Abstract
Previous studies have demonstrated that the mRNAs encoding the prolamine and glutelin storage proteins are localized to morphologically distinct membranes of the endoplasmic reticulum (ER) complex in developing rice (Oryza sativa L.) endosperm cells. To gain insight about this mRNA localization process, we investigated the association of prolamine polysomes on the ER that delimit the prolamine protein bodies (PBs). The bulk of the prolamine polysomes were resistant to extraction by 1% Triton X-100 either alone or together with puromycin, which suggests that these translation complexes are anchored to the PB surface through a second binding site in addition to the well-characterized ribosome-binding site of the ER-localized protein translocation complex. Suppression of translation initiation shows that these polysomes are bound through the mRNA, as shown by the simultaneous increase in the amounts of ribosome-free prolamine mRNAs and decrease in prolamine polysome content associated with the membrane-stripped PB fraction. The prolamine polysome-binding activity is likely to be associated with the cytoskeleton, based on the association of actin and tubulin with the prolamine polysomes and PBs after sucrose-density centrifugation.
Unlike most cereals that accumulate predominantly one type of storage protein, rice (Oryza sativa L.) seeds synthesize both major classes of storage proteins, the prolamines and the glutelins (globulins). Both storage protein types are initially synthesized on ER membranes but are then stored as PBs in different intracellular compartments (Muench and Okita, 1997). Prolamines are translocated into the ER lumen, where they are sequestered, forming a protein-inclusion granule delimited by the rough ER. Newly synthesized glutelins are also transported into the ER lumen but are subsequently sorted via the Golgi apparatus and deposited in the vacuole (Tanaka et al., 1980; Krishnan et al., 1986). Because of the cellular events leading to prolamine PB biogenesis, the rough ER can be viewed as being composed of morphologically distinct subdomains, the PB-ER and the C-ER (Li et al., 1993a). These subdomains are also functionally distinct in that prolamine and glutelin mRNAs are differentially sorted and translated on the PB-ER and C-ER, respectively, as shown by biochemical, in vitro, and in situ hybridization experiments (Li et al., 1993a). Therefore, rice is able to accommodate the synthesis and packaging into separate PBs of both major classes of storage proteins by translating these mRNAs on different ER subdomains. The mechanism responsible for the localization of the storage protein mRNA on these ER membrane types has yet to be identified.
mRNA localization has been extensively characterized in a wide variety of animal cells, many of which are polarized cell types (St. Johnston, 1995). The cytoskeleton has a direct role in the transport and anchoring of several of these mRNAs to specific intracellular locations. Both the microtubule and the microfilament networks are known to function together or independently in the localization of these mRNAs (Yisraeli et al., 1990; Sundell and Singer, 1991; St. Johnston, 1995). Actin mRNAs in the brown algae of the genus Fucus are localized to the cell plate in the developing embryo (Bouget et al., 1996). This RNA localization, however, is insensitive to cytoskeleton inhibitors and therefore may depend on some other mechanism for localization.
In addition to mRNA localization, the cytoskeleton has also been implicated in translation. Most of the polysomes in the cytosol are thought to be associated with the microfilament component of the detergent-resistant cytoskeleton scaffolding network (Pachter, 1992; Hesketh, 1994). Much of this evidence has come from electron microscopy, in situ hybridization, and biochemical fractionation studies of detergent-treated cell extracts and cells treated with cytoskeleton-destabilizing agents such as cytochalasin. These results suggest that many polysomes that were once considered “free” in the cytosol are in fact associated with the cytoskeleton, and adds to the view that the cytosol is divided into metabolically distinct compartments formed by the orderly association of metabolites and macromolecules (Pachter, 1992). Although much of this evidence is from animal systems, recent evidence indicates that polysomes in plant cells are also associated with the cytoskeleton. Biochemical evidence for cytoskeleton-bound polysomes has come from studying pea stems and roots and corn endosperm (Davies and Abe, 1989; Davies et al., 1993; Ito et al., 1994). In addition to cytoskeleton-bound polysomes in plants, there is also evidence for the association of polysomes to both the cytoskeleton and the membranes (Davies et al., 1993; Stankovic et al., 1993; Ito et al., 1994). Isolated maize PBs are enmeshed in F-actin, and nonionic detergent treatment removes all of the fluorescently labeled ER membrane and all of the phospholipids but retains almost all of the polysomes and the cytoskeleton components (Davies et al., 1993; Stankovic et al., 1993). Treatment with the ionic detergent deoxycholate solubilizes the cytoskeleton and releases the polysomes. These polysome types, termed cytomatrix-bound polysomes (Ito et al., 1994), are also found in animal cells (Zambetti et al., 1990b).
Although it is clear that many polysomes are bound to the cytoskeleton, the nature of this interaction is only poorly understood. In plant cells there is some evidence for the role of the ribosome or perhaps the nascent polypeptide chain in anchoring the polysome to the cytoskeleton (You et al., 1992; Davies et al., 1993; Stankovic et al., 1993). Treatment of animal cells with inhibitors of translation such as fluoride results in an increase in ribosome-free mRNAs, but fails to release these mRNAs from the cell matrix (Howe and Hershey, 1984; Bag and Pramanik, 1987). This suggests that some polysomes are anchored not through the ribosome but via the mRNA. In addition, many membrane-bound polysomes are not released by treatment with cytochalasin or puromycin alone, but when the inhibitors are used simultaneously, these membrane-bound polysomes are released (Zambetti et al., 1990b). This observation suggests that membrane-bound polysomes are anchored at two sites, one associated with the cytoskeleton, and the other with the well-characterized polypeptide translocation complex receptor that binds the ribosome (Rapoport, 1992).
In an effort to understand the mechanism responsible for prolamine and glutelin mRNA localization in rice endosperm cells, we used a biochemical approach to study the interaction among the prolamine polysomes, the PB-ER, and the cytoskeleton. Here we present evidence for the existence of a second prolamine polysome-binding activity in addition to the ribosome receptor of the polypeptide translocation complex of the ER. We also show that this second prolamine polysome-binding activity is likely to be associated with the cytoskeleton and that it interacts not with the nascent polypeptide chain but most likely with the mRNA itself.
MATERIALS AND METHODS
Buffers and Seed Extract Preparation
Seeds from rice (Oryza sativa L. var M 201) were harvested 15 d after flowering (mid-developing), frozen in liquid nitrogen, and stored at −80°C. For experimental analysis, 1 g of seed was typically ground with a mortar and pestle in liquid nitrogen and then in 3 mL of CSB (5 mm Hepes adjusted to pH 7.5 with 3.2 mm KOH, 10 mm MgOAc, 2 mm EGTA, 1 mm PMSF, 1 mm DTT, 1 unit mL−1 RNase inhibitor [Inhibit-Ace, 5 Prime→3 Prime, Inc., Boulder, CO], and 200 mm Suc [modified from Abe and Davies, 1991]). Other buffers used were CSB containing either 1% Triton X-100 or 1% PTE, or buffer U consisting of 200 mm Tris-HCl, pH 8.5, 50 mm KOAc, 25 mm MgOAc, 2 mm EGTA, 100 μg mL−1 heparin, 2% PTE, and 1% sodium deoxycholate (Davies et al., 1993). The crude seed extract was then filtered through one layer of Miracloth (Calbiochem). All steps were carried out on ice or at 4°C. A PB fraction was obtained by first removing most of the large starch grains by centrifuging the extract at 100g for 5 s, and then centrifuging the resulting supernatant at 500g for 10 min to obtain the PB fraction. The PB fraction was then analyzed directly or after pelleting and resuspension in other buffers.
Suc-Density Centrifugation
Aliquots of crude extracts or PB fractions were analyzed by Suc-density centrifugation using either 15 to 60% or 20 to 80% Suc gradients containing 5 mm Hepes adjusted to pH 7.5 with 3.2 mm KOH, and 10 mm MgOAc. The 15 to 60% gradients were generated as described previously (Davies and Abe, 1995) and were centrifuged at 250,000g in a SW-50 rotor (Beckman) for 60 min. The 20 to 80% gradients were generated by overlayering 80:60:40:20% Suc solutions in a 1:2:2:1 volume ratio, respectively, and allowing them to equilibrate into a continual gradient overnight at 4°C. The 20 to 80% gradients were centrifuged at either 300,000g in a SW-55 rotor (Beckman) for 60 min or 300,000g in a SW-41 rotor (Beckman) for 150 min. After centrifugation an absorbance profile at 254 nm was obtained and fractions were collected using a UV5 monitor and a gradient fractionator (model 185, Isco, Lincoln, NE).
Membrane Solubilization and Analysis
The solubilization of membranes in PB fractions or Suc-density gradient fractions was performed by incubating the samples in CSB containing 1% Triton X-100 or 1% PTE, or in buffer U for 10 min on ice. The efficiency of membrane solubilization was determined by analyzing the FA composition in untreated and detergent-treated PB fractions using GC, or by analyzing the release of the ER membrane protein calnexin (see “Protein and RNA Blotting”). The GC analysis was similar to that described previously (Miquel and Browse, 1992). A PB fraction was isolated as described above, and resuspended in CSB containing Triton X-100. The detergent-treated fraction was then centrifuged to obtain a PB pellet and supernatant. The pellet was resuspended in an equal volume of chloroform:methanol:formic acid solution (10:10:1), whereas 1.5 volumes of chloroform:methanol:formic acid solution (10:10:1) was added to the supernatant followed by the addition of 1.5 volumes of methanol. After incubation at −20°C overnight, the samples were then centrifuged at high speed in a clinical centrifuge at 4°C and the supernatant resuspended in one-fourth volume of Hajras solution (1 m KCl, 0.2 m H3PO4). After shaking briefly, the sample was centrifuged and the lower organic phase isolated, dried under nitrogen, and resuspended in 100 to 500 μL of chloroform. To an aliquot of the chloroform sample was added 10 μL of a 17:0 FA standard (0.25 mg mL−1) as an internal control. The sample was dried and resuspended in 1 mL of sulfuric acid solution and incubated at 80°C. Water (1.5 mL) and hexane (250 μL) were added and the sample was vigorously agitated and then centrifuged. Approximately 3 μL of the upper hexane phase was analyzed for FA composition on a gas chromatograph (model 5890A, Hewlett-Packard). The GC profile data were normalized to the internal 17:0 FA standard.
Inhibitor Treatment
Treatment of rice seeds with 100 μm cytochalasin B, 100 μm cytochalasin D, 100 μm nocodazole, or 25 mm NaF was performed using either intact rice panicles bearing mid-developing rice seeds or half-sectioned seeds. For panicle treatment, panicles were cut under water and transferred to a 15-mL test tube containing Murashige and Skoog medium supplemented with 2% Suc and a mixture of 21 amino acids (Donovan and Lee, 1977). Inhibitors were added to the medium and the tubes placed in a growth chamber under 11 h of light and 13 h of dark (30 h for the cytochalasin B and D and nocodazole treatments and 6 h for the NaF treatment). For half-seed treatments, seeds were sectioned longitudinally with a razor blade and placed in the same Murashige and Skoog medium with the appropriate drug and incubated in the dark at 26°C (6 h for the cytochalasin B and D treatment and 4 h for the NaF treatment). Seeds from these treatments were then frozen in liquid nitrogen and stored at −80°C and later processed as described above.
Protein and RNA Blotting
Protein and RNA were extracted from the same Suc- gradient fraction after the addition of one volume of phenol. For RNA extraction, the aqueous phase was extracted with phenol:chloroform:isoamyl alcohol (24:24:1, v/v) and the RNA in the aqueous phase was precipitated by the addition of 0.1 volume of sodium acetate and 2 volumes of 95% ethanol. The sample was placed at −20°C for at least 1 h, and the RNA was then pelleted in a microcentrifuge, washed with 70% ethanol, and resuspended in TE buffer (10 mm Tris-HCl, pH 7.5, and 1 mm Na2EDTA). For protein extraction, 3 to 5 volumes of methanol containing 0.77% ammonium acetate was added to the organic phase and the protein samples were incubated overnight at −20°C. The protein was pelleted in a microcentrifuge, washed in acetone, and resuspended in protein gel-loading buffer (Sambrook et al., 1989).
RNA samples were dot blotted to nitrocellulose along with known amounts of prolamine cDNA as standards and hybridized with radiolabeled prolamine or glutelin cDNA overnight at 65°C and washed under stringent conditions. RNA levels were quantified either by densitometric measurement of the radiographic signals or by phosphor imager (Bio-Rad) analysis. Direct comparison of these two methods showed that the results agreed within 5%. The protein samples were boiled for 5 min and then run on 5 to 15% SDS-PAGE gradient gels with a 4% stacking gel and transferred to nitrocellulose. The blots were probed with rice prolamine antisera, soybean calnexin antisera, or commercially available mouse anti-β-tubulin monoclonal antibody (T-9026, Sigma) and mouse anti-actin monoclonal antibody (N350, Amersham; 691001, ICN). Secondary antibodies were conjugated to horseradish peroxidase and were visualized by chemiluminescent detection (Super Signal chemiluminescence kit, Pierce).
RESULTS
Detergent and Salt Treatment of PB Fractions Indicates the Presence of Non-ER/Polysome-Binding Activity
Differential centrifugation of endosperm tissue extracted in CSB was used to isolate an enriched PB fraction. After filtration of the crude extract, most of the starch grains were removed by centrifugation at 100g for 5 min and an enriched PB fraction was obtained by centrifugation at 500g for 10 min. This fraction, although enriched with PBs, also contained a significant amount of C-ER membranes, as visualized by electron microscopy (Li et al., 1993a). The membranes in this PB fraction were solubilized by the addition of 1% Triton X-100 in CSB and the extract was then recentrifuged. The supernatant was removed and buffer U was added to the pellet to release the remaining bound polysomes. Polysomes were isolated from the pellet and the supernatant of the detergent extract by centrifugation at 200,000g for 3 h through a 60% Suc pad. The polysome pellets were solubilized in buffer U and quantified by A260. Only 22% of the total polysomes were released from the PB fraction by detergent treatment (Table I).
Table I.
Percentage release of polysomes and FAs from the detergent-washed PB fraction
| Fraction | Polysomes | FA Type
|
|||
|---|---|---|---|---|---|
| 16:1 | 18:1 | 18:2 | 18:3 | ||
| % | |||||
| Pellet | 78.0 | 4.2 | 4.8 | 3.8 | 2.8 |
| Supernatant | 22.0 | 95.8 | 95.2 | 96.2 | 97.2 |
A PB fraction was isolated in CSB as described in “Materials and Methods,” and then washed in CSB plus Triton X-100 and recentrifuged at 500g for 10 min to yield a pellet and a supernatant fraction.
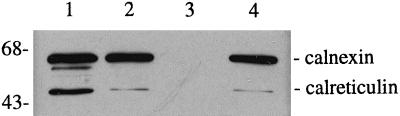
To determine the effectiveness of detergent treatment in solubilizing the membrane from the PB pellet, FA analysis was performed using GC. This analysis demonstrated that greater than 95% of each of the FAs (16:0, 18:1, 18:2, and 18:3) were found in the supernatant fraction of the detergent extract (Table I). These FAs constitute greater than 90% of the total FAs in the PB fraction and, therefore, are the major FA components of the membrane in this fraction. Protein gel-blot analysis and immunodetection of the ER membrane-bound chaperone calnexin demonstrated that calnexin is effectively released from the PB pellet after 1% Triton X-100 treatment (Fig. 1). This treatment, therefore, would also solubilize the ribosome receptor of the ER membrane translocation complex (Rapoport, 1992), thus releasing the polysomes from their ER membrane attachment site. Retention of the majority of polysomes in the detergent-washed pellet indicates that most of the polysomes in the PB fraction are associated with the PB surface at a second site, in addition to the ribosome-binding site on the ER membrane translocation complex.
Figure 1.
Protein gel blot demonstrating the release of calnexin from the detergent-washed PB fraction. A crude extract was obtained by grinding 1 g of seeds in 3 mL of CSB (lane 1). The extract was centrifuged at 500g, and the pellet was resuspended in 3 mL of CSB plus 1% Triton X-100 (the detergent extract, lane 2). The detergent extract was centrifuged to produce the pellet (resuspended in 3 mL of CSB plus Triton X-100; lane 3) and the supernatant (lane 4). Equal volumes of sample were loaded in each lane. Protein gel blots were probed with calnexin antisera. The upper bands correspond to the ER membrane-bound chaperone calnexin, and the lower bands correspond to the ER lumenal chaperone calreticulin, with which the calnexin antisera cross-reacts.
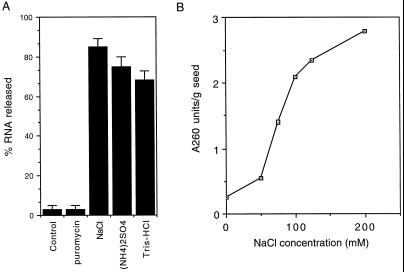
To determine if the anchoring of polysomes to the PB fraction is mediated by the nascent polypeptide or the RNA component of the polysomes, the PB fraction was washed twice with 1% Triton X-100 and then incubated at 25°C for 15 min in CSB containing 1 mm puromycin, a drug that mediates premature release of the polypeptide chain from the ribosome. We have previously shown that 1 mm puromycin effectively releases the nascent polypeptide chain from polysomes in rice seed extracts under similar conditions (Li et al., 1993b). Puromycin had no effect on the release of polysomes or mRNA from detergent-treated PB fraction (Fig. 2A), indicating that the interaction between the polysomes and the PBs is not mediated by the nascent prolamine polypeptide. Polysomes associated with the membrane-stripped PB fraction were, however, released by various solutions of relatively high ionic strength, including 200 mm Tris-HCl, pH 7.5, 2% ammonium sulfate, or 200 mm NaCl (Fig. 2A).
Figure 2.
Release of RNA and polysomes from the detergent-treated PB fraction. A, The detergent-treated PB fraction was incubated at 4°C for 15 min in the absence (control) or in the presence of 200 mm NaCl, 2% (NH4)2SO4, or 200 mm Tris-HCl, pH 7.5, and then centrifuged at 500g for 10 min. The RNA in the pellet and supernatant was isolated by phenol:chloroform extraction and ethanol precipitated, and then quantified by A260. The 1 mm puromycin-treated PBs were incubated in the absence of high salt concentrations. Bars indicate se. B, Release of polysomes from membrane-stripped PBs by NaCl. PBs were treated as above but in the presence of varying concentrations of NaCl, and then the released polysomes were fractionated through a 60% Suc pad, resuspended in buffer U, and quantified by A260. The amount of polysome release is estimated from A260 readings per gram of seed.
To further characterize the effectiveness of salt on the release of the polysomes from the PB fraction, we determined the concentration of NaCl required to release the polysomes. An enriched PB fraction was treated with varying concentrations of NaCl and then clarified by centrifugation at 500g. At least 75 to 100 mm NaCl was required to release 50% of the total released polysomes (Fig. 2B). The 100 mm NaCl-released polysome fraction was analyzed on a Suc-density gradient, and the absorbance profile revealed that the polysomes were intact, containing up to seven ribosomes per transcript (data not shown). The above data suggest that the polysomes are bound to the PB surface by a detergent-resistant, salt-sensitive binding site either through the mRNA or ribosome, but not through the nascent polypeptide chain.
Evidence for mRNA-Binding Activity
To elucidate whether the binding activity was interacting with the mRNA or the ribosomes of the polysome complex, we determined whether ribosome-free prolamine mRNAs could be associated with membrane-stripped PBs. As a strategy to increase the amount of cellular mRNAs lacking ribosomes, developing seeds were first treated with 25 mm NaF before the mRNA composition of the membrane-stripped PBs was analyzed. NaF has been used in both animal (Hoerz and McCarty, 1971; Lenk et al., 1977) and plant (García-Hernández et al., 1994) cells to dissociate polysome complexes by selectively inhibiting the initiation but not elongation step of protein synthesis. To test the efficiency of NaF on translation initiation, we monitored the incorporation of [3H]Leu into protein after 8, 12, 24, 36, and 48 h. In untreated seeds the incorporation rate was linear for up to 36 h, whereas seeds treated with NaF showed only a slight increase in incorporation after 8 h (data not shown).
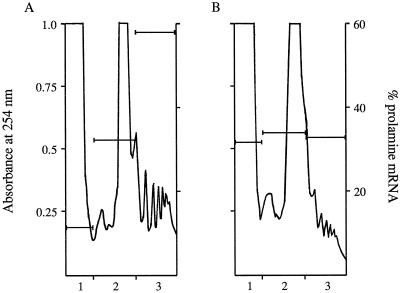
Membrane-stripped PB fractions isolated from control and 8-h NaF-treated seeds were prepared and extracted with buffer U to release the polysomes and ribosome-free mRNA from the PB fraction. The extract was then subjected to high-speed centrifugation to pellet the PBs. The clarified extract containing polysomes and ribosome-free mRNA was loaded onto a 15 to 60% Suc gradient and then centrifuged at 250,000g for 60 min (Fig. 3A). Fractions corresponding to soluble components, ribosomal subunits and monosomes, and polysomes were isolated. Total RNA was isolated from these fractions and the prolamine mRNA content was measured. A majority (90% of the total) of the prolamine mRNA in the control profile was present in the polysome (57%) and monosome (33%) fractions, with only a small amount (10%) present as ribosome-free species in the top soluble fraction (Fig. 3A). NaF treatment caused a decrease in the amount of polysomes and an increase in the amount of monosomes (Fig. 3B).
Figure 3.
Suc-gradient profiles of PB fractions from control seeds (A) or seeds treated with NaF (B). Seeds were treated with or without 25 mm NaF as described in the text. Membrane-stripped PBs were prepared and then washed in buffer U and repelleted. Equal volumes of the supernatant from control and NaF-treated seeds were layered onto 15 to 60% gradients. Bars represent percent prolamine mRNA content in each of the fractions as determined by dot-blot analysis. A se < 2.5% was routinely obtained in quantifying mRNA levels. Fractions correspond to soluble fraction (1) ribosomal subunit and monosome fraction (2), and polysome fraction (3).
This change in the polysome-monosome profile, however, had no effect on the total prolamine mRNA content, which was nearly equivalent (within 4.5%) to the control profile. Consistent with the change in the polysome-monosome profile, the distribution of prolamine mRNA was altered, especially in the polysome fraction of the Suc gradient. Prolamine mRNA content decreased approximately 25% in the polysome fraction compared with the control. However, a corresponding increase in prolamine mRNA content was evident in the soluble fraction, so that all three fractions contained nearly the same levels of prolamine mRNA. Hence, the prolamine mRNA percentages found in the NaF-treated samples represent an authentic shift in prolamine mRNA from the polysomes to ribosome-free mRNA rather than an artifact attributable to the degradation of mRNA. Similar results were seen when mid-developing seeds were sectioned longitudinally and then treated with 25 mm NaF in nutrient solution for 4 h. The increase in prolamine mRNA in the soluble fraction of the NaF gradient indicates that ribosome-free prolamine mRNA remains associated with membrane-stripped PBs. These results suggest that ribosome-free prolamine mRNA interacts with a putative RNA-binding activity near or on the PB surface.
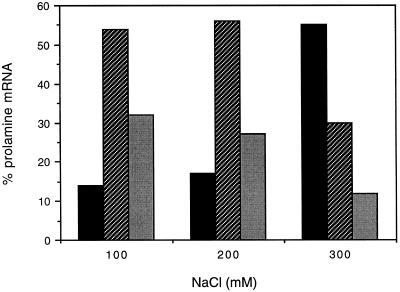
To further characterize the RNA-binding activity, membrane-stripped PBs from NaF-treated seeds were extracted successively with 100, 200, and 300 mm NaCl and centrifuged to remove the PBs. The supernatant of each of these treatments was then fractionated through 15 to 60% Suc gradients and then assessed for prolamine mRNA levels in the soluble fraction, ribosomal subunit/monosome fraction, and polysome fraction (Fig. 4). Because of the much longer incubation periods in this experiment, the polysomes were partially degraded, which altered the distribution of ribosome-associated prolamine RNAs in the Suc-density gradients. The bulk of the prolamine mRNA was associated with the ribosomal subunit/monosome fraction in the 100 and 200 mm salt extracts instead of the polysome fraction, as seen in Figure 3. Despite the apparent partial degradation of polysomes, the patterns of released prolamine mRNAs in each of the fractions from the three salt extractions were different (Fig. 4). Prolamine mRNA that was associated with monosomes predominated in the 100 and 200 mm salt extracts, but were a smaller component of the 300 mm extract. Prolamine mRNA that was associated with polysomes decreased in each of the successive salt-extraction steps, whereas ribosome-free prolamine mRNA was most efficiently released in the 300 mm salt fraction. Overall, these results indicate the presence of a prolamine mRNA-binding activity that requires relatively high ionic-strength levels to release the RNA from the PBs.
Figure 4.
Salt release of prolamine mRNA from NaF-treated seeds. The membrane-stripped PB fraction from NaF-treated seeds was extracted successively with 100, 200, and finally 300 mm NaCl. The total supernatant from each extract was fractionated onto a 15 to 60% Suc gradient. RNA was isolated from each of the fractions and prolamine mRNA quantified by dot-blot analysis. Black bars, Soluble; hatched bars, monosome; and shaded bars, polysome.
Isolation of PB Populations
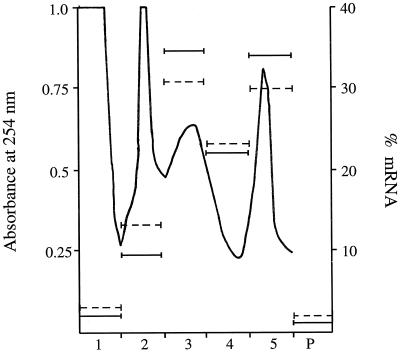
The resistance of the prolamine polysome-PB interaction to Triton X-100 detergent treatment but sensitivity to solutions of high ionic strength suggests that the prolamine polysomes may be anchored to the cytoskeleton. This type of detergent-resistant, salt-sensitive interaction exists for other cytoskeleton-bound polysomes (Davies et al., 1991, 1993; Pachter, 1992). To better characterize the specific interaction of the prolamine polysomes with the C-ER and PB-ER membranes and their possible interaction with the cytoskeleton, experiments were initiated to isolate enriched fractions of these two membrane types. Mid-developing rice seeds were ground in CSB, filtered through Miracloth, and layered onto a 20 to 80% Suc gradient and centrifuged at 300,000g for 60 min. Four major peaks were visible on the UV absorbance profile (Fig. 5), and consisted of: an upper peak corresponding to the soluble protein fraction (fraction 1); a narrow, second peak corresponding to the 80S monosome fraction (fraction 2); a broad, third peak (fractions 3 and 4); and a narrow, fourth peak (fraction 5).
Figure 5.
Suc-gradient (20–80%) profile of crude rice seed tissue extracted in CSB. One gram of seed was extracted in 3 mL of CSB and 250 μL of this was layered onto the gradient. The gradient was separated into six fractions, the last fraction corresponding to the dense pellet at the bottom of the gradient. Solid bars represent the percent prolamine mRNA content and dashed bars represent the percent glutelin mRNA content in each of the fractions as determined by dot-blot analysis.
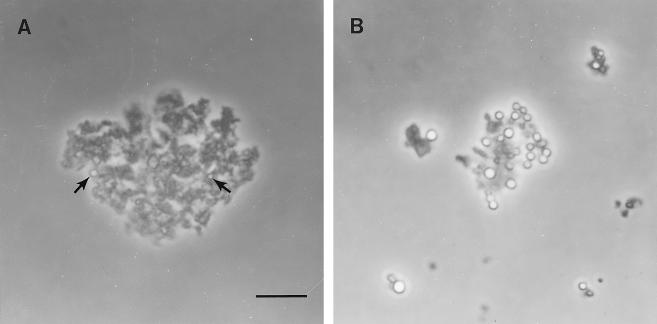
The positions of peaks 3 and 4 in the profile initially suggested that they corresponded to the C-ER and PB-ER, respectively, because the dense PBs would sediment lower in the gradient. Biochemical evidence, however, indicates that peaks 3 and 4 were qualitatively similar in composition; they both contained abundant levels of prolamine mRNAs in approximately equal amounts (Fig. 5). Prolamine polypeptides co-fractionated with prolamine mRNAs and were readily detectable in fractions corresponding to peak 3, although they were most abundant in peak 4 (see Fig. 7, A and B, top panels). In addition, glutelin mRNAs were not enriched in fractions corresponding to peak 3, but were found in peak 4 (Fig. 5). Phase-contrast microscopy of peaks 3 and 4 demonstrate that the PBs in these peaks differed in their size and abundance (Fig. 6). Peak 3 contained fewer, smaller PBs compared with the larger, more abundant PBs of peak 4. Overall, these observations indicate that peaks 3 and 4 are membrane fractions consisting of a complex of C-ER and PB-ER, which appear distinguishable by the size and abundance of the PBs. Peaks 3 and 4, therefore, will be referred to as light and heavy membrane fractions, respectively.
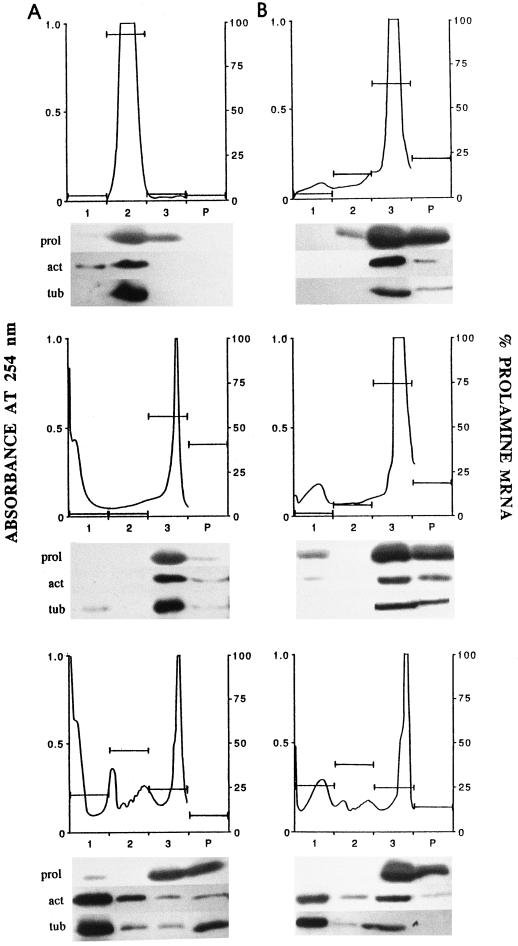
Figure 7.
Suc-density gradient UV-absorbance profiles of light (A) and heavy (B) membrane fractions. Light and heavy membrane fractions were isolated onto a 20 to 80% Suc-density gradient, divided into three aliquots, and then the aliquots were treated with CSB (top panels), CSB plus 1% PTE detergent (middle panels), or high-ionic-strength detergent solution (buffer U, bottom panels). These were then refractionated onto 20 to 80% Suc gradients as shown here. The numbers and letters at the bottom of each panel are fractions of the Suc-density gradient that correspond to soluble fraction (1), light membrane fraction (2), heavy membrane fraction (3), and pellet fraction (P). Bars represent percent prolamine mRNA content as in Figure 5. Below each Suc-density gradient profile are immunoblots of protein extracted from each fraction. Blots were probed with anti-prolamine (prol), anti-actin (act), and anti-tubulin (tub) antibodies.
Figure 6.
Phase-contrast microscopy of peak 3 (A, the light membrane fraction) and peak 4 (B, the heavy membrane fraction) showing representative complexes of membranes and PBs. Arrows indicate the smaller, less-abundant PBs among membranes in peak 3. Bar = 10 μm.
The Association of Prolamine mRNA, ER Membranes, and the Cytoskeleton
The light and heavy membrane fractions were isolated and individually incubated in low-ionic-strength CSB, CSB plus 1% PTE (a nonionic detergent), or high-ionic-strength buffer U at 4°C for 10 min, and then refractionated through a 20 to 80% Suc-density gradient (Fig. 7). The profiles of these gradients were divided into four fractions: fraction 1 corresponds to the supernatant; fraction 2 corresponds to the light membrane fraction; fraction 3 corresponds to the heavy membrane fraction; and fraction 4 (pellet fraction) consists of the dense pellet that was resuspended from the bottom of the centrifuge tube and, therefore, does not have a measured absorbance (Fig. 7). Both the light and heavy membrane fractions in CSB sedimented as expected when compared with their profile positions in Figure 5 (Fig. 7, A and B, top panels). Dot-blot hybridization and protein gel-blot analysis showed that the majority of prolamine mRNA, prolamine protein, actin, and tubulin co-fractionated with the light and heavy membrane fractions (Fig. 7, A and B, top panels). When the light membrane fraction was incubated in CSB plus 1% PTE to dissolve the membranes and then refractionated on the Suc-density gradient, the sedimentation profile changed markedly. The major absorbance peak increased in buoyant density and banded at the bottom of the gradient near the heavy membrane fraction (Fig. 7A, middle panel). Most of the prolamine mRNA, prolamine protein, actin, and tubulin was found to be associated with the densely sedimenting absorbance peak (Fig. 7A, middle panel), although there was a significant amount of mRNA and protein that pelleted to the bottom of the gradient. When the heavy membrane fraction was detergent treated and refractionated in the same manner, the major peak sedimented toward the bottom of the gradient with a slight increase in density compared with the peak observed in CSB alone (Fig. 7B, middle panel). Much of the prolamine mRNA, prolamine protein, actin, and tubulin co-sedimented with this major absorbance peak (Fig. 7B, middle panel).
Treatment of both the light and heavy membrane fractions with buffer U resulted in the release of monosomes and polysomes (Fig. 7, A and B, bottom panels). Prolamine mRNA was most abundant in the monosome/polysome fraction (fraction 2), whereas prolamine protein remained primarily associated with the heavy membrane fraction (fraction 3) in both profiles. Buffer U treatment of both the light and heavy fractions solubilized much of the actin and tubulin. However, some of these proteins were resistant to this treatment and remained in the lower fractions (Fig. 7, A and B, bottom panels). These experiments indicate that most of the prolamine polysomes are complexed not only with the ER membranes but with the cytoskeleton as well. In addition, because solubilization of the membrane with nonionic detergent did not release the prolamine mRNA, this would suggest that the prolamine polysomes are bound to the cytoskeleton, either directly or indirectly. It is interesting that glutelin polysomes behaved in a manner similar to prolamine polysomes under these conditions (data not shown), suggesting that a common association may exist between the prolamine and glutelin polysomes and their respective ER domains.
The Effect of Cytoskeleton Inhibitors on the Association of Prolamine mRNA and PBs
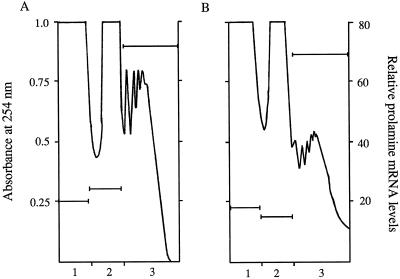
In an effort to determine if there is a functional association of the cytoskeleton with prolamine mRNA, the effects of cytoskeleton-destabilizing drugs on polysome release were studied. Cytochalasin B and nocodazole were used to destabilize microfilaments and microtubules, respectively. Panicles bearing mid-developing seeds were immersed in nutrient solution alone or solution containing 100 μm cytochalasin B or 100 μm nocodazole for 30 h. Seeds were ground in buffer U to solubilize the membranes and cytoskeleton, and the extract was fractionated on a polysome gradient. The polysome profiles of control and cytochalasin-treated seed extracts are shown in Figure 8. The cytochalasin B-treated seed profile showed a reduced amount of polysomes compared with the control seeds. However, no significant decrease in prolamine mRNA in the cytochalasin B-treated versus the control samples was observed (Fig. 8).
Figure 8.
Suc-gradient profiles from control seeds (A) or seeds treated with 100 μm cytochalasin B (B) for 30 h. Seeds (500 mg) were ground in 2 mL of buffer U and filtered before layering 200 μL onto a 15 to 60% gradient. Bars represent relative prolamine concentrations as determined from dot-blot scans.
To determine if a certain percentage of prolamine polysomes in the cytochalasin-treated seeds were in fact released from the cytoskeleton but not immediately degraded, we determined the prolamine mRNA content in a PB/cytoskeleton fraction to eliminate any free polysomes. Control and cytochalasin B-treated seeds were extracted in CSB plus 1% Triton X-100 and the extract was fractionated through a 20 to 80% Suc gradient. The major absorbance peak contained the PB/cytoskeleton fraction. Total RNA was isolated from this peak from both the control and the cytochalasin B-treated samples. Dot-blot analysis using a prolamine probe showed that there was only 3.5 ± 2% less prolamine mRNA associated with this fraction in cytochalasin B-treated than in control plants. Similar results were seen when 100 μm cytochalasin D was used in place of cytochalasin B, and when half-sectioned seeds were treated for 6 h with these cytoskeleton-destabilizing drugs rather than panicles. These results suggest that the microfilament network that is associated with the PB surface is not involved in anchoring prolamine polysomes or mRNA, or that this network is resistant to the destabilizing effects of cytochalasin. Nocodazole treatment showed a very similar profile and prolamine mRNA content in each of the fractions compared with the control (data not shown), suggesting that the microtubule component of the cytoskeleton does not play a role in binding of cellular polysomes to the cytoskeleton.
DISCUSSION
The association of mRNAs and polysomes with the cytoskeleton has been well documented in several animal cell types, including Drosophila and Xenopus spp. oocytes, and certain types of cultured cells such as HeLa, fibroblasts, and oligodendrocytes (for review, see Hesketh, 1994; St. Johnston, 1995). One of the functions of this interaction is to localize mRNAs as a mechanism for achieving translation in specific locations within the cell. In plants the association of polysomes with the cytoskeleton has been demonstrated in pea roots and stems and in maize endosperm (Davies and Abe, 1989; Davies et al., 1993; Ito et al., 1994). The microfilament component of the cytoskeleton in maize endosperm cells is largely concentrated in spheres surrounding the PB (Abe et al., 1991; Clore et al., 1996). Polysomes are localized to this cytoskeleton and are released by solutions of high ionic strength and other reagents that depolymerize actin, but not PTE (Davies et al., 1993). The percentage of release of polysomes from the maize PBs by increasing concentrations of salt is proportional to the number of ribosomes associated with the polysome, suggesting that the ribosome anchors the polysome to the cytoskeleton (Davies et al., 1993). We show that, as in maize, in rice endosperm PB fractions most of the polysomes remain associated with the cytoskeleton after extraction in nonionic detergent and are subsequently released after treatment with high-ionic-strength detergent (Figs. 2 and 7). These data suggest that F-actin seems to be the scaffold upon which other components (i.e. membranes, microtubules, and polysomes) are associated (Davies et al., 1993). Our results differ from those seen in maize with respect to the type of interaction of the prolamine polysomes with the cytoskeleton. The results of the NaF treatments (Figs. 3 and 4) demonstrate that the prolamine mRNA cytoskeleton interaction in rice endosperm is mediated by the RNA, but do not rule out the additional role of ribosomes in this interaction. The ribosome-free prolamine mRNA appears to bind more tightly to the cytoskeleton than do polysomes because higher concentrations of salt are required to release these ribosome-free mRNAs (Fig. 4). In animal systems there is evidence for the association of the mRNA and the ribosome with the cytoskeleton, and it is suggested that both have an anchoring role (Hesketh, 1994).
Crude rice endosperm extracts can be resolved into two membrane fractions on Suc-density gradients. The buoyant densities of these two membrane fractions (peaks 3 and 4; Fig. 5) initially led us to believe that the C-ER membranes were enriched in peak 3 and the PB-ER membranes were enriched in peak 4. However, the presence of equal amounts of prolamine and glutelin mRNAs in each of the fractions suggests that both the C-ER and PB-ER membranes were present in both fractions, since our previous work demonstrated that glutelin mRNA is enriched on the C-ER and prolamine mRNA is enriched on the PB-ER (Li et al., 1993a). The distinguishing feature between these two fractions is that they differ in PB size and abundance (Fig. 6). The fact that both membrane fractions are associated with the cytoskeleton (Fig. 7) may provide an explanation for the finding that both ER types are found in the light and heavy membrane fractions. As shown previously in maize, F-actin appears to hold zein PBs into aggregates in crude extracts prepared in CSB (Abe et al., 1991). In rice F-actin also surrounds the prolamine PBs with longer, less densely staining filaments running throughout the cytoplasm (D.G. Muench and T.W. Okita, unpublished data). Perhaps this cytoskeleton network also associates with the C-ER network, thus forming a C-ER/PB-ER complex held together by the cytoskeleton. Complexes containing large PBs would then have a greater density and would sediment to the lower membrane fraction, whereas those ER membranes having very small, developing PBs, or none at all, would be less dense and would sediment to the light membrane fraction. The membrane component of the light membrane fraction is responsible for its buoyancy, since treatment of this fraction with PTE resulted in a dramatic shift to the lower portion of the gradient (Fig. 7A, top and middle panels). The large PBs in the heavy membrane fraction counter the buoyancy of the membrane in this fraction; however, upon PTE treatment there was a small shift in the sedimentation of this peak (Fig. 7B, top and middle panels).
Several researchers have shown that fragments of insolubilized ER membranes remain after cells are treated with nonionic detergent (Dang et al., 1983; Hesketh and Pryme, 1991; Vedeler et al., 1991). These insolubilized membrane remnants may be responsible for associating membrane-bound polysomes with the detergent-resistant fraction, even though these polysomes may not be associated with the cytoskeleton. Other researchers claim that nonionic detergents adequately dissolve the membrane and allow for the release of membrane-bound polysomes with high-ionic-strength solutions (Lenk et al., 1977; Adams et al., 1983; Fey et al., 1986). For example, maize PB fractions treated with nonionic detergent completely dissolved the membrane, as determined by lipid analysis and loss of fluorescent membrane staining (Abe et al., 1991; Ito et al., 1994). We have shown that Triton X-100 is very efficient in solubilizing the membrane and membrane-associated proteins in the PB pellet (Table I; Fig. 1). Although it is possible that a very small amount of membrane remains in our extracts, the fact that ribosome-free prolamine mRNAs are still associated with the cytoskeleton (Fig. 3) demonstrates that ribosome-membrane interactions are not required for the anchoring of prolamine mRNA to the PB.
Cytochalasin B treatment caused a reduction in the amount of total polysomes in the cell; however, it does not release prolamine mRNA from the cytoskeleton (Fig. 8). Although this was an unexpected result, it may be that prolamine mRNA is bound to a population of F-actin that has some resistance to the effects of cytochalasin treatment. When rice endosperm sections are treated with cytochalasin B or D and then F-actin is visualized by Texas red-phalloidin staining, there is no visible effect on the quantity or morphology of the actin around the PBs, even though the cells showed actin aggregation around the nucleus (D.G. Muench and T.W. Okita, unpublished data), which is a common result of cytochalasin treatment (Spector et al., 1989). This result differs from what was seen in maize, in which treatment of endosperm sections with cytochalasin D resulted in obvious changes in actin organization when visualized by indirect immunofluorescence (Clore et al., 1996).
Alternatively, it may be that prolamine mRNAs are not complexed with actin or tubulin, but with a different component of the ER-associated cytoskeleton. An example of an mRNA-cytoskeleton interaction such as this was shown by a specific membrane-bound mRNA in HeLa cells (Zambetti et al., 1990a). In that study cytochalasin D and puromycin together did not release the mRNA from the membrane. Because microtubules did not fractionate with these membranes, the authors suggested that another component of the cytoskeleton is binding the mRNAs, perhaps the intermediate filaments, and they raised the possibility that mRNAs are associated with the cytoskeleton in a heterogeneous manner. Results from other studies are also consistent with this view. In Fucus spp. actin mRNA is localized to the cell plate in dividing embryos; however, drugs that affect the integrity of the cytoskeleton do not affect the localization of the actin mRNA (Bouget et al., 1996). Also, microfilament-destabilizing drugs did not affect the association of actin mRNA and histone mRNA with the cytoskeleton in ascidian eggs (Jeffery, 1984).
Okita et al. (1994) proposed three possible models for the nonrandom localization of rice storage protein mRNAs to the ER in endosperm cells. One model suggests that there are different populations of signal-recognition particles that associate with specific subdomains of the ER, so that prolamine and glutelin mRNAs are docked to different domains via their encoded signal peptides. The second model proposes that the long-lasting interaction of BiP to the nascent and mature prolamine polypeptide and to the prolamine PB (Li et al., 1993b; Muench et al., 1997) could enrich prolamine mRNAs around the PB because of a longer retention time on the PB-ER. The third model proposes a direct RNA-sorting mechanism as a result of an RNA signal that would specifically target prolamine mRNA to the PB-ER in association with the cytoskeleton (Okita et al., 1994). Although we cannot argue for or against any of these mechanisms as being responsible for the enrichment of prolamine mRNA to the PB-ER, it appears that this mRNA associates with the cytoskeleton that surrounds the prolamine PBs. Because glutelin polysomes are also cytoskeleton bound, the anchoring mechanism on the surface of the ER may be the same between these two polysome types. Therefore, the differential sorting of prolamine and glutelin mRNAs may occur before their anchoring at the surface of the respective ER membranes. The fact that ribosome-free prolamine mRNA retains the ability to bind to the cytoskeleton suggests that it may be localized before translation in a manner similar to that by which transcripts such as bicoid or oskar are localized in Drosophila spp., or myelin basic protein are localized in oligodendrocytes (St. Johnston, 1995). Our current efforts are focused on determining the mechanism that mediates the localization of storage protein mRNAs in rice.
ACKNOWLEDGMENT
We thank Dr. Anders Carlsson for his assistance with the FA analysis.
Abbreviations:
- C-ER
cisternal ER
- CSB
cytoskeleton-stabilizing buffer
- FA
fatty acid
- PB
protein body
- PTE
polyoxyethylene 10-tridecyl
- X:Y
a fatty acyl group containing X carbon atoms and Y cis double bonds
Footnotes
This work was supported by U.S. Department of Agriculture National Research Initiative Competitive Grants Program Award no. 94-37304-1174 to T.W.O.
LITERATURE CITED
- Abe S, Davies E. Isolation of F-actin from pea stems: evidence from fluorescence microscopy. Protoplasma. 1991;162:51–61. [Google Scholar]
- Abe S, You W, Davies E. Protein bodies in corn endosperm are enclosed by and enmeshed in F-actin. Protoplasma. 1991;165:139–149. [Google Scholar]
- Adams A, Fey E, Pike S, Taylorson C, White H, Rabin E. Preparation and properties from a complex of rat liver polyribosomes with components of the cytoskeleton. Biochem J. 1983;216:215–226. doi: 10.1042/bj2160215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bag J, Pramanik S. Attachment of mRNA to the cytoskeletal framework and translational control of gene expression in rat L6 muscle cells. Biochem Cell Biol. 1987;65:565–575. doi: 10.1139/o87-073. [DOI] [PubMed] [Google Scholar]
- Bouget FY, Gertulla S, Shaw SL, Quatrano RS. Localization of actin mRNA during the establishment of cell polarity and early cell divisions in Fucus embryos. Plant Cell. 1996;8:189–201. doi: 10.1105/tpc.8.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore A, Dannenhoffer J, Larkins B. eF1A is associated with a cytoskeleton network surrounding protein bodies in maize endosperm cells. Plant Cell. 1996;8:2003–2014. doi: 10.1105/tpc.8.11.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C, Yang D, Pollard T. Association of methionyl-tRNA synthetase with detergent-insoluble components of the rough endoplasmic reticulum. J Cell Biol. 1983;96:1138–1147. doi: 10.1083/jcb.96.4.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies E, Abe S. Isolation and role for cytoskeleton-bound polysomes in a variety of plants (abstract no. 425) Plant Physiol. 1989;89:S-71. [Google Scholar]
- Davies E, Abe S. Method for isolation and analysis of polyribosomes. Methods Cell Biol. 1995;50:209–221. doi: 10.1016/s0091-679x(08)61032-8. [DOI] [PubMed] [Google Scholar]
- Davies E, Comer EC, Lionberger JM, Stankovic B, Abe S. Cytoskeleton-bound polysomes in plants. III. Polysome-cytoskeleton-membrane interactions in corn endosperm. Cell Biol Int. 1993;17:331–340. [Google Scholar]
- Davies E, Fillingham BD, Oto Y, Abe S. Evidence for the existence of cytoskeleton-bound polysomes in plants. Cell Biol Int Rep. 1991;15:975–981. doi: 10.1016/0309-1651(91)90147-b. [DOI] [PubMed] [Google Scholar]
- Donovan G, Lee J. The growth of detached wheat heads in liquid culture. Plant Sci Lett. 1977;9:107–113. [Google Scholar]
- Fey E, Ornells D, Penman S. Association of RNA with the cytoskeleton and the nuclear matrix. J Cell Sci. 1986;5:99–120. doi: 10.1242/jcs.1986.supplement_5.6. [DOI] [PubMed] [Google Scholar]
- García-Hernández M, Davies E, Staswick PE. Arabidopsis p40 homologue: a novel acidic protein associated with the 40S subunit of ribosomes. J Biol Chem. 1994;269:20744–20749. [PubMed] [Google Scholar]
- Hesketh J. Translation and the cytoskeleton: a mechanism for targeted protein synthesis. Mol Biol Rep. 1994;19:233–243. doi: 10.1007/BF00986965. [DOI] [PubMed] [Google Scholar]
- Hesketh J, Pryme I. Interaction between mRNA, ribosomes and the cytoskeleton. Biochem J. 1991;277:1–10. doi: 10.1042/bj2770001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerz W, McCarty KS. Initiation of protein synthesis in a rabbit reticulocyte system. Biochim Biophys Acta. 1971;228:526–535. doi: 10.1016/0005-2787(71)90058-x. [DOI] [PubMed] [Google Scholar]
- Howe JG, Hershey JWB. Translational initiation factor and ribosome association with the cytoskeletal framework fraction from HeLa cells. Cell. 1984;37:85–93. doi: 10.1016/0092-8674(84)90303-9. [DOI] [PubMed] [Google Scholar]
- Ito Y, Abe S, Davies E. Co-localization of cytoskeleton proteins and polysomes with a membrane fraction from peas. J Exp Bot. 1994;45:253–259. [Google Scholar]
- Jeffery W. Spatial distribution of messenger RNA in the cytoskeletal framework of ascidian eggs. Dev Biol. 1984;103:482–492. doi: 10.1016/0012-1606(84)90335-x. [DOI] [PubMed] [Google Scholar]
- Krishnan HB, Franceschi VR, Okita TW. Immunochemical studies on the role of the Golgi complex in protein body formation in rice seeds. Planta. 1986;169:471–480. doi: 10.1007/BF00392095. [DOI] [PubMed] [Google Scholar]
- Lenk R, Ransom L, Kaufmann Y, Penman S. A cytoskeletal structure with associated polyribosomes obtained from HeLa cells. Cell. 1977;10:67–78. doi: 10.1016/0092-8674(77)90141-6. [DOI] [PubMed] [Google Scholar]
- Li X, Franceschi VR, Okita TW. Segregation of storage protein mRNAs on the rough endoplasmic reticulum membranes of rice endosperm cells. Cell. 1993a;72:869–879. doi: 10.1016/0092-8674(93)90576-c. [DOI] [PubMed] [Google Scholar]
- Li X, Wu Y, Zhang D-Z, Gillikin JW, Boston R, Franceschi VR, Okita TW. Rice prolamine protein body biosynthesis: a BiP-mediated process. Science. 1993b;262:1054–1056. doi: 10.1126/science.8235623. [DOI] [PubMed] [Google Scholar]
- Miquel M, Browse J. Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis. J Biol Chem. 1992;267:1502–1509. [PubMed] [Google Scholar]
- Muench DG, Okita TW. The storage proteins of rice and oat. In: Larkins BA, Vasil IK, editors. Cellular and Molecular Biology of Plant Development. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 289–330. [Google Scholar]
- Muench DG, Wu Y, Zhang Y, Li X, Boston R, Okita TW. Molecular cloning, expression and subcellular localization of a BiP homolog from rice endosperm tissue. Plant Cell Physiol. 1997;38:404–412. doi: 10.1093/oxfordjournals.pcp.a029183. [DOI] [PubMed] [Google Scholar]
- Okita TW, Li X, Roberts MW. Targeting of mRNAs to domains of the endoplasmic reticulum. Trends Cell Biol. 1994;4:91–96. doi: 10.1016/0962-8924(94)90181-3. [DOI] [PubMed] [Google Scholar]
- Pachter JS. Association of mRNA with the cytoskeletal framework: its role in the regulation of gene expression. Critical Reviews in Eukaryotic Gene Expression. 1992;2:1–18. [PubMed] [Google Scholar]
- Rapoport TA. Transport of proteins across the endoplasmic reticulum membrane. Science. 1992;258:931–936. doi: 10.1126/science.1332192. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Spector I, Shochet N, Blasberger D, Kashman Y. Latrunculins: novel marine macrolides that disrupt microfilament organization and affect cell growth. I. Comparison with cytochalasin D. Cell Motil Cytoskeleton. 1989;13:127–144. doi: 10.1002/cm.970130302. [DOI] [PubMed] [Google Scholar]
- Stankovic B, Abe S, Davies E. Co-localization of polysomes, cytoskeleton, and membranes with protein bodies from corn endosperm. Protoplasma. 1993;177:66–72. [Google Scholar]
- St. Johnston D. The intracellular localization of messenger RNAs. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- Sundell CL, Singer RH. Requirement of microfilaments in sorting of actin messenger RNA. Science. 1991;253:1275–1277. doi: 10.1126/science.1891715. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Sugimoto T, Ogawa M, Kasai Z. Isolation and characterization of two types of protein bodies in the rice endosperm. Agric Biol Chem. 1980;44:1633–1639. [Google Scholar]
- Vedeler A, Pryme I, Hesketh J. The characterization of free, cytoskeletal and membrane-bound polysomes in Kregs II ascites and 3T3 cells. Mol Cell Biochem. 1991;100:183–193. doi: 10.1007/BF00234167. [DOI] [PubMed] [Google Scholar]
- Yisraeli JK, Sokol S, Melton DA. A two step model for the localization of maternal mRNA in Xenopus oocytes: involvement of microtubules and microfilaments in the translocation and anchoring of vg1 mRNA. Development. 1990;108:289–298. doi: 10.1242/dev.108.2.289. [DOI] [PubMed] [Google Scholar]
- You W, Abe S, Davies E. Cosedimentation of pea root polysomes with the cytoskeleton. Cell Biol Int Rep. 1992;16:663–673. doi: 10.1016/s0309-1651(06)80008-1. [DOI] [PubMed] [Google Scholar]
- Zambetti G, Fey E, Penman S, Stein J, Stein G. Multiple types of mRNA-cytoskeleton interactions. J Cell Biochem. 1990a;44:177–187. doi: 10.1002/jcb.240440306. [DOI] [PubMed] [Google Scholar]
- Zambetti G, Wilming L, Fey EG, Penman S, Stein J, Stein G. Differential association of membrane-bound and non-membrane bound polysomes with the cytoskeleton. Exp Cell Res. 1990b;191:246–255. doi: 10.1016/0014-4827(90)90011-x. [DOI] [PubMed] [Google Scholar]