Abstract
Cotesia vestalis is an endoparasitic wasp that attacks larvae of the diamondback moth (Plutella xylostella), a herbivore of cruciferous plants. Females of C. vestalis use herbivore-induced plant odorants released from plants infested by P. xylostella as a host-searching cue. Transcriptome pyrosequencing was used to identify genes in the antennae of C. vestalis adult females coding for odorant receptors (ORs) and odorant binding proteins (OBPs) involved in insect olfactory perception. Quantitative gene expression analyses showed that a few OR and OBP genes were expressed exclusively in the antenna of C. vestalis adult females whereas most other classes of genes were expressed in the antennae of both males and females, indicating their diversity in importance for the olfactory sensory system. Together, transcriptome profiling of C. vestalis genes involved in the antennal odorant-sensory system helps in detecting genes involved in host- and food-search behaviors through infochemically-mediated interactions.
Introduction
In insect sensory systems antennae, maxillary palps and/or labial palps are the main olfactory organs. In these specific parts, olfactory sensory neurons (OSNs) are enclosed in hair-like cuticular structures with multiple pores, called sensilla. The OSN dendrites are submersed in the sensillum lymph that contains water soluble odorant binding proteins (OBPs) and chemosensory proteins (CSPs) [1]. Some enzymes, specifically expressed in the antennae [pheromone degrading enzymes (PDE) or carboxylesterase (CCE)] are also secreted and thought to be involved in signal inactivation [2], [3]. The two large gene families expressing OBPs and odorant receptors (ORs), however, characterize the molecular basis of insect olfaction and are assumed to be exclusive to this group of animals [4].
OBPs are small (10 to 30 kDa) globular and highly abundant proteins. They are thought to be involved in the uptake of volatile, hydrophobic compounds from the environment and their translocation to odorant receptors located in the ORN membranes [5]–[8]. Additionally, OBPs have been suggested to filter or purify odorants [8], [9], to act as activator factors of ORs (after conformational change) [10] or as carriers expressed in non-olfactory tissue [11], [12].
Insect ORs have recently been recognized as ligand-gated ion channels causing an inward cationic current [13], [14]. In vivo and in vitro assays show that ORs are active in a heteromeric complex of unknown stoichiometry composed of the OR and the universal co-receptor (ORCO) [14]–[17].The ORs are thought to be essential for the recognition of a variety of odorant compounds [18], [19] and to be involved in the pore construction and current determination [20], whereas ORCO is required for neuronal cell-surface targeting and proper signal transduction [21], [22].
Olfaction is important for fundamental behaviors such as feeding, mating and interaction with a prey or host. Cotesia vestalis (Haliday) (Hymenoptera: Braconidae) is an endoparasitic wasp that attacks larvae of the diamondback moth (DBM), Plutella xylostella L. (Lepidoptera: Yponomeutidae), an oligophagous herbivore of cruciferous plants. Female C. vestalis use herbivore-induced plant volatiles (HIPVs) from plants infested by P. xylostella larvae as host-searching cues [23]. A mixture of four HIPVs (n-heptanal, α-pinene, sabinene and (Z)-3-hexenyl acetate) released from P. xylostella-infested cabbage plants triggers innate chemotaxis in naive parasitoids, whereas the individual compounds by themselves do not [24]. Therefore, it is not a single HIPV but rather a blend of different HIPVs that is recognized by the natural enemies of herbivores [24]–[26]. However, little is known about the sensory mechanisms underlying complicated HIPV recognition.
We surveyed sensory genes, especially for ORs and OBPs, expressed in the antennae of C. vestalis adult females by Roche 454 transcriptome pyrosequencing to partially evaluate the odorant-sensory systems. To date, transcriptome pyrosequencing is the most widely used system for de novo sequencing and analysis of transcriptomes especially in non-model organisms. In addition, the OR and some of OBP genes identified were examined in order to evaluate their tissue and gender-specific patterns of expression.
Materials and Methods
Plants and Insects
Cabbage plants (Brassica oleracea L. cv. Shikidori) were cultivated in a growth chamber at 25°C with a photoperiod of 16 h (natural+supplemental light). DBM (P. xylostella L. [Lepidoptera: Yponomeutidae]) were collected originally from fields of Kyoto Prefecture, Japan, in 2001. No specific permits were required for the field studies. The land was not privately owned or under any statutory protection and the field studies did not involve endangered or protected species. DBM collected were mass-reared on potted cabbage plants in a climate-controlled room (25°C, 16 h photoperiod). C. vestalis (Hymenoptera: Braconidae) were obtained from the above field-collected, parasitized larvae of DBM. To obtain further generations of C. vestalis, adult wasps were placed with DBM larvae on cabbage plants in plastic-glass cages (25×35×30 cm; three windows covered by nylon gauze and one door for introducing plants and wasps). These cages were maintained in a climate-controlled room (25°C, 16 h photoperiod) for 4 days. The parasitized DBM larvae were isolated and held in a plastic container (1.6 L) with a few cabbage leaves for a week. Every C. vestalis cocoon was transferred into a plastic cage (0.2 L) and incubated until the adults emerged (4–7 days). The emerged adult wasps were collected within 24 h and temporarily stored at −80°C. This amounted to 720 females and 680 males sampled wasps. Each part of C. vestalis (antenna, head and body) was dissected on dry ice and collected in an Eppendorf tube. The tubes contained either 60 antennae, 30 heads or 30antennae and each tube constituted a unit sample. All tubes were stored at −80°C until processed.
RNA Preparation, cDNA Library Construction and 454 Sequencing
Total RNA from 6 unit samples (a total of 360 female antennae) was isolated using a Qiagen RNeasy Mini Kit and an RNase-Free DNase Set (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Total RNA (4.5 µg) was then taken through mRNA purification using a RiboMinus Eukaryote Kit for RNA-Seq (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions (yielding 600 ng mRNA). Total RNA and mRNA quality were assessed by an Agilent Bioanalyzer RNA 6000 Pico Assay (Agilent Technologies, Santa Clara, CA, USA) and quantified by a Quant-iT RiboGreen Assay (Invitrogen).
Some of the odorant-sensory genes were predicted to be expressed at very low levels. In order to evenly mine genes expressed in the antenna at widely dissimilar levels, we prepared 3 libraries constructed using three types of cDNA sources: amplified cDNAs from total RNA (library 1), amplified cDNAs from mRNA (library 2), and non-amplified cDNAs from mRNA (library 3). The cDNAs were synthesized and amplified from mRNA or total RNA (50 ng each) with the Ovation RNA-Seq System (NuGEN, San Carlos, CA, USA) according to the manufacturer’s instructions, and the amplified double-stranded cDNA fragments were used in the following steps. For the preparation of non-amplified cDNAs, the mRNA (200 ng) was fragmented in 10 mM ZnCl2 and 10 mM Tris-HCl (pH 7.0) at 70°C for 30 s, and the double-stranded cDNA was synthesized using the cDNA Synthesis System Kit with random hexamer primers (Roche Applied Science, Indianapolis, IN, USA). The cDNA fragments were subjected to ligation to the sequencing adaptors provided with the GS FLX Titanium Rapid Library Preparation Kit (Roche Applied Science), and small fragments were removed with AMPure XP (Beckman Coulter, Fullerton, CA, USA). Sequencing was performed on a GS FLX platform with Titanium chemistry (Roche/454) using a Medium/Small region of a PicoTiterPlate (PTP) per library, following the manufacturer’s instructions. Libraries 1 and 2 were sequenced using one region on the plate and library 3 using two.
454 de novo Transcriptome Assembly and Analysis
A pool of the processed sequence reads from the cDNA libraries was clustered and assembled using the TGICL pipeline version 2.1 [27] with a minimum sequence overlap of 49 nt and a minimum percentage overlap identity of 80%. The primary assembling eventually resulted in 17 328 contigs from the valid sequence reads. To annotate the dataset, both the contigs and singletons were searched for sequence similarity using BLASTX [28] against the NCBI RefSeq protein database (http://www.ncbi.nlm.nih.gov/RefSeq/), and the Gene Ontology (GO) analysis was conducted using the UniProt-GOA database (http://www.ebi.ac.uk/GOA/). In order to specifically annotate OBP and OR genes, the contigs and singletons were also searched for sequence similarity using BLASTX against the reported OBP and OR sequences of Nasonia vitripennis [29], [30] and the UniProtKB/TrEMBL database (http://www.ebi.ac.uk/uniprot/). Contigs and singletons with an E-value of 1E-5 or lower, on the BLAST hits, were used.
Quantitative Reverse Transcriptase (RT)-PCR
Five independent replications of total RNA samples were prepared from about 30 body, 120 head or 480 antenna tissues of C. vestalis males or females, using a Qiagen RNeasy Mini Kit and an RNase-Free DNase Set (Qiagen) following the manufacturer’s protocol. First-strand cDNA was synthesized using a PrimeScript RT reagent Kit (Takara, Otsu, Japan), and 0.5 µg of total RNA at 37°C for 15 min. Real-time PCR was done on an Applied Biosystems 7300 Real-Time PCR System (Foster City, CA, USA) using Power SYBR Green Master Mix (Applied Biosystems), cDNA (1 µl from 20 µl of each RT product pool), and 300 nM primers. The PCR program was: initial polymerase activation 2 min at 50°C, and 10 min at 95°C, 40 cycles of 15 s at 95°C, and 60 s at 60°C. PCR conditions were chosen by comparing threshold values in a dilution series of the RT product, followed by non-RT template control and non-template control for each primer pair. Relative RNA levels were calibrated and normalized with the level of those of 60S ribosomal protein L10 (Cv_000471) for C. vestalis. Primers for real-time PCR were designed using the Primer-BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) for a length of the resulting PCR product of approximately 150 bp. Primers used for this study are shown in Table S1.
Results
An Overview of the Transcriptome of the Antennae of Adult C. vestalis Female
We constructed three libraries using three different types of cDNA sources: amplified cDNAs from total RNA (library 1), amplified cDNA from mRNA (library 2), and two non-amplified cDNAs from mRNA (libraries 3-1 and 3-2). From these the pyrosequencing yielded 76 892, 110 009, 122 436 and 133 578 valid reads, respectively, for a total of 442 915 reads (Table 1). However, after excluding ribosomal RNA (rRNA), the number of valid reads strongly decreased to 247 361 and showed that only library 2 was not contaminated with rRNA (93 009 valid reads minus rRNA). This was probably due to the enrichment of mRNA through two rounds of mRNA purification: the first using the RiboMinus Eukaryote Kit (Invitrogen) after purification; and the second using the Ovation RNA-Seq System (NuGEN), in which Oligo dT primers were used for RT reaction (see Materials and Methods). After cleaning the data, the high quality reads were assembled into 17 328 contigs and 31 921 singletons (Table 1; DDBJ accession number: DRA000551). The contigs had an average size of 549 base pairs (bp).
Table 1. Summary of the materials and results of pyrosequencing.
| Category | Library 1 | Library 2 | Library 3-1 | Library 3-2 | Total |
| RNA source | TotalRNA | mRNA | mRNA | mRNA | |
| cDNA amplification | NuGEN | NuGEN | Roche Std. | Roche Std. | |
| Run regions | 1/8 | 1/8 | 1/8 | 1/8 | |
| Valid reads | 76 892 | 110 009 | 122 436 | 133 578 | 442 915 |
| Valid reads(minus rRNA) | 32 093 | 93 009 | 56 899 | 65 360 | 247 361 |
| Number ofcontigs | 17 328 | ||||
| Number ofsingletons | 31 921 | ||||
| Average sizeof contigs | 549 | ||||
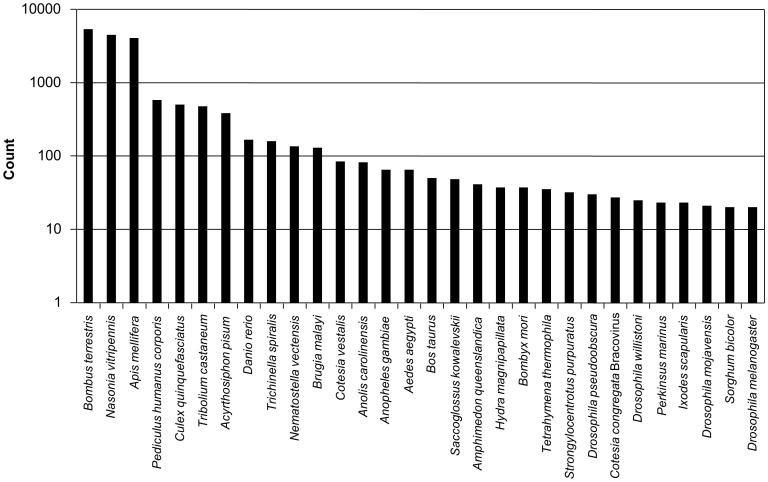
The BLAST searches of contigs/singletons against the UniProtKB/TrEMBL database found the C. vestalis sequences to be similar to amino acid sequences from two bee species Apis mellifera (4051 hits with E-values ≤1E-5) and Bombus terrestris (5346 hits) and the parasitoid wasp Nasonia vitripennis (4454 hits) (Figure 1). Sequences from some other insects such as Pediculus humanus corporis, Culex quinquefasciatus, Tribolium castaneum, and Acyrthosiphon pisum were also similar to C. vestalis contigs/singletons, whereas the model insect Drosophila sp. showed low-hit similarities.
Figure 1. Species distribution of top hits for the pyrosequencing contigs/singletons from C. vestalis adult female antennae (n = 17 597; 35.7%, BLAST hits).
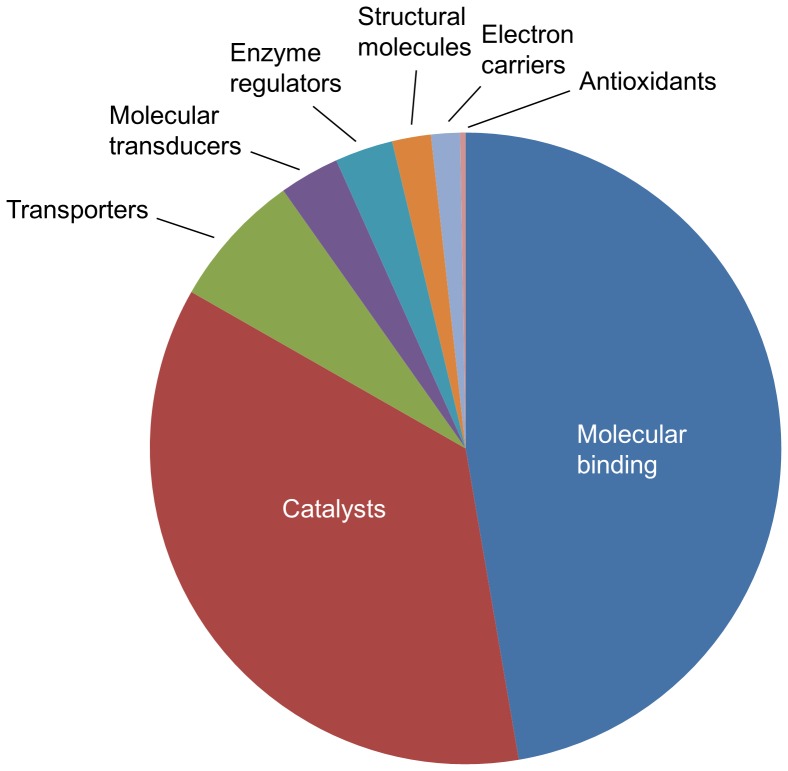
Molecular function distributions from the Gene Ontology (GO) analyses showed that in C. vestalis the genes expressed in the antennae were mostly linked to molecular binding activity (e.g., nucleotide, ion and odorant binding; in 47% of the total scores) or categorized as catalysts (e.g., hydrolase and oxidoreductase; in 36% of the total scores) (Figure 2). The contigs/singletons involved in molecular transporters and transducers (including odorant receptors) constitute the next most abundant categories amounting to 7 and 3%, respectively.
Figure 2. Distribution of transcriptome pyrosequencing from contigs/singletons.
The major categories of level 2 molecular functions from a Gene Ontology (GO) analysis are shown (n = 10 642).
Annotation of a Part of OR and OBP Genes
Contigs and singletons with at least one match to ORs and OBPs and with an E-value of 1E-5 or lower were selectively annotated. BLAST search indicated that ORs were predicted in 64 contigs and 99 singletons, and that OBPs were predicted in 74 contigs and 71 singletons (Tables 2, 3, S2 and S3). The number of predicted OBP contigs regenerated from at least 10 reads were 22 (Table 3), but only six were regenerated from OR contigs (Table 2 ). Therefore, in the antennae of female C. vestalis, OBP genes appeared to be expressed more abundantly than OR genes. OBP1 and OBP2 in particular were expressed at remarkably high levels.
Table 2. The part of contigs from Cotesia vestalis with similarity to OR genes.
| Name | Contig | Read count | Length (bp) | Homology | E-value* |
| OR1 | Cv_002063 | 293 | 1999 | Olfactory receptor [Microplitis mediator] | 0 |
| OR2 | Cv_000919 | 24 | 842 | Odorant receptor 44 [Nasonia vitripennis] | 2E−44 |
| OR3 | Cv_000252 | 15 | 992 | Putative odorant receptor 13a [Harpegnathos saltator] | 7E−32 |
| OR4 | Cv_005941 | 14 | 688 | Odorant receptor 265 [N. vitripennis] | 7E−23 |
| OR5 | Cv_006402 | 10 | 1005 | Odorant receptor 37 [Tribolium castaneum] | 8E−15 |
| OR6 | Cv_006618 | 10 | 1236 | Putative odorant receptor 13a [H. saltator] | 3E−45 |
Contigs which are regenerated at least from 10 reads are listed. *We identified genes whose E-values exhibited at least 1E-5 or less, with the BLAST searches.
Table 3. The part of contigs from Cotesia vestalis with similarity to OBP genes.
| Name | Contig | Read count | Length (bp) | Homology | E-value* |
| OBP1 | Cv_001562 | 2184 | 1276 | Odorant-binding protein 3 [Microplitis mediator] | 5E−15 |
| OBP2 | Cv_002750 | 1998 | 1139 | Pheromone-binding protein 1 [M. mediator] | 4E−61 |
| OBP3 | Cv_000569 | 770 | 952 | Odorant-binding protein 10 [M. mediator] | 1E−65 |
| OBP4 | Cv_002480 | 430 | 1212 | Odorant-binding protein 2 [M. mediator] | 1E−57 |
| OBP5 | Cv_003043 | 399 | 1186 | Odorant-binding protein 4 [M. mediator] | 6E−34 |
| OBP6 | Cv_001669 | 230 | 1082 | Odorant-binding protein 4 [M. mediator] | 7E−23 |
| OBP7 | Cv_001286 | 162 | 1326 | Odorant-binding protein 6 [M. mediator] | 3E−61 |
| OBP8 | Cv_001766 | 123 | 1013 | Odorant-binding protein 3 [M. mediator] | 2E−28 |
| OBP9 | Cv_003903 | 74 | 642 | Odorant-binding protein 3 [M. mediator] | 1E−35 |
| OBP10 | Cv_001246 | 71 | 980 | Odorant-binding protein 18 [Nasonia vitripennis] | 9E−32 |
| OBP11 | Cv_000714 | 63 | 970 | Odorant-binding protein 10 [M. mediator] | 5E−62 |
| OBP12 | Cv_002207 | 41 | 1736 | Odorant-binding protein 1 [M. mediator] | 7E−54 |
| OBP13 | Cv_000843 | 27 | 594 | Pheromone-binding protein 1 [M. mediator] | 1E−16 |
| OBP14 | Cv_000016 | 26 | 590 | Odorant-binding protein 10 [M. mediator] | 4E−47 |
| OBP15 | Cv_000232 | 23 | 599 | Pheromone-binding protein 1 [M. mediator] | 8E−42 |
| OBP16 | Cv_001034 | 17 | 551 | Odorant-binding protein 3 [M. mediator] | 6E−10 |
| OBP17 | Cv_005637 | 16 | 804 | Odorant-binding protein 6 [M. mediator] | 3E−19 |
| OBP18 | Cv_005027 | 15 | 501 | Odorant-binding protein 4 [M. mediator] | 6E−8 |
| OBP19 | Cv_001050 | 14 | 714 | Odorant-binding protein 3 [M. mediator] | 1E−20 |
| OBP20 | Cv_006168 | 12 | 632 | Odorant-binding protein 71 [N. vitripennis] | 5E−7 |
| OBP21 | Cv_001120 | 12 | 896 | Odorant-binding protein 2 [M. mediator] | 6E−29 |
| OBP22 | Cv_001116 | 10 | 737 | Odorant-binding protein 10 [M. mediator] | 8E−23 |
Contigs which are regenerated at least from 10 reads are listed. *We identified genes whose E-values exhibited at least 1E-5 or less, with the BLAST searches.
Expression Patterns of C. vestalis OR and OBP Genes
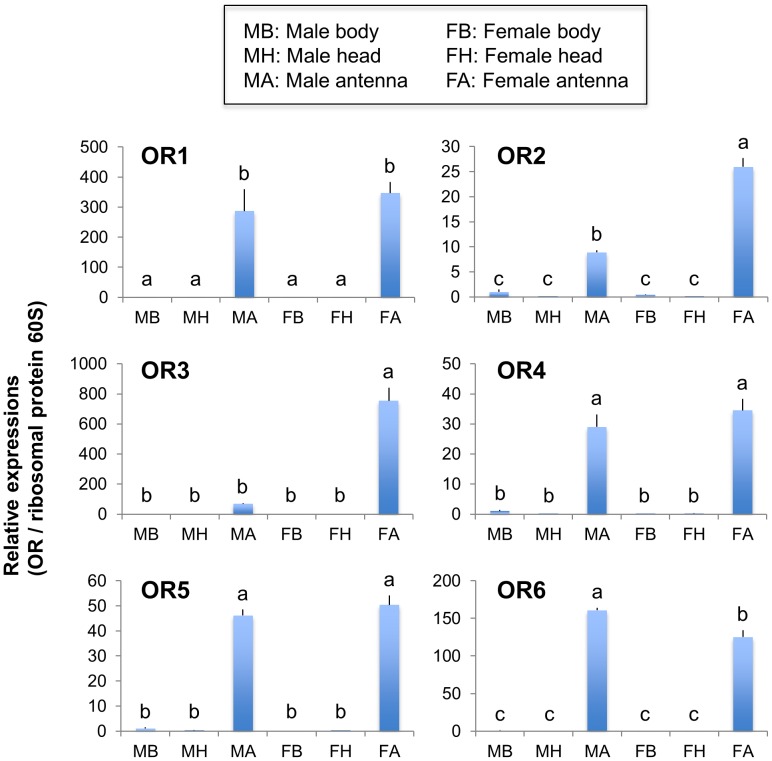
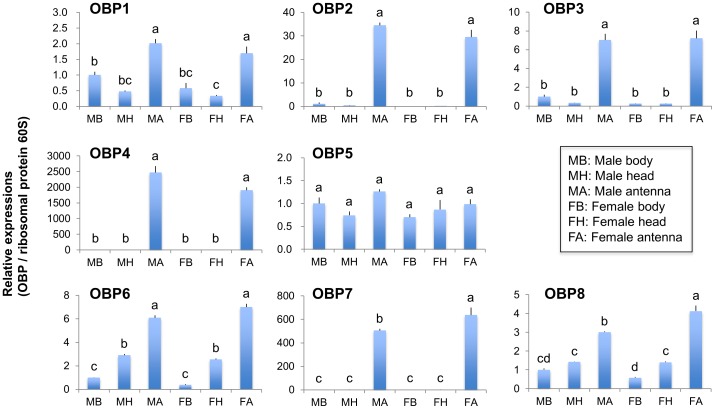
We conducted quantitative RT-PCR analyses in different tissues (bodies, heads and antennae) of adult males and females to assess the expression of C. vestalis OR1 to 6 and OBP1 to 10. These contigs are regenerated from at least 100 and 10 sequence reads, respectively. Analyses showed that all the OR genes analyzed were expressed in an antenna-specific manner (Figure 3). The expression of genes for OR2 and OR3 in the antennae of female wasps was much higher than in the antennae of male wasps, while an OR6 gene was expressed at a slightly lower level in females than in males. OBP genes had three different expression patterns: i) ubiquitously expressed (OBP5), ii) barely antenna-specific (OBP1, 6 and 8), and iii) highly antenna-specific (OBP2, 3, 4 and 7) (Figure 4). Moreover, genes for OBP7 and 8 were expressed at slightly higher levels in the antennae of female wasps than in those of males.
Figure 3. Tissue and genderspecific expressions of OR genes in C. vestalis adults.
Transcription levels of genes for OR1 to 6 were normalized to those of C. vestalis 60S ribosomal protein L10 (Cv_000471), and expressed relative to the normalized transcript levels in the body of C. vestalis males. Data represent the mean and standard errors (n = 5). Means followed by different small letters are significantly different (P<0.05, Tukey-Kramer HSD test).
Figure 4. Tissue and genderspecific expressions of OBP genes in C. vestalis adults.
Transcription levels of genes for OBP1 to 10 were normalized to those of C. vestalis 60S ribosomal protein L10 (Cv_000471), and expressed relative to the normalized transcript levels in the body of C. vestalis males. Data represent the mean and standard errors (n = 5). Means followed by different small letters are significantly different (P<0.05, Tukey-Kramer HSD test).
Discussion
Quality-checked transcriptome pyrosequencing reads of genes expressed in the antennae of C. vestalis adult females assembled into 17 328 contigs, including 64 OR and 74 OBP contigs. The parasitoid wasp, Nasonia vitripennis, which has similar genetic traits as C. vestalis (Figure 1), has a total of 301 OR genes, of which 225 are intact genes and 76 are pseudogenes [29]. Similarly, honey bee genome analyses revealed a major expansion of the OR family to 174 genes, encoding 163 potentially functional receptors, at least twice as many as those found in Drosophila melanogaster [29]. The total number of OR contigs and singletons found in the pyrosequences of C. vestalis antennal genes is 163 (Table S2). This large repertoire of OR genes might enable the wasp to sense the wide range of pheromones, floral scents, HIPVs and other olfactory cues. However, low expression levels of some OR genes prevented us from mining all members of this superfamily by transcriptomes. For a more comprehensive analysis, genomic analyses of C. vestalis should be conducted. In contrast, OBP genes were likely to be expressed at much higher levels in C. vestalis antennae, resulting in a total of 74 contigs and 71 singletons from large numbers of valid sequence reads (e.g., 2184 reads for OBP1 and 1998 reads for OBP2, Tables 3 and S3). The number of contigs obtained is comparable to that found in other insect species, such as the 66 putative OBP genes found in the mosquito Aedes aegypti [31]. While OR expression is generally restricted to neurons of the olfactory sensilla, OBP expression is associated with both olfactory and gustatory sensilla [32]. This may in part account for the different expression thresholds between OR and OBP genes.
There were trends in the antennal-specific transcription pattern of OR and OBP genes ( Figures 3 and 4). Predominantly, OR2, OR3, OBP7 and OBP8 genes were expressed in female antennae. These genes could play a role in odorant perception of certain HIPVs or sex pheromone in the specific host-searching and oviposition behavior [23] of female C. vestalis. Microplitis mediator OBP6, the homologue of C. vestalis OBP7 (Figure S1), has been reported to bind specifically to several plant odorants [33]. For ORs, however, little is known about odorant ligands and most of the valid data have been obtained from Drosophila spp. [34], [35]. Female C. vestalis are attracted selectively to a specific blend of DBM-induced cabbage volatiles including (Z)-3-hexenol, n-heptanal, α-pinene, sabinene, (Z)-3-hexenyl acetate, limonene, (E)-4,8-dimethyl-1,3,7-nonatriene and camphor [24]. These host-searching cues can be specifically recognized by antennal-specific ORs and OBPs. However, there is little to suggest chemosensory difference between adult male and female antennae or between adult male and female maxillary palps in mosquitoes. Neither tissue shows significant sexual dimorphism with respect to the types of sensilla they bear [36] or stimuli they respond to [37]. If the same holds true for parasitoid wasps, all antennal-specific OBPs and ORs, expressed irrespective of their sex and not only by females, may function in odorant recognition. Otherwise, the interplay of several OBPs and ORs is likely to be important for the recognition of general odorants and specific HIPVs. Functional characterization of these odorant-sensory proteins, with the extensive cloning of some full-length cDNAs, and in situ localization analysis will help to understand C. vestalis chemoreception mechanisms.
Supporting Information
Phylogenetic alignment of the amino acid sequence of OBPs. The OBPs derived from five different Hymenoptera species were aligned and organized in a phylogenic tree: Microplitis mediator (Mm); Nasonia vitripennis (Nv); Vepsa crabro (Vc), Polistes dominulus (Pd) and the current study [Cotesia vestalis (Cv)]. Alignments were conducted with ClustalW and MUCLE. Neighbor-joining tree was constructed using MEGA5 and presented with 50% cut-off bootstrap value of 1 000 bootstrap replicates sampled. The numbers at the branching points are bootstrap values (%). GenBank accession numbers of protein sequences are: MmOBP1, ABM05968; MmOBP2, ABM05969; MmOBP3, ABM05970; MmOBP4, ABM05971; MmOBP5, ABM05972; MmOBP6, ABO015559; MmOBP7, ABM05973; MmOBP10, AEO27860; MmCSP1, ABO15560; NvOBP, XP_001601068; NvOBP, XP_001603472.2; NvOBP71, XP_001601290; NvOBP70, CCD17839; NvOBP1, XP_001606873; DmOBP56e, ABW77913; PdOBP1, AAP55718; PdCSP1, AAP55719; VcCSP1, AAV68929.
(TIF)
RT-PCR primers used for this study.
(XLS)
All the sets of contigs and singletons from Cotesia vestalis with similarity to OR genes.
(XLS)
All the sets of contigs and singletons from Cotesia vestalis with similarity to OBP genes.
(XLS)
Acknowledgments
We gratefully acknowledge Mr. Masaki Yamamoto and Dr. Takeshi Shimoda for rearing insects, Dr. Nobuyuki Fuse and Ms. Minako Izutsu for tissue collection, Ms. Eri Kawaguchi for the cDNA library construction and 454 sequencing, and Dr. Rika Ozawa for statistical analysis.
Funding Statement
This work was financially supported in part by Global COE Program A06 of Kyoto University; the Core-to-Core Program from the Japan Society for the Promotion of Science (JSPS) (No.20004); and a Grant-in-Aid for Scientific Research from JSPS (No. 24770019) to GA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sánchez-Gracia A, Vieira FG, Rozas J (2009) Molecular evolution of the major chemosensory gene families in insects. Heredity 103: 208–216. [DOI] [PubMed] [Google Scholar]
- 2. Ishida Y, Leal WS (2008) Chiral discrimination of the Japanese beetle sex pheromone and a behavioral antagonist by a pheromone-degrading enzyme. Proc Natl Acad Sci USA 105: 9076–9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Durand N, Carot-Sans G, Chertemps T, Bozzolan F, Party V, et al. (2010) Characterization of an antennal carboxylesterase from the pest moth Spodoptera littoralis degrading a host plant odorant. PloS One 5: e15026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hansson BS, Stensmyr MC (2011) Evolution of insect olfaction. Neuron 72: 698–711. [DOI] [PubMed] [Google Scholar]
- 5. Foret S, Maleszka R (2006) Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera). Genome Res 16: 1404–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Swarup S, Williams TI, Anholt RRH (2011) Functional dissection of Odorant binding protein genes in Drosophila melanogaster . Genes Brain Behav 10: 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Syed Z, Ishida Y, Taylor K, Kimbrell DA, Leal WS (2006) Pheromone reception in fruit flies expressing a moth’s odorant receptor. Proc Natl Acad Sci USA 103: 16538–16543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou JJ (2010) Odorant-binding proteins in insects. Vitam Horm 83: 241–272. [DOI] [PubMed] [Google Scholar]
- 9. Pelosi P, Maida R (1990) Odorant-binding proteins in vertebrates and insects: similarities and possible common function. Chem Senses 15: 205–215. [Google Scholar]
- 10. Laughlin JD, Ha TS, Jones DNM, Smith DP (2008) Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell 133: 1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pelosi P, Zhou JJ, Ban LP, Calvello M (2006) Soluble proteins in insect chemical communication. Cell Mol Life Sci 63: 1658–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vieira FG, Rozas J (2011) Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: origin and evolutionary history of the chemosensory system. Genome Biol Evol 3: 476–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, et al. (2008) Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452: 1007–1011. [DOI] [PubMed] [Google Scholar]
- 14. Sato K, Pellegrino M, Nakagawa T, Vosshall LB, Touhara K (2008) Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452: 1002–1006. [DOI] [PubMed] [Google Scholar]
- 15. Benton R (2006) On the ORigin of smell: odorant receptors in insects. Cell Mol Life Sci 63: 1579–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Benton R, Sachse S, Michnick SW, Vosshall LB (2006) Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo . PLoS Biol 4: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vosshall LB, Hansson BS (2011) A unified nomenclature system for the insect olfactory coreceptor. Chem Senses 36: 497–498. [DOI] [PubMed] [Google Scholar]
- 18. Carey AF, Wang G, Su CY, Zwiebel LJ, Carlson JR (2010) Odorant reception in the malaria mosquito Anopheles gambiae . Nature 464: 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nichols AS, Chen S, Luetje CW (2011) Subunit contributions to insect olfactory receptor function: channel block and odorant recognition. Chem Senses 36: 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pask GM, Jones PL, Rützler M, Rinker DC, Zwiebel LJ (2011) Heteromeric anopheline odorant receptors exhibit distinct channel properties. PloS One 6: e28774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones PL, Pask GM, Rinker DC, Zwiebel LJ (2011) Functional agonism of insect odorant receptor ion channels. Proc Natl Acad Sci USA 108: 8821–8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, et al. (2004) Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction . Neuron 43: 703–714. [DOI] [PubMed] [Google Scholar]
- 23. Shiojiri K, Takabayashi J, Yano S, Takafuji A (2000) Flight response of parasitoids toward plant-herbivore complexes: a comparative study of two parasitoid-herbivore systems on cabbage plants. Appl Entomol Zool 35: 87–92. [Google Scholar]
- 24. Shiojiri K, Ozawa R, Kugimiya S, Uefune M, van Wijk M, et al. (2010) Herbivore-specific, density-dependent induction of plant volatiles: honest or “cry wolf” signals? PloS One 5: e12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Arimura G, Matsui K, Takabayashi J (2009) Chemical and molecular ecology of herbivore-induced plant volatiles: proximate factors and their ultimate functions. Plant Cell Physiol 50: 911–923. [DOI] [PubMed] [Google Scholar]
- 26. Kessler A, Baldwin IT (2001) Defensive function of herbivore-induced plant volatile emissions in nature. Science 291: 2141–2144. [DOI] [PubMed] [Google Scholar]
- 27. Pertea G, Huang X, Liang F, Antonescu V, Sultana R, et al. (2003) TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19: 651–652. [DOI] [PubMed] [Google Scholar]
- 28. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robertson HM, Gadau J, Wanner KW (2010) The insect chemoreceptor superfamily of the parasitoid jewel wasp Nasonia vitripennis . Insect Mol Biol 19 Suppl 1 121–136. [DOI] [PubMed] [Google Scholar]
- 30. Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, et al. (2010) Functional and evolutionary insights from the genomes of three parasitoid Nasonia species . Science 327: 343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou JJ, He XL, Pickett JA, Field LM (2008) Identification of odorant-binding proteins of the yellow fever mosquito Aedes aegypti: genome annotation and comparative analyses. Insect Mol Biol 17: 147–163. [DOI] [PubMed] [Google Scholar]
- 32. Bohbot J, Vogt RG (2005) Antennal expressed genes of the yellow fever mosquito (Aedes aegypti L.); characterization of odorant-binding protein 10 and takeout. Insect Biochem Mol Biol 35: 961–979. [DOI] [PubMed] [Google Scholar]
- 33. Zhang S, Chen LZ, Gu SH, Cui JJ, Gao XW, et al. (2011) Binding characterization of recombinant odorant-binding proteins from the parasitic wasp, Microplitis mediator (Hymenoptera: Braconidae). J Chem Ecol 37: 189–194. [DOI] [PubMed] [Google Scholar]
- 34. Hallem EA, Carlson JR (2006) Coding of odors by a receptor repertoire. Cell 125: 143–160. [DOI] [PubMed] [Google Scholar]
- 35. Su CY, Menuz K, Carlson JR (2009) Olfactory perception: receptors, cells, and circuits. Cell 139: 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McIver S (1971) Comparative studies on the sense organs on the antennae and maxillary palps of selected male culicine mosquitoes. Can J Zool 49: 235–239. [DOI] [PubMed] [Google Scholar]
- 37. Kellogg FE (1970) Water vapour and carbon dioxide receptors in Aedes aegypti . J Insect Physiol 16: 99–108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic alignment of the amino acid sequence of OBPs. The OBPs derived from five different Hymenoptera species were aligned and organized in a phylogenic tree: Microplitis mediator (Mm); Nasonia vitripennis (Nv); Vepsa crabro (Vc), Polistes dominulus (Pd) and the current study [Cotesia vestalis (Cv)]. Alignments were conducted with ClustalW and MUCLE. Neighbor-joining tree was constructed using MEGA5 and presented with 50% cut-off bootstrap value of 1 000 bootstrap replicates sampled. The numbers at the branching points are bootstrap values (%). GenBank accession numbers of protein sequences are: MmOBP1, ABM05968; MmOBP2, ABM05969; MmOBP3, ABM05970; MmOBP4, ABM05971; MmOBP5, ABM05972; MmOBP6, ABO015559; MmOBP7, ABM05973; MmOBP10, AEO27860; MmCSP1, ABO15560; NvOBP, XP_001601068; NvOBP, XP_001603472.2; NvOBP71, XP_001601290; NvOBP70, CCD17839; NvOBP1, XP_001606873; DmOBP56e, ABW77913; PdOBP1, AAP55718; PdCSP1, AAP55719; VcCSP1, AAV68929.
(TIF)
RT-PCR primers used for this study.
(XLS)
All the sets of contigs and singletons from Cotesia vestalis with similarity to OR genes.
(XLS)
All the sets of contigs and singletons from Cotesia vestalis with similarity to OBP genes.
(XLS)