal host-virus interactions. Curr Opin Microbiol. 2011;14(4):449–457. doi: 10.1016/j.mib.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Bratbak G, Wilson W, Heldal M. Viral control of Emiliania huxleyi blooms? J Mar Syst. 1996;9(1–2):75–81. [Google Scholar]
- 24.Castberg T, et al. Isolation and characterization of a virus that infects Emiliania huxleyi (Haptophyta) J Phycol. 2002;38(4):767–774. [Google Scholar]
- 25.Jacquet S, et al. Flow cytometric analysis of an Emiliania huxleyi bloom terminated by viral infection. Aquat Microb Ecol. 2002;27:111–124. [Google Scholar]
- 26.Evans C, Malin G, Wilson WH, Liss PS. Infectious titers of Emiliania huxleyi virus 86 are reduced by exposure to millimolar dimethyl sulfide and acrylic acid. Limnol Oceanogr. 2006;51(5):2468–2471. [Google Scholar]
- 27.Evans C, et al. The relative significance of viral lysis and microzooplankton grazing as pathways of dimethylsulfoniopropionate (DMSP) cleavage: An Emiliania huxleyi culture study. Limnol Oceanogr. 2007;52(3):1036–1045. [Google Scholar]
- 28.Frada MJ, Bidle KD, Probert I, de Vargas C. In situ survey of life cycle phases of the coccolithophore Emiliania huxleyi (Haptophyta) Environ Microbiol. 2012;14(6):1558–1569. doi: 10.1111/j.1462-2920.2012.02745.x. [DOI] [PubMed] [Google Scholar]
- 29.Lynch DV, Dunn TM. An introduction to plant sphingolipids and a review of recent advances in understanding their metabolism and function. New Phytol. 2004;161(3):677–702. doi: 10.1111/j.1469-8137.2004.00992.x. [DOI] [PubMed] [Google Scholar]
- 30.Van Mooy BAS, et al. Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature. 2009;458(7234):69–72. doi: 10.1038/nature07659. [DOI] [PubMed] [Google Scholar]
- 31.Irie A, Kubo H, Hoshi M. Glucosylceramide having a novel tri-unsaturated long-chain base from the spermatozoa of the starfish, Asterias amurensis. J Biochem. 1990;107(4):578–586. doi: 10.1093/oxfordjournals.jbchem.a123089. [DOI] [PubMed] [Google Scholar]
- 32.Ohashi Y, et al. Squid nerve sphingomyelin containing an unusual sphingoid base. J Lipid Res. 2000;41(7):1118–1124. [PubMed] [Google Scholar]
- 33.Coolen MJL. 7000 years of Emiliania huxleyi viruses in the Black Sea. Science. 2011;333(6041):451–452. doi: 10.1126/science.1200072. [DOI] [PubMed] [Google Scholar]
- 34.Pagarete A, Allen MJ, Wilson WH, Kimmance SA, de Vargas C. Host-virus shift of the sphingolipid pathway along an Emiliania huxleyi bloom: Survival of the fattest. Environ Microbiol. 2009;11(11):2840–2848. doi: 10.1111/j.1462-2920.2009.02006.x. [DOI] [PubMed] [Google Scholar]
- 35.Evans C, Malin G, Mills GP, Wilson WH. Viral infection of Emiliania huxyleyi (Prymnesiophyceae) leads to elevated production of reactive oxygen species. J Phycol. 2006;42(5):1040–1047. [Google Scholar]
- 36.Jacobson MD. Reactive oxygen species and programmed cell death. Trends Biochem Sci. 1996;21(3):83–86. [PubMed] [Google Scholar]
- 37.Bidle KD, Kwityn CJ. Assessing the role of metacaspase expression and caspase activity on viral susceptibility of the coccolithophore, Emiliania huxleyi. J Phycol. 2012;48(5):1079–1089. doi: 10.1111/j.1529-8817.2012.01209.x. [DOI] [PubMed] [Google Scholar]
- 38.Vercammen D, Declercq W, Vandenabeele P, Van Breusegem F. Are metacaspases caspases? J Cell Biol. 2007;179(3):375–380. doi: 10.1083/jcb.200705193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engel A, et al. Transparent exopolymer particles and dissolved organic carbon production by Emiliania huxleyi exposed to different CO2 concentrations: A mesocosm experiment. Aquat Microb Ecol. 2004;34:93–104. [Google Scholar]
- 40.Passow U, et al. Origin of transparent exopolymer particles (TEP) and their role in the sedimentation of particulate matter. Cont Shelf Res. 2001;21:327–346. [Google Scholar]
- 41.Berman-Frank I, Rosenberg G, Levitan O, Haramaty L, Mari X. Coupling between autocatalytic cell death and transparent exopolymeric particle production in the marine cyanobacterium Trichodesmium. Environ Microbiol. 2007;9(6):1415–1422. doi: 10.1111/j.1462-2920.2007.01257.x. [DOI] [PubMed] [Google Scholar]
- 42.Kahl LA, Vardi A, Schofield O. Effects of phytoplankton physiology on export flux. Mar Ecol Prog Ser. 2008;354:1–16. [Google Scholar]
- 43.Joassin P, et al. Carbon and nitrogen flows during a bloom of the coccolithophore Emiliania huxleyi: Modeling a mesocosm experiment. J Mar Syst. 2011;85:71–85. [Google Scholar]
- 44.Brussaard CPD, Bratbak G, Baudoux A-C, Ruardij P. Phaeocystis and its interaction with viruses. Biogeochemistry. 2007;83:201–215. [Google Scholar]
- 45.Joint I, et al. Cell-to-cell communication across the prokaryote-eukaryote boundary. Science. 2002;298(5596):1207. doi: 10.1126/science.1077075. [DOI] [PubMed] [Google Scholar]
- 46.Mari X, Kerros M-E, Weinbauer MG. Virus attachment to transparent exopolymeric particles along trophic gradients in the southwestern lagoon of New Caledonia. Appl Environ Microbiol. 2007;73(16):5245–5252. doi: 10.1128/AEM.00762-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D. Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature. 2011;474(7353):604–608. doi: 10.1038/nature10172. [DOI] [PubMed] [Google Scholar]
- 48.Frada M, Probert I, Allen MJ, Wilson WH, de Vargas C. The “Cheshire Cat” escape strategy of the coccolithophore Emiliania huxleyi in response to viral infection. Proc Natl Acad Sci USA. 2008;105(41):15944–15949. doi: 10.1073/pnas.0807707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thyrhaug R, Larsen A, Thingstad TF, Bratbak G. Stable coexistence in marine algal host-virus systems. Mar Ecol Prog Ser. 2003;254:27–35. [Google Scholar]
- 50.Sorensen G, Baker AC, Hall MJ, Munn CB, Schroeder DC. Novel virus dynamics in an Emiliania huxleyi bloom. J Plankton Res. 2009;31(7):787–791. doi: 10.1093/plankt/fbp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martínez JM, Schroeder DC, Larsen A, Bratbak G, Wilson WH. Molecular dynamics of Emiliania huxleyi and cooccurring viruses during two separate mesocosm studies. Appl Environ Microbiol. 2007;73(2):554–562. doi: 10.1128/AEM.00864-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinez-Martinez J, Schroeder DC, Wilson WH. Dynamics and genotypic composition of Emiliania huxleyi and their co-occurring viruses during a coccolithophore bloom in the North Sea. FEMS Microbiol Ecol. 2012;81(2):315–323. doi: 10.1111/j.1574-6941.2012.01349.x. [DOI] [PubMed] [Google Scholar]
- 53.Egge JK, Heimdal BR. Blooms of Emiliania huxleyi (Haptophyta) in mesocosm experiments: Effects of nutrient supply in different N:P ratios. Sarsia. 1994;79(4):333–348. [Google Scholar]
- 54.Sturt HF, Summons RE, Smith K, Elvert M, Hinrichs K-U. Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry: New biomarkers for biogeochemistry and microbial ecology. Rapid Commun Mass Spectrom. 2004;18(6):617–628. doi: 10.1002/rcm.1378. [DOI] [PubMed] [Google Scholar]
- 55.Passow U, Alldredge AL. A dye-binding assay for the spectrophotometric measurement of transparent exopolymer particles (TEP) in the ocean. Limnol Oceanogr. 1995;40(7):1326–1335. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.