Abstract
Background
Human papillomavirus (HPV) types 16 and 18 are the high-risk, sexually transmitted infectious causes of most cervical intraepithelial neoplasias (CIN) or cancers. While efficacious vaccines to reduce the sexual acquisition of these high-risk HPVs have recently been introduced, no virus-targeted therapies exist for those already exposed and infected. Considering the oncogenic role of the transforming (E6 and E7) genes of high-risk HPVs in the slow pathogenesis of cervical cancer, we hypothesize that timely disruption or abolition of HPV genome expression within pre-cancerous lesions identified at screening may reverse neoplasia. We aimed to derive model zinc finger nucleases (ZFNs) for mutagenesis of the genomes of two high-risk HPV (types 16 & 18).
Methods and results
Using ZiFiT software and the complete genomes of HPV types16 and 18, we computationally generated the consensus amino acid sequences of the DNA-binding domains (F1, F2, & F3) of (i) 296 & 327 contextually unpaired (or single) three zinc-finger arrays (sZFAs) and (ii) 9 & 13 contextually paired (left and right) three- zinc-finger arrays (pZFAs) that bind genomic DNA of HPV-types 16 and 18 respectively, inclusive of the E7 gene (s/pZFAHpV/E7). In the absence of contextually paired three-zinc-finger arrays (pZFAs) that bind DNA corresponding to the genomic context of the E6 gene of either HPV type, we derived the DNA binding domains of another set of 9 & 14 contextually unpaired E6 gene-binding ZFAs (sZFAE6) to aid the future quest for paired ZFAs to target E6 gene sequences in both HPV types studied (pZFAE6). This paper presents models for (i) synthesis of hybrid ZFNs that cleave within the genomic DNA of either HPV type, by linking the gene sequences of the DNA-cleavage domain of the FokI endonuclease FN to the gene sequences of a member of the paired-HPV-binding ZFAs (pZFAHpV/E7 + FN), and (ii) delivery of the same into precancerous lesions using HPV-derived viral plasmids or vectors.
Conclusions
With further optimization, these model ZFNs offer the opportunity to induce target-mutagenesis and gene-therapeutic reversal of cervical neoplasia associated with HPV types 16 & 18.
Background
High-risk human papillomaviruses (HPVs) as oncogenic agents associated with cervical neoplasia
Human papillomaviruses (HPVs) are circular double stranded (ds) deoxyribonucleic acids (DNA) viruses of the genus alphapapillomavirus and family papillomavirus. Productive infection with HPVs only occurs within the stratified epithelium of the skin or mucous membranes [1]. Although there are over 200 HPV types, and 30 to 40 of these are sexually transmitted, only types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58 and 59 are denoted ‘high-risk’ HPVs [2,3]. Infection with these HPV types may lead to the development of cervical, penile or anal intraepithelial neoplasia. This is because most HPV infections are temporary, being eventually cleared by the immune system (70% and 90% in the first and second year following infection, respectively) with little or no long-term clinical significance [4]. In some individuals (representing 5% to 10% of infected women), however, the infection persists and could increase the risk for development of pre-cancerous lesions of the cervix, which can progress to invasive cervical cancer [4]. Cervical cancer is the cause of substantial morbidity and mortality among women world-wide. Each year, an estimated 490,000 cases of cervical cancer occur, resulting in approximately 270,000 deaths [1,2]. High-risk HPVs induce malignant transformation (neoplasia) and promote tumor growth through their early oncogenes E6 (HpVgp1) and E7 (HpVgp2). On the one hand, E6, which has a close relationship with the cellular protein E6-AP (E6-associated protein), mediates ubiquitin binding to the p53 protein, thereby flagging it for proteosomal degra-dation [5]. Degradation of p53, a protein that primarily prevents cell growth and stimulates apoptosis in the presence of DNA damage, promotes neoplasia. When present in normal levels, p53 also upregulates the p21 protein, which blocks the formation of the cyclin D/Cdk4 complex. This prevents the phosphorylation of retinoblastoma protein (pRB) and in turn halts cell cycle progression by preventing the activation of the eukaryotic transcription factor E2F. In short, p53 is either absent or its levels are greatly reduced within cervical cancer cells [6,7]. On the other hand, the E7 oncoprotein induces neoplasia by binding to and acting on multiple functional partners, notably retinoblastoma protein (pRB) and the class I histone deacetylases HDAC1 and HDAC2 [8-10]. Specifically, E7 destabilizes pRB levels through cullin 2–mediated proteasomal degradation [8]. E7 also indirectly binds to HDAC1 and HDAC2 as well as the reverse transcriptase (hTERT) region of telomerase via sequences in its zinc-finger domain [9]. While mutations within the zinc-finger domain of E7 do not affect its propensity to bind to and degrade pRB, they do abrogate its ability to immortalize cells, suggesting that both activities of E7 are required for immortalization [7-10]. Overall, cervical cancer cells can be said to be addicted to and or to thrive on the expression of the E6 and E7 proteins of high-risk HPVs.
Limitation of existing treatment and prevention modalities for ‘high-risk’ HPV
The slow neoplastic process following infection with high-risk HPVs, which usually takes 15–20 years, provides many opportunities for early detection and treatment of the pre-cancerous lesion. Specifically, progression to invasive cancer can almost always be prevented when standard prevention strategies are applied. In most countries, cervical screening using a Papanicolaou (Pap) test [11] or liquid-based cytology [12] is used to detect abnormal cells that could develop into cancer. In addition, targeted screening for HPVs or diagnostic testing (detecting E6, E7, or p16 mRNA) is now available [13-15]. If and when abnormal cells are found, women are invited to have a colposcopy with visual inspection under acetoacetic acid (VIUA). During a colposcopic inspection, biopsies can be taken and abnormal areas removed with simple procedures, typically by wedge- biopsy, cauterizing loop, freezing (cryotherapy), or chemo-/radio-therapy [16]. Treating abnormal cells in this way can halt the initiated development of cervical cancer [17]. However, only a few high-risk women, particularly within the developing world, ever get the timely test to detect abnormal cervical lesions. In addition, only a minority of those who are tested and are positive ever receive the therapy required to arrest disease. This may be attributed to inadequate follow-up and poor referral systems within poorly-resourced country health centers. Recent development of HPV vaccines (Cervarix and Gardasil), which when effectively used can prevent primary infection with the HPV types (16 and 18) that cause 70% of cervical cancers, has generated optimism that the global incidence of cervical cancer may be subdued [18,19]. It is nevertheless important to acknowledge that (i) coverage of HPV vaccination remains low within resource-limited yet cervical cancer-high incidence settings, and (ii) the current HPV vaccines are only effective when offered to girls who are not yet sexually active and have therefore never been exposed to high-risk HPVs.
Overall, the picture described above underscores the relevance of investing in development of specifically HPV-targeted treatments as a complementary public health strategy for controlling the global incidence of cervical cancer. More recently, Lin et al. [20] have proposed and experimentally tested the use of proteosome and histone de-acetylase (HDAC) inhibitors as novel HPV-targeted treatments for cervical cancer.
The alternate option of directly disrupting or abolishing HPV gene expression
Bacteria endowed with the restriction modification (R-M) system exhibit amazing resistance to tropism by the xenogeneic DNA of bacteria-infecting viruses, bacteriophages. RM systems can be said to be a primitive form of the many restriction factors that have recently been found to possess innate antiviral properties [21-23]. I [21] previously proposed using the anti-phage DNA machinery inherent within bacterial RM systems as a model for devising eukaryotic virus gene therapies. Towards this goal, I and colleagues [22,23] identified several bacterially-derived restriction enzymes with potential to cleave the DNA of human-infecting viruses, including frequency and site mapping of HIV-1, HIV-2 and several other SIV gene cleavages using a proviral DNA model [23]. However, most naturally-occurring bacterial restriction enzymes (aka REases) previously found to possess high potency (to slice or disrupt) against human infecting viruses in this manner were observed to cleave at palindromic sites that are short and equally prevalent within the human host genome, thereby raising concerns about in-situ safety. Our failure to address this priority milestone limited the use of those REase-therapeutic precursors to modeling and the extracellular space [22]. The long-term goal in the field of RM enzymatic therapeutics has been to generate synthetic restriction endonucleases with longer recognition sites specific only for the eukaryotic virus by mutating or engineering existing enzymes. Zinc finger nucleases (ZFNs) are artificial, hybrid restriction enzymes created by covalently linking a DNA-binding zinc finger domain (composed of three to six finger arrays) to the non-specific DNA cleavage domain (or simply FN) of the Flavobacterium okeanokoites restriction endonuclease FokI[24]. ZFNs have recently emerged as a potentially powerful tool for fighting eukaryotic viruses, either by primarily editing host genomes, or secondarily targeting incoming viral genomes [24-32]. On the one hand, Perez et al. [29], using engineered ZFNs targeting human CCR5, previously demonstrated the establishment of HIV-1 resistance in CD4+ T cells through generation of a double-strand break (DSB) at predetermined sites in the CCR5 coding region upstream of the natural CCR5D32 mutation. More recently, Holt et al. [30] demonstrated control of HIV-1 infection within NSG-mice transplanted with human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeting CCR5. On the other hand, with the intent of disrupting incoming viral genomes, we recently identified DNA binding domains of ZFNs to use as gene-therapeutics against proviral HIV DNA and HSV-II [31,32].
In the light of the above literature, we hypothesized that target mutagenesis of those high-risk forms of HPVs using ZFNs may possess potential for genetic therapy and reversal of cervical cancer in-situ.
The aim of this study was to identify and model DNA-binding domains of zinc finger arrays (ZFAs) as precursors for (i) synthesizing ZFNs for target mutagenesis of two high-risk HPV (types 16 & 18) –genomes, and development of a novel gene-therapeutic armament for reversing the primary oncogenic processes leading to cervical neoplasia.
Methods and results
(a) Database of DNA-binding domains of contextually unpaired three zinc-finger arrays targeting HPV types 16 and 18 genomic DNA
First, using Context-Dependent Assembly (CoDA) inherent in the ZiFiT-ZFA software of the zinc finger consortium [33-36] and the complete genomes of HPV types 16 and 18, we computationally generated the consensus amino acid sequences of the DNA binding domains or F1, F2 & F3 helices of 296 (see, Additional file 1 and Figure 1) and 327 (see, Additional file 2 and Figure 2) unpaired (single, either left or right) three zinc- finger (Zif) arrays (sZFAs) for targeting genomic DNA of HPV types 16 and 18, respectively. These ZFAs are henceforth denoted sZFAHpV16 and sZFAHpV18 respectively, or simply sZFAHpV. In principle, the FASTA formats of each virus type’s complete genomic DNA sequences (NCBI Accession #: |NC_001526.2| and |NC_001357.1| or IDs: 5607 and 5608, respectively) were individually fed into the user-interface preset to assemble three finger arrays. Although the HPV genomes do not contain intronic sequences, the exon/intron case-sensitivity algorithm was turned to its ON-mode throughout these experiments; this algorithm is necessary to distinguish between intron and exon sequences by denoting exons in uppercase and introns in lowercase. Overall, for either virus type, we derived DNA-binding domains of unpaired or single three-ZFAs (sZFAs) capable of specifically recognizing and binding to consecutive 9 bp sequences across the entire genome (see Figures 1 and 2, respectively). Analyses of the issuing cleavage patterns revealed that the highest incidence of unpaired ZFA-binding HPV type 16 genomic DNA is situated within the first and last 1,581 bp (20% of 7,905 bp) of the complete HPV type 16 genome (genomic context: before 0.2 and after 0.8, respectively) (see Figure 1). According to Zheng and Baker [37], the HPV genome can be organized into three major regions: early, late, and a long control region (LCR or non-coding region [NCR]), which are separated by two polyadenylation (pA) sites: early pA (AE) and late pA (AL). The early region of a papillomavirus genome occupies over 50% of the genome from its 5′ end and encodes six common open reading frames (E1, E2, E4, E5, E6 and E7) that translate their respective proteins. Two additional ORFs, E3 and E8, were assigned to this region initially. However, only the E8 ORF in bovine papilloma virus-1 and HPV-31 have so far been proven to encode a spliced E8^E2C fusion protein, which functions as a negative regulator of viral transcription and replication. The late region of every papillomavirus genome, covering almost 40% of the genome, lies downstream of the early region and encodes L1 and L2 ORFs for translation of a major (L1) and a minor (L2) capsid protein. The long control region (LCR), a segment of about 850 bp (10% of the HPV genome), has no protein-coding function, but bears the origin of replication as well as multiple transcription factor binding sites that are important in regulation of RNA polymerase II-initiated transcription from viral early as well as late promoters. On the basis of this genome structure, those ZFAs binding to the 5′ end 20% would fall under the early region, while the 3′ end 20% falls both under the late and LCR region. For HPV type 18, on the other hand, despite an occurrence of ZFAs with DNA-binding domains able to target 9 bp sequences distributed randomly across almost all its genome, the highest incidence of ZFAs with HPV-type 18 genomic DNA binding-domains was found to be situated within the last 1,571 bp (20% of the total genomic 7,857 bp) of the complete genome (genomic context: 0.8-1.00) (see Figure 2). In the context of the HPV genome structure provided by Zheng and Baker [37], this 3′ end falls under the late and long control region (LCR) of all papillomavirus genomes, and encodes L1 and L2 ORFs for translation of a major (L1) and a minor (L2) capsid protein, alongside the LCR regional origin of replication plus multiple transcription factors [37]. Each of these contextually unpaired or single zinc finger arrays (sZFAs) is composed of three zinc fingers (Zif or simply ZF). Zifs or ZFs are protein motifs that each possesses two beta strands and an alpha helix stabilized by coordination of a zinc ion mediated by pairs of conserved cysteine and histidine (C2H2) residues [38-40]. Residues 1 to 6 (numbered relative to the start) of the alpha-helix mediate target binding and recognition of triplets of DNA sequences through the formation of base-specific contacts in the major groove of the double-stranded target DNA (for illustration, see Additional file 1 and Additional file 2) [38-41]. Several Zifs/ZFs can be linked together, as is the case in the sZFAHpV16 and sZFAHpV18 databases, in order to yield a contextually unpaired multi-finger array capable of recognizing a longer and thereby preferentially unique sequence in any target double-stranded genomic DNA, aside from the host. As already shown in Additional file 1 and Additional file 2, several such contextually unpaired ZFAs were uncovered with target-binding potency across the entire genomic contexts of either HPV type 16 or 18 DNA.
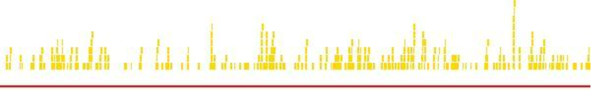
Figure 1.
Schematics of zinc finger arrays targeting HPV type 16 genomic DNA. This figure offers a diagrammatic illustration of loci of action of zinc finger arrays that specifically bind to 9 bp nucleotide sequences within the genomic DNA context of HPV type 16. Hits (blue, green, and gold bars) represent targets along the gene (red bars). ZFAs hits in the graphic are color-coded on the basis of spacer size (5 bp = Blue; 6 bp = Green; 7 bp = Gold).
Figure 2.
Schematics of zinc finger arrays binding to HPV type 18 genomic DNA. This figure offers a diagrammatic illustration of loci of action of zinc finger arrays that specifically bind to 9 bp nucleotide sequences within the genomic DNA context of HPV type 18. Hits (blue, green, and gold bars) represent targets along the gene (red bars). ZFA hits in the graphic are color-coded on the basis of spacer size (5 bp = Blue; 6 bp = Green; 7 bp = Gold).
(b) Database of DNA-binding domains of contextually paired three zinc-finger arrays targeting HPV types 16 and 18 genomic DNA specific zinc finger nucleases
Second, using Context-Dependent Assembly (CoDA) inherent in the ZiFiT-ZFN software of the zinc finger consortium [33-36] and the complete genomes of HPV types 16 and 18, we computationally compiled the amino acid sequences of the alpha-helical DNA-binding domains of 9 and 13 paired three zinc-finger arrays targeting HPV types 16 and 18 genomic DNA, respectively (see Additional file 3 and Additional file 4, respectively). Throughout our assembly of the DNA-binding domains of these paired ZFA (pZFAs), all ZiFiT-ZFN algorithms were pre-set as they were for derivation of the unpaired ZFAs (sZFAs) above, except that a 5, 6, or 7 base-pair (bp) overlapping sequence was selected in addition. Because ZFNs function as dimers, it is these paired ZFAs (pZFAs) assembled in this section of the results that are intended for engineering ZFNs that cleave the genomes of the study HPV types, as modeled further below [38]. These paired ZFA (pZFAs) are henceforth denoted pZFAHpV16 and pZFAHpV18 respectively, or simply pZFAHpV. Overall, pZFAHpV with demonstrable in-silico ability to bind to target sequences at positions 0.45 (~3,557 bp), 0.75 (~5,929 bp), and across 0.85 to 0.90 within the HPV type 16 genomic DNA context were derived. These genomic contextual regions approximately correspond to sequences between the early region’s hypothetical protein HpV16gp5 (context 3332-3619 bp) and the late region’s major L1 capsid protein (5,560-7,155 bp) (see Figure 3). In contrast, pZFAHpV capable of binding at positions 0.1 (~786 bp), 0.25 (~1,964 bp), 0.45 (~3, 535 bp), 0.65 (~5,107 bp), 0.75 (~5,892 bp) and 0.85 (~6,679 bp) respectively within the HPV type 18 genomic DNA context were derived. These regions correspond to the genomic contexts of the genes E7, E1, E2, E3, E4, L2 and L1, respectively (see Figure 4). It is important to note that, while we have generated pZFAHpV that are precursors for synthesizing HPV-specific ZFNs, engineering of the actual ZFNs can only be achieved in-vitro as is further modeled below (see section d of results).
Figure 3.
Schematics of zinc finger nucleases cleaving HPV type 16 genomic DNA. This figure offers a diagrammatic illustration of loci of action of zinc finger nucleases that target and cleave >18 bp (9x2 + 5, 6, or 7) sequences within the genomic DNA context of HPV type 16. Hits (blue, green, and gold bars) represent targets along the gene (red bars). ZFN hits in the graphic are color-coded on the basis of spacer size (5 bp = Blue; 6 bp = Green; 7 bp = Gold).
Figure 4.
Schematics of zinc finger nucleases cleaving HPV type 18 genomic DNA. This figure offers a diagrammatic illustration of loci of action of zinc finger nucleases that target and cleave >18 bp (9x2 + 5, 6, or 7) sequences within the genomic DNA context of HPV type 18. Hits (blue, green, and gold bars) represent targets along the gene (red bars). ZFN hits in the graphic are color-coded on the basis of spacer size (5 bp = Blue; 6 bp = Green; 7 bp = Gold).
(c) Repository of DNA-binding domains of unpaired zinc finger arrays targeting HPV types 16 and 18 E6 genes
Lastly, in the absence of any paired ZFA (pZFAs) that are precursors for engineering ZFNs targeting within the genomic context corresponding to the E6 transforming gene of either HPV type 16 (>gi|310698439:83-559|) or 18 (>gi|9626069:105–581), we focused our efforts on assembling another two databases of 9 (Figure 5) and 14 (Figure 6) DNA-binding domains of unpaired or single ZFAs (sZFAs) that target this early gene variant from HPV types 16 and 18, respectively. These sZFA databases offer us a basis for creating novel E6-targeting pZFAs precursors of ZFNs, by (i) sequential pairing of those neighboring sZFAs lying within the context of a desired pZFA target, and (ii) in-vitro optimization [42]. Alternatively, those E6 gene binding sZFAs may be used to enhance delivery of existing HPV-targeted treatments for cervical cancer such as the proteosome and histone deacetylase (HDAC) inhibitors previously described by Lin et al. [20].
Figure 5.
Schematics of zinc finger arrays binding to HPV type 16 E6 gene. This figure offers a diagrammatic illustration of loci of action of zinc finger arrays that specifically bind to 9 bp nucleotide sequences within the DNA context of the HPV type 16 transforming gene, E6. Hits (blue, green, and gold bars) represent targets along the gene (red bars). ZFA hits in the graphic are color-coded on the basis of spacer size (5 bp = Blue; 6 bp = Green; 7 bp = Gold).
Figure 6.
Schematics of zinc finger arrays binding to HPV type 18 E6 gene. This figure offers a diagrammatic illustration of loci of action of zinc finger arrays that specifically bind to 9 bp nucleotide sequences within the DNA context of the HPV type 18 transforming gene, E6. Hits (blue, green, and gold bars) represent targets along the gene (red bars). ZFA hits in the graphic are color-coded on the basis of spacer size (5 bp = Blue; 6 bp = Green; 7 bp = Gold).
(d) Models for synthesis and delivery of ZFNs targeting HPV types 16 and 18
The following text describes the generation of models for (i) synthesis of hybrid ZFNs to cleave within either HPV type’s genomic DNA by linking the gene sequences of the DNA-cleavage domain of the FokI endonuclease-FN to gene sequences coding for members of the paired-HPV-binding ZFAs (pZFAHpV/E7 + FN) and (ii) delivery of the same into precancerous lesions using HPV-derived viral plasmids or vectors:
First, using an approach similar to that described by Kim et al. (1996), employing the gene sequences of the DNA-cleavage domain of the Fok I endonuclease-FN (derived from Flavobacterium okeanokoites and belonging to the type IIS class) and members of a pair of ZFAs (pZFAs) (see Additional file 3 and Additional file 4, respectively) in our databases provided in section b above, it is possible to fuse the two sequences to yield 9 and 13 hybrid, chimeric ZFNs (pZFAHpV + FN) with ability to cleave the genomic DNAs of HPV types 16 and 18, respectively (for the predicted corresponding ZFN-cleavage sites, see Figures 3 and 4) [38]. The two individual components of the model ZFNs (pZFAHpV and FN) are molecularly cloned into ZFN expression vectors in-vitro using unique XbaI/NotI restriction sites [33-36,38-41]. Specifically, PCR amplification of gene-sequences of each individual component is done using primers that introduce XbaI/NotI sites as a strategy to enable a pair of the pZFAHpV + FN complex to be inserted into alternative sites of Zinc Finger Consortia plasmids capable of recognizing target sites with a 7 bp spacer [33,34]. In principle, since the desired effect of the ZFN is achieved in-vivo by two paired zinc finger arrays (pZFAs) each fused to a nuclease domain, two members of a pair (pZFAs) are sub-cloned. Dimerization of the FokI nuclease occurs in-vivo when both members of the pZFA bind their target sequence, thereby ensuring that the two FokI nucleases attach to the target DNA in a particular configuration in order to introduce a double strand break (DSB). Following actual synthesis, it may be necessary to incorporate further improvements, say by (i) modular analysis to add one, two or even three other ZFA on to our currently three-paired ZFAs (pZFAs) so as to enhance specificity and avoid off-target genome toxicity [33-36]. In-vitro assembly and testing for efficacy of the desired target-cleavage need to be done pre-clinically say by (ii) using either a bacteria-one hybrid (B1H) or yeast one-hybrid (Y1B) system, so as to inform and select the best ZFNs to use in-vivo[42]. Elsewhere, (iii) the Fok1 cleavage domain has been modified as a strategy for generating a hybrid capable of functionally interrogating the ZFN dimer interface so as to prevent homodimerization while still enhancing the efficiency of cleavage [42].
Second, an exact approach toward gene delivery and transduction of the ‘pre-cancerous lesion’ cells with these ZFHpV + FN complexes in-vivo is theoretically proposed. Specifically, we propose the use of human papillomavirus plasmids capable of episomal replication in human cell lines, to deliver the diploid or paired copy of the high-risk HPV-specific ZFN genotype, pZFHpV + FN. The pZFHpV + FN genotype may be sub-cloned using similar PCR-primers to the ones proposed above for consortia plasmids, with tailored modifications to HPV plasmids or PsV. Sverdrup et al. [43] have previously generated such human papillomavirus (HPV) plasmids containing the viral E1 and E2 genes (or the E1 gene alone) and an origin of replication, which they simultaneously demonstrated to replicate to significant levels in the transfected human cervical carcinoma C-33A cell line. Alternatively, HPV PsV encapsidation of the zinc finger consortium’s plasmids carrying the pZFHpV + FN (see results of modeling above) may suffice. Graham et al. [44], using such HPV PsV-encapsidation of DNA plasmids, produced mucosal vaccine vectors expressing an experimental antigen derived from the M and M2 proteins of respiratory syncytial virus. The same were shown to evoke 10,000-fold higher CD8+ T-cell and antibody responses in mice than naked DNA. Moreover, on the basis of light emission and immunofluorescence microscopy, it was shown by immunization with HPV PsV-encapsidated luciferase- and red fluorescent protein (RFP)-expressing plasmids that the HPV PsV-encapsidated plasmids induce antigen expression restricted to the vaginal epithelium (an ideal target), although this expression is only transient and further modifications of these HPV plasmids are needed to ensure stable expression of the exogenous pZFHpV + FN genotype [45].
(e) Software and databases
The National Center for Biotechnology Information (NCBI) HPV type 16 and 18 genome databases used are available at the following URLs:
• http://www.ncbi.nlm.nih.gov/genome?term=NC_001526.2
The two Zinc Finger Consortium’s CoDA-ZiFiT software used are available at the following URL: http://zifit.partners.org/ZiFiT/ChoiceMenu.aspx.
Discussion
I present here databases of paired ZFAs (pZFA) that are precursors for engineering ZFNs to target and cleave the genomic DNA of the two high-risk HPV types most associated with cervical cancer. With the appropriate in-vivo gene-delivery and transduction plasmids or vectors, models of ZFNs synthesized from these pZFA may offer us the means for targeted HPV mutagenesis and therapeutic reversal of the primary oncogenic processes driving cervical neoplasia. Considering the oncogenic role of the transforming genes (E6/HpVgp1 or E7/HpVgp2) of high-risk types of HPVs in the slow pathogenesis of cervical cancer, therefore, we hypothesized that timely in-situ disruption or abolition of HPV genome expression within detected high-risk lesions may reverse cervical neoplasia. To this end, we identified DNA-binding domains of ZFAs for engineering ZFNs for targeted mutagenesis of these two high-risk HPV genomes as precursors for developing a novel gene-therapeutic armament for reversing the primary oncogenic processes leading to cervical neoplasia. As shown in Additional file 1 and Additional file 2, plus Additional file 3 and Additional file 4, respectively, multiple unpaired/single and paired ZFAs (Figures 1, 2, 3 and 4) were initially identified. The paired ZFAs (pZFAs) were used to model ZFNs that bind and cleave HPV type 16 and 18 genomic DNA (Figures 3 and 4). Because the modeled ZFNs could only target sequences of the E7 gene of either virus type studied, we focused our additional efforts on the E6 transforming gene, resulting in another set of two databases of 9 and 14 single ZFAs (sZFAs) that bind to sequences of this early gene from both types of HPV studied (16 and 18, respectively) (Figures 5 and 6).
Many previous studies have explored the therapeutic potential of ZFN technology against DNA viruses that infect humans, primarily through editing host genomes [27-30], secondarily by targeting the infective viral genomes [31,32]. ZFNs generally act by introducing a DSB into the target (say, HPV) genome which in the absence of a template for homologous recombination (HR) would lead to repair, if any, by non-homologous end-joining (NHEJ) [38-41]. NHEJ is therefore a major mechanism by which our modeled ZFNs are expected to induce gene architectural disruptions and functional distortions in the HPVs, although complete HPV gene deletion through frame-shift mutations is another possibility [33-36,38-41,46]. Towards a similar purpose [31,32], our model HPV-binding ZFNs would be usable first to cleave and disrupt HPV type 16 genomic DNA at the contextual positions corresponding to about 0.45 (~3,557 bp), 0.75 (~5,929 bp), and across 0.85 to 0.90. Disabling of these regions, which correspond to sequences between the early region’s hypothetical protein HpV16gp5 (context 3332-3619 bp) and the major L1 capsid protein (5,560-7,155 bp) (see Figure 3), may abrogate or reduce HPV type 16 survival or replication fitness. In contrast, HPV type 18 would be cleaved at regions approximately corresponding to the genomic contextual positions 0.1 (~786 bp), 0.25 (~1,964 bp), 0.45 (~3, 535 bp), 0.65 (~5,107 bp), 0.75 (~5,892 bp) and 0.85 (~6,679 bp); or simply the early-gene region (E7, E1, E2, E3, E4), late region (L2 and L1) and the LCR region (containing the origin of replication and multiple transcriptional factors) is predicted to be especially susceptible to similar abrogation or reduction in survival and replication fitness (Figure 4) [37]. Considering the addictive and oncogenic role of the HPV genes E6 and E7 in the pathogenesis of cervical dysplasia and/or neoplasia [6-10,20], it should be therapeutically adequate to target only these genes, explaining our further exploration of single ZFAs (sZFAs) for the E6 gene of either HPV type studied (Figures 5 and 6). Secondly, any two of the modular ZFNs cleaving at the extreme 5′ and 3′ ends of a viral genome could further be modified and optimized [42] to non-specifically target and delete most (i.e. >90%) of the genomic DNAs of the HPVs studied. In view of the currently evidenced low transduction and genome-modification rates of existing vectors and ZFN-technology, it is important that the success rates of effecting such changes in HPVs within precancerous lesions of the cervix are evaluated in-vitro and in-vivo, say by using HeLa cell lines or humanized mouse models [42,47-49]. Thirdly, it is rational to propose use of those single HPV-genome targeting ZFAs (sZFAHpV) as magnetic drivers for novel HPV-DNA targeting therapeutics such as transcriptional repressors or the proteosomal/histone deacetylase (HDAC) inhibitors discussed by Lin et al. [20].
This study has a number of limitations. First, the work has been limited to sequence analyses and is not accompanied by in-vitro studies. This can be attributed to the limited resource capacity of our laboratory. However, these same methods [33-36] have previously been employed successfully to assemble pZFAs that were used to engineer ZFNs, which have been experimentally proven to be safe and effective [33-44,46]. However, preclinical studies in-vitro using either a bacterial or yeast hybrid system as described in the modeling results in section d of our results need to be conducted to determine the efficacy of the model ZFNHpV before their trial-use in the clinic. Experiments such as those recently conducted by Wilen et al. [47] for the Ad5/F35 vector carrying CXCR4-specific zinc-finger nucleases used to engineer HIV-resistant human CD4+ T cells may be conducted in humanized mouse models. Secondly, inclusion of ZFAs and ZFNs targeting other high risk HPVs, say types 31 and 45, may be a necessary strategy to increase the projected efficacy of this method from 70% (contribution of HPV types 16 and 18 to global cervical dysplasia) to nearly 100%. Thirdly, it is unlikely that the entire genomes of these high-risk HPVs will be excised through frame-shift mutations. Therefore, the already error-prone NHEJ-repair of the DSBs introduced in the HPV genomes could facilitate rare and random yet possibly effective recombinations that may yield either wild-type or mutant HPVs still capable of infecting and transforming cervical epithelia to cancer. This further underlines the need for the above proposed safety and efficacy studies. Fourthly, it must be asked how these modeled HPV-plasmids carrying and transducing ZFNs targeting HPV types 16 and 18 will be used in the clinic. Our translational projection or proposition is to achieve this via direct subdermal or intra-epithelial injection into those sites identified by currently existing screening protocols as pre-cancerous [11-15]. If instituted at the right time, this would essentially either eliminate the need to use invasive techniques with adverse side effects (cone-biopsies, cauterizations, cryotherapies, chemo-/radio-therapy) or would at least supplement them [16,17]. In other words, it would offer a unique opportunity for medically treating women who did not receive the HPV-vaccine as girls and have never used potentially effective microbicidal agents such as carrageen [50,51], but have been diagnosed with pre-cancerous lesions at screening. However, this proposed mode of administration is not without its limitations, including uncertainties regarding the cells that will be targeted using the modeled 2xZFN-gene delivery and transduction, let alone the efficacy of ZFN-expression and or target-genome modification by the therapeutic proteins (ZFNHpV). Also, the delivery plasmids or vectors are potentially prone to attack by cells of the host immune-system, and concomitant local or systemic immunosuppressive (say steroidal) therapy might be needed to avoid this. Lastly, recent unbiased genome-wide analyses have shown that ZFNs may exhibit off-target effects in-vivo that might not be predicted by in-silico approaches such as the ones employed here [48,49].
In conclusion, we present databases of paired ZFAs targeting HPV-genomic-DNA (pZFAHpV) and their model ZFNHpV for targeting two high-risk HPVs (types 16 and 18). With the appropriate optimization, modification of these modeled ZFNHpV and in-vivo delivery HPV PsV encapsidation plasmids or vectors, a translational possibility of therapeutically reversing the primary HPV-induced oncogenic processes driving cervical neoplasia is proposed.
Competing interests
WM is Chief Scientific Officer at Restrizymes Biotherapeutics (U) Ltd, and member of the steering committee of the Young and early careers’ investigators (YECI) of the Global HIV Vaccine Enterprise.
Authors’ contribution
WM concieved the idea for this study, conducted the experiments, analysed the data, and wrote the final manuscript.
Supplementary Material
List of zinc finger arrays targeting HPV type 16 genomic DNA. This file offers a detailed list and loci of action of zinc finger arrays that specifically bind to 9 bp nucleotide sequences within the genomic DNA context of HPV type 16.
List of zinc finger arrays binding to HPV type 18 genomic DNA. This file offers a detailed list and loci of action of zinc finger arrays that specifically bind to 9 bp nucleotide sequences within the genomic DNA context of HPV type 18.
List of zinc finger nucleases cleaving HPV type 16 genomic DNA. This file offers a detailed list and loci of action of zinc finger nucleases that target and cleave >18 bp (9x2 + 5, 6, or 7) sequences within the genomic DNA context of HPV type 16.
List of zinc finger nucleases cleaving HPV type 18 genomic DNA. This file offers a detailed list and loci of action of zinc finger nucleases that target and cleave >18 bp (9x2 + 5, 6, or 7) sequences within the genomic DNA context of HPV type 18.
Acknowledgements
I am grateful to Dr. Henry Kajumbula (MakCHS, Medical Microbiology) and Prof. Wilson Byarugaba (KIU-WC, Biochemistry) for their interest, guidance and collaboration on prior related-work. WM is supported by Grant Number 5R24TW008886 OGAC, NIH and HRSA. The content of this paper is solely the responsibility of the author(s) and does not necessarily represent the official views of the supporting offices.
References
- Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118(12):3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- Antonsson A, Forslund O, Ekberg H, Sterner G, Hansson BG. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. J Virol. 2000;74(24):11636–11641. doi: 10.1128/JVI.74.24.11636-11641.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt RJ. Human papillomaviruses: Diseases, diagnosis, and a possible vaccine. Clinical Microbiology Newsletter. 2005;27(18):139–145. doi: 10.1016/j.clinmicnews.2005.09.001. [DOI] [Google Scholar]
- Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Muñoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Munger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M, Grace M, Huh K. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78:11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248(4951):76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- Liu X, Dakic A, Zhang Y, Dai Y, Chen R, Schlegel R. HPV E6 protein interacts physically and functionally with the cellular telomerase complex. Proc Natl Acad Sci USA. 2009;106(44):18780–18785. doi: 10.1073/pnas.0906357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger K, Werness BA, Dyson N, Phelps WC, Harlow E, Howley PM. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longworth MS, Laimins LA. The binding of histone deacetylases and the integrity of zinc finger-like motifs of the E7 protein are essential for the life cycle of human papillomavirus type 31. J Virol. 2004;78:3533–35341. doi: 10.1128/JVI.78.7.3533-3541.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helt AM, Galloway DA. Destabilization of the retinoblastomaretinoblastoma tumor suppressor by human papillomavirus type 16 E7 is not sufficient to overcome cell cycle arrest in human keratinocytes. J Virol. 2001;75:6737–6747. doi: 10.1128/JVI.75.15.6737-6747.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldie SJ, Kuntz KM, Weinstein MC, Freedberg KA, Palefsky JM. Cost-effectiveness of screening for anal squamous intraepithelial lesions and anal cancer in human immunodeficiency virus-negative homosexual and bisexual men. Am J Med. 2000;108(8):634–641. doi: 10.1016/S0002-9343(00)00349-1. [DOI] [PubMed] [Google Scholar]
- Syrjänen S. Current concepts on human papillomavirus infections in children. APMIS: Acta pathologica, microbiologica, et immunologica Scandinavica. 2010;118(6–7):494–509. doi: 10.1111/j.1600-0463.2010.02620.x. [DOI] [PubMed] [Google Scholar]
- Dunne EF, Nielson CM, Stone KM, Markowitz LE. Giuliano AR: Prevalence of HPV infection among men: A systematic review of the literature. J Infect Dis. 2006;194(8):1044–1057. doi: 10.1086/507432. [DOI] [PubMed] [Google Scholar]
- Molden T, Kraus I, Skomedal H, Nordstrøm T, Karlsen F. PreTect HPV-Proofer: real-time detection and typing of E6/E7 mRNA from carcinogenic human papillomaviruses. J Virol Methods. 2007;142(1–2):204–212. doi: 10.1016/j.jviromet.2007.01.036. [DOI] [PubMed] [Google Scholar]
- Wentzensen N. von Knebel Doeberitz M: Biomarkers in cervical cancer screening. Dis Markers. 2007;23(4):315–330. doi: 10.1155/2007/678793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffman M, Castle PE. The promise of global cervical-cancer prevention. N Engl J Med. 2005;353(20):2101–2104. doi: 10.1056/NEJMp058171. [DOI] [PubMed] [Google Scholar]
- Gilbert LK, Alexander L, Grosshans JF, Jolley L. Answering frequently asked questions about HPV. Sex Transm Dis. 2003;30(3):193–194. doi: 10.1097/00007435-200303000-00002. [DOI] [PubMed] [Google Scholar]
- WHO. Human papillomavirus vaccines: WHO position paper. Wkly Epidemiol Rec. 2009;84(15):118–131. [PubMed] [Google Scholar]
- Roden R, Wu TC. How will HPV vaccines affect cervical cancer? Nat Rev Cancer. 2006;6:753–763. doi: 10.1038/nrc1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Bazzaro M, Wang M-C, Chan KC, Peng S, Roden RSB. Combination of Proteasome and HDAC Inhibitors for Uterine Cervical Cancer Treatment. Clin Cancer Res. 2009;15:570. doi: 10.1158/1078-0432.CCR-08-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayengera M. HIV and Gene Therapy: The proposed [R-M enzymatic] model for a gene therapy against HIV. Makerere Med J. 2003;38:28–30. [Google Scholar]
- Wayengera M, Kajumbula H, Byarugaba W. Identification of restriction endonuclease with potential ability to cleave the HSV-2 genome: inherent potential for biosynthetic versus live microbicides. Theor Biol Med Model. 2008;5:18. doi: 10.1186/1742-4682-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayengera M, Byarugaba W, Kajjumbula H. Frequency and site mapping of HIV-1/SIVcpz, HIV-2/SIVsmm and other SIV gene sequence cleavage by various bacteria restriction enzymes: Precursors for a novel HIV inhibitory product. Afr J Biotechnol. 2007;6(10):1225–1232. [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol Ther. 2008;16(7):1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- Guo J, Gaj T, Barbas CF. Directed Evolution of an Enhanced and Highly Efficient FokI Cleavage Domain for Zinc Finger Nucleases. J Mol Biol. 2010;1:96. doi: 10.1016/j.jmb.2010.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33(18):5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23(8):967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- Perez E, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, Guschin DY, Rupniewski I, Waite AJ, Carpenito C, Carroll RG, Orange JS, Urnov FD, Rebar EJ, Ando D, Gregory PD, Riley JL, Holmes MC, June CH. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26(7):808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, Cannon PM. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28(8):839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayengera M. Proviral HIV-genome-wide and pol-gene specific Zinc Finger Nucleases: usability for targeted HIV gene therapy. Theor Biol Med Model. 2011;8:26. doi: 10.1186/1742-4682-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayengera M. Identity of zinc finger nucleases with specificity to herpes simplex virus type II genomic DNA: novel HSV-2 vaccine/therapy precursors. Theor Biol Med Model. 2011;8:23. doi: 10.1186/1742-4682-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell JG, Barbas CF. Zinc finger tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:516–523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Zaback PZ, Joung JK, Voytas DF, Dobbs D. Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 2007;35:W599–W605. doi: 10.1093/nar/gkm349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462–W468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, Pierick CJ, Hoffman E, Maeder ML, Khayter C, Reyon D, Dobbs D, Langenau DM, Stupar RM, Giraldez AJ, Voytas DF, Peterson RT, Yeh JR, Joung JK. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat Methods. 2011;8(1):67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng ZM, Baker CC. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci. 2006;11:2286–2302. doi: 10.2741/1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93(3):11556–11560. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee Y-L, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(2):646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Dreier B, Segal DJ, Barbas CF. Insights into the molecular recognition of the 5 '-GNN-3 ' family of DNA sequences by zinc finger domains. J Mol Biol. 2000;303(4):489–502. doi: 10.1006/jmbi.2000.4133. [DOI] [PubMed] [Google Scholar]
- Moore M, Klug A, Choo Y. Improved DNA binding specificity from polyzinc finger peptides by using strings of two-finger units. Proc Natl Acad Sci USA. 2001;98:1437–1441. doi: 10.1073/pnas.98.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8(1):74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- Sverdrup FM, Sheahan LC, Khan SA. Development of human papillomavirus plasmids capable of episomal replication in human cell lines. Gene Therapy. 1999;6:1317–1321. doi: 10.1038/sj.gt.3300957. [DOI] [PubMed] [Google Scholar]
- Graham BS, Kines RC, Corbett KS, Nicewonger J, Johnson TR, Chen M, LaVigne D, Roberts JN, Cuburu N, Schiller JT, Buck CB. Mucosal delivery of human papillomavirus pseudovirus-encapsidated plasmids improves the potency of DNA vaccination. Mucosal Immunol. 2010;3(5):475–486. doi: 10.1038/mi.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SA, Sverdrup FM. Methods for the Construction of Human Papillomavirus Vectors. Methods in Molecular Medicine, Gene Therapy Protocols. 1997;7:117–125. doi: 10.1385/0-89603-484-4:117. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- Wilen CB, Wang J, Tilton JC, Miller JC, Kim KA, Rebar EJ, Sherrill-Mix SA, Patro SC, Secreto AJ, Jordan AP, Lee G, Kahn J, Aye PP, Bunnell BA, Lackner AA, Hoxie JA, Danet-Desnoyers GA, Bushman FD, Riley JL, Gregory PD, June CH, Holmes MC, Doms RW. Engineering HIV-Resistant Human CD4+ T Cells with CXCR4-Specific Zinc-Finger Nucleases. PLoS Pathog. 2011;7(4):e1002020. doi: 10.1371/journal.ppat.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Ramirez CL, Joung KJ, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods. 2011;8(9):765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang J, Friedman G, Holmes MC, Gregory PD, Glimm H, Schmidt M, Naldini L, von Kalle C. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011;29(9):816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- Buck C, Thompson C, Roberts J, Müller M, Lowy D, Schiller J. Carrageenan is a potent inhibitor of papillomavirus infection. PLoS pathog. 2006;2(7):69. doi: 10.1371/journal.ppat.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J, Buck C, Thompson C, Kines R, Bernardo M, Choyke P, Lowy DR, Schiller JT. Genital transmission of HPV in a mouse model is potentiated by nonoxynol-9 and inhibited by carrageenan. Nature Med. 2007;13(7):857–861. doi: 10.1038/nm1598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of zinc finger arrays targeting HPV type 16 genomic DNA. This file offers a detailed list and loci of action of zinc finger arrays that specifically bind to 9 bp nucleotide sequences within the genomic DNA context of HPV type 16.
List of zinc finger arrays binding to HPV type 18 genomic DNA. This file offers a detailed list and loci of action of zinc finger arrays that specifically bind to 9 bp nucleotide sequences within the genomic DNA context of HPV type 18.
List of zinc finger nucleases cleaving HPV type 16 genomic DNA. This file offers a detailed list and loci of action of zinc finger nucleases that target and cleave >18 bp (9x2 + 5, 6, or 7) sequences within the genomic DNA context of HPV type 16.
List of zinc finger nucleases cleaving HPV type 18 genomic DNA. This file offers a detailed list and loci of action of zinc finger nucleases that target and cleave >18 bp (9x2 + 5, 6, or 7) sequences within the genomic DNA context of HPV type 18.