Abstract
A cytokine/stress signaling kinase Tak1 (Map3k7) deficiency is known to impair hematopoietic progenitor cells. However, the role of TAK1 signaling in the stem cell function of the hematopoietic system is not yet well defined. Here we characterized hematopoietic stem cells (HSCs) harboring deletion of Tak1 and its activators, Tak1 binding proteins 1 and 2 (Tab1 and Tab2) using a competitive transplantation assay in a mouse model. Tak1 single or Tab1/Tab2 double deletions completely eliminated the reconstitution activity of HSCs, whereas Tab1 or Tab2 single deletion did not cause any abnormality. Tak1 single or Tab1/Tab2 double deficient lineage-negative, Sca-1+, c-Kit+ (LSK) cells did not proliferate and underwent cell death. We found that Tnfr1 deficiency restored the reconstitution activity of Tak1 deficient bone marrow cells for 6–18 weeks. However, the reconstitution activity of Tak1- and Tnfr1-double deficient bone marrow cells declined over the long term, and the number of phenotypically identified long-term hematopoietic stem cells were diminished. Our results indicate that TAB1- or TAB2-dependent activation of TAK1 is required for maintenance of the hematopoietic system through two mechanisms: one is prevention of TNF-dependent cell death and the other is TNF-independent maintenance of long-term HSC.
Introduction
Hematopoiesis is maintained by self-renewal of hematopoietic stem cells (HSCs) and differentiation and proliferation of HSC-derived hematopoietic progenitors [1]–[3]. Actively proliferating bone marrow cell populations include short-term repopulating HSCs (ST-HSCs) and multipotent progenitors, MPPs. Phenotypically both populations are lineage negative, Sca1+ c-Kit+, (LSK), and CD34+, while ST-HSCs are Flt3− and MPPs are Flt3+. Although ST-HSCs and MPPs can provide multilineage hematopoietic cells for a short term [4], hematopoietic pluripotency is maintained by the less frequently self-renewed population of HSCs: long-term multilineage reconstituting hematopoietic stem cells (LT-HSCs) that are phenotypically comprised of CD34−, LSK or CD150+, LSK [1], [5]. Several recent studies have revealed that the protection from stress-mediated cell damage is critical for HSC maintenance [6]. For example, the FOXO family of transcriptional factors, which regulate stress responses, are essential for HSC maintenance [7]. Deficiency of DNA damage sensor, Atm1, abolishes HSC function [8]. Recently, chromatin remodeling factor, Bmi1, was found to be indispensable for HSC function by modulating DNA damage signaling and oxidative stress [9]. A protein kinase LKB is reported to play a unique role in maintenance of HSCs by preventing oxidative stress-induced mitochondrial dysfunction [10]–[12]. While the importance of stress responses in HSCs is clear, the signaling pathways to protect HSCs from cell damage are still largely unknown.
TAK1 (MAP3K7) is a member of mitogen-activated protein kinase kinase kinases (MAPKKK), and activated by inflammatory cytokines and stress conditions. TAK1 and TAK1 binding proteins 1 and 2 (TAB1 and 2) are major components of the TAK1 complex, which can activate MAPK cascades as well as the transcription factor NF-κB. TAB1 and TAB2 are structurally unrelated and bind to TAK1 at different sites [13], [14]. TAB2 and its closely related protein, TAB3, confer ubiquitin binding domains and tethers between TAK1 and the polyubiquitin chain, resulting in activation of TAK1 in cytokine signaling pathways [15]–[19]. TAB2 and TAB3 can redundantly function in cytokine signaling pathways, but TAB2 is indispensable during development [20]. We recently demonstrated that TAB1 is involved in stress-dependent TAK1 activation [21] and the basal activity of TAK1 in epithelial tissues [22]. TAK1 is critical in modulating reactive oxygen species and preventing cell death in epithelial cells in vivo [23]–[26]. Thus, the stress- and cytokine-activated TAK1 complex is one of the pathways to protect cells from reactive oxygen species at least in the epithelium. Based on this, we hypothesized that the TAK1 complex participates in protection of HSC and may be involved in HSC maintenance. Inducible Tak1 deficiency in hematopoietic cells and hepatocytes has recently been reported to cause cell death within 12 days after induction of gene deletion through a TNF-dependent mechanism [27], [28]. This short term assay demonstrates that TAK1 is important for preventing cell death in ST-HSCs and MPPs. However, the involvement of TAK1 signaling in stem cell function, specifically the ability of long-term reconstitution, has not yet been determined.
In this study, we investigated the role of TAK1, TAB1 and TAB2 in LT-HSCs. We defined the capacity for long-term multipotency as well as the number of LT-HSCs in the competitively transplanted Tak1-, Tab1- and Tab2-deficient bone marrow cells. We also assessed whether the TNF pathway contributes to the failure of Tak1-deficient ST-HSC and LT-HSC maintenance by employing bone marrow cells with both Tak1 and Tnfr1 deficiency.
Materials and Methods
Mice
Tak1-floxed (Tak1flox/flox or Tak1FF), Tab1-floxed (Tab1flox/flox or Tab1FF), Tab2-floxed (Tab2flox/flox or Tab2FF) mice were backcrossed for a minimum of five generations to C57BL/6 mice [20], [29], [30]. Tnfr1-deficient (Tnfr1−/−) (C57/BL6 backcrossed), Rosa26-CreERT (mixed background) mice were obtained from The Jackson Laboratory [31], [32]. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All animal experiments were conducted with the approval of the North Carolina State University Institutional Animal Care and Use Committee (Approval number: 10-114B). All efforts were made to minimize animal suffering.
Competitive Reconstitution Assay
Seven- to nine-week-old C57BL6.SJL congenic mice were lethally irradiated (7–10 Gy) and intravenously infused with a mixture of bone marrow mononuclear (BMN) cells that were obtained from untreated test (CD45.2+) and competitor (CD45.1+) donor mice at five to six weeks of age. Each recipient received either 2×105 test and 2×105 competitor BMN cells (1∶ 1 ratio) or 1×106 test and 2×105 competitor BMN cells (5∶ 1) as indicated. At six to seven weeks post-transplantation, the peripheral blood (PB) samples from the recipient mice were analyzed to confirm successful reconstitution, and then mice were administered 160 mg/kg tamoxifen by intraperitoneal (i.p). injection for three consecutive days. PB samples were collected and analyzed every three weeks post-tamoxifen injection to determine the chimerism of circulating myeloid (CD11b+ or Gr-1+), B (B220+) and T (CD3ε+) cells.
Flow Cytometric Analysis and Cell Sorting
Whole bone marrow cells were flushed from the femurs and tibias of six- to nine-week-old mice with Hank’s Balanced Salt Solution without magnesium and calcium [HBSS(–)]. The bone marrow cells were then suspended in 0.83% ammonium chloride (NH4Cl) for 5 min at room temperature to lyse red blood cells and washed with HBSS(–). BMN cells were resuspended in HBSS(–) and filtered through a 35-µm cell strainer (BD Biosciences) to obtain single cell suspension. The cells were incubated for 20 min on ice with anti-CD16/32 antibody to block FcγRII/III, followed by incubation with fluorochrome-conjugated antibodies against cell surface antigens as described below. After labeling, cells were washed once with HBSS(–), resuspended in HBSS(–) and analyzed on FACS LSRII (BD Biosciences). BMN cells or splenocytes were sorted using a FACSAria (BD Biosciences) or MoFlo (Beckman). Specific monoclonal antibodies against the following antigens were used for flow cytometric analysis: CD3ε (145-2C11), CD4 (RM4-5), CD8a (53-6.7), CD11b (M1/70), Gr-1 (RB6-8C5), CD19 (6D5), B220 (RA3-6B2), Ter-119 (TER-119), Sca-1 (E13-161.7), c-Kit (2B8), IL-7Rα (A7R34), CD34 (RAM34), CD41 (MWReg30), CD48 (HM48-1), CD150 (TC15-12F12.2), CD45.1 (A20), CD45.2 (104), and CD16/32 (93). A cocktail of monoclonal antibodies against CD4, CD8a, B220, CD19, CD11b, Gr-1 and Ter-119 was used as a lineage marker (Lineage). To collect PB samples, mice were bled from the right mandibular vein using a 5 mm animal lancet (Goldenrod, Mineola, NY), and the blood were collected into microcentrifuge tubes containing HBSS(–) buffer supplemented with EDTA at a final concentration of approximately 1 mM. Anticoagulated PB samples were treated with 0.83% NH4Cl, stained with antibodies against CD45.1, CD45.2, CD3ε, B220, CD11b and Gr-1, and analyzed on FACS LSRII (BD Biosciences).
HPC Proliferation Assay
Mice were administered 160 mg/kg tamoxifen by i.p. injection for three consecutive days, sacrificed at day 4, and BMN cells were isolated. BMN cells were seeded into serum-free StemSpan SFEM medium (StemCell Technologies, Cat# 09600) supplemented with 10 mg/ml heparin (Calbiochem, Cat#375095), 100 µg/ml mSCF (Peprotech, Cat#250-03), 500 µg/ml mTPO (Peprotech, Cat#315-14), 100 µg/ml mIGF2 (R&D, Cat#792-MG), 100 µg/ml hFGF1 (Peprotech, Cat#100-17A), and 100 µg/ml mANGPTL3 (R&D, Cat#136-AN) [33] in 24-well plates; 2.5×106 cells/well, 2×106 cells/well and 1×106 cells/well were plated for 3-, 6- and 9-day culture, respectively.
Quantitative Real Time PCR Analysis
For gene expression analysis, LT-HSC (CD34−, Flt3−, LSK), ST-HSC (CD34+, Flt3−, LSK), MPP (CD34+, Flt3+, LSK), MP (Lineage−, c-Kit+, Sca-1−) and Lineage+ cells were sorted from WBM cells using a FACSAria cell sorter. Total RNA was isolated from these cells using an RNeasy kit (QIAGEN) and transcribed into cDNA using SuperScript VILO cDNA Synthesis Kit (Life Technologies). Expression levels of Tak1, Tab1 and Tab2 were determined by quantitative real time PCR (qPCR) and normalized to the level of Actb. The following primers were used: Tak1-forward, 5′-CGTCTTCTGCCAGTGAGATG-3′; Tak1-reverse, 5′-ATCTTTTGCTCTCCACTTAGCTT-3′; Tab1-forward, 5′-ACCCTGCTGGTGAGGAACT-3′; Tab1-reverse, 5′-AGGGACAGAGTCACACTAGTCTT-3′; Tab2-forward, 5′-GGATAGAATAAGCGAAGCCCGGAA-3′; Tab2-reverse, 5′-CTCTTTGAAGCCGTTCCATCCT-3′; Actb-forward, 5′- CATCCGTAAAGACCTCTATGCCAAC-3′; and Actb-reverse, 5′-ATGGAGCCACCGATCCACA-3′. Data are shown as values relative to LT-HSCs.
For the verification of gene deletion, genomic DNA was isolated from BMN cells using a DNeasy Blood & Tissue Kit (QIAGEN) according to the manufacturer’s instructions. Using qPCR, the relative copy number was determined by generating a standard curve. The following primers were used: Tak1-forward, 5′-AGGTTGTCGGAAGAGGAGCT-3′; Tak1-reverse, 5′-CTCCACAATGAAAGCCTTCC-3′; Tab1-forward, 5′-ACCCTGTTTCTGTGCCCTACTCAA-3′; Tab1-reverse, 5′-ACTGTGAGAGCCGTTGACCATCT-3′; Tab2-forward, 5′-TGCGCTGTTCTCTCTCAGGA-3′; and Tab2-reverse, 5′-CAAGTCCAAGTTGAGAGAAG-3′. Data are shown as values relative to Tak1flox/flox BMN cells.
Western Blotting
BMN cells, splenocytes or thymocytes were lysed in extraction buffer containing 20 mM HEPES (pH7.4), 150 mM NaCl, 1.5 mM MgCl2, 0.5 mM DTT and 0.5% Triton X-100 supplemented with a protease inhibitor cocktail (ThermoScientific) and phosphatase inhibitor cocktail (Nacalai Tesque). Cell debris and nuclei were pelleted by centrifugation at 20,000×g for 10 min at 4°C, and the resulting supernatant was used for Western blotting analysis.
Statistical Analysis
Data were analyzed using two-tailed Student’s t test. Values are expressed as means ± S.D. Differences were considered significant at p<0.05.
Results and Discussion
The TAK1 Complex is Required for LT-HSC Function
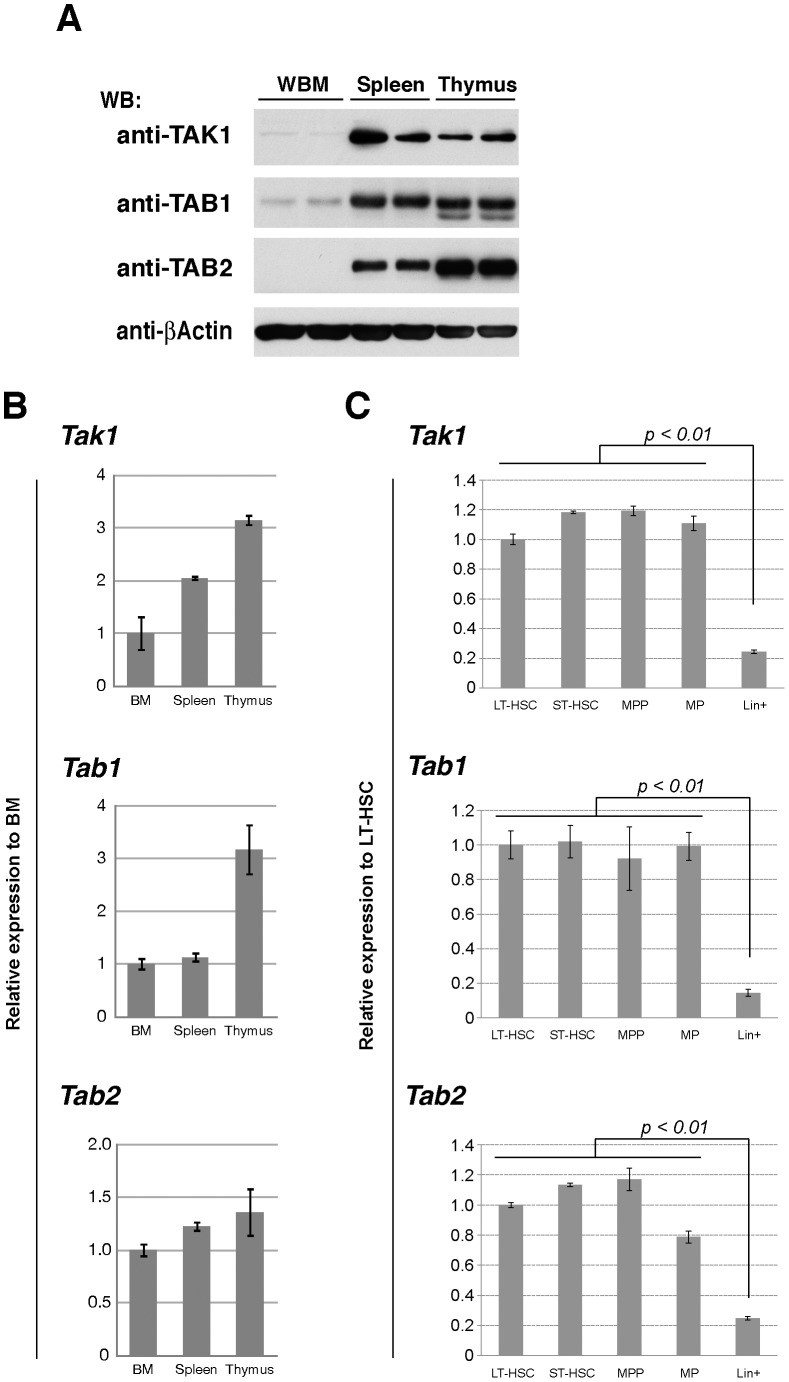
To determine the importance of the TAK1 complex in HSCs, we first examined the protein levels of TAK1, TAB1 and TAB2 in bone marrow cells (Figure 1A). To our surprise, protein levels of TAK1, TAB1 and TAB2 were all very low in bone marrow cells, when compared to the levels in the spleen and thymus. We also found that the levels of mRNA of Tak1, Tab1 and Tab2 were also lower in bone marrow cells, but the differences among the organs were much less pronounced, compared to the differences in the protein levels (Figure 1B). These protein levels may also be post-transcriptionally modulated. Among bone marrow cells, the mRNA levels of Tak1, Tab1 and Tab2 were significantly higher in the undifferentiated populations including LT-HSCs and progenitor cells compared to differentiated bone marrow cells (Figure 1C). This raises the possibility that TAK1, TAB1 and TAB2 may play an important role in HSCs. Therefore, we characterized bone marrow cells in adult mice having deletions of Tak1, Tab1 or Tab2 genes using the ubiquitously-expressed inducible Cre recombinase system, Rosa26.CreERT deleter mice [31]. Rosa26.CreERT Tak1flox/flox (referred to as Tak1iKO), Rosa26.CreERT Tab1flox/flox (referred to as Tab1iKO), and Rosa26.CreERT Tab2flox/flox (referred to as Tab2iKO) were compared with littermate or age-matched controls including Rosa26.CreERT Tak1flox/+ (referred to as Tak1Het or het), Rosa26.CreERT Tab1flox/+ (referred to as Tab1Het or het), Rosa26.CreERT Tab2flox/+ (referred to as Tab2Het or het), no-Cre (Tak1flox/flox, Tab1flox/flox and Tab2flox/flox) and Cre-alone (Rosa26.CreERT). Tamoxifen was intraperitoneally injected once per day for three consecutive days (the first day of tamoxifen injection is designated as day 1). Intact Tak1, Tab1 and Tab2 genes were greatly decreased in Tak1iKO, Tab1iKO and Tab2iKO in bone marrow cells at day 4 (Figure S1), indicating that these genes were efficiently deleted four days after the start of tamoxifen treatment. Since Tak1 deficiency causes damage to multiple tissues within four days, which will be reported elsewhere, the reduction in the protein levels was confirmed by immunoblots of TAB1 and TAB2 in splenocytes at day 4 (Figure S2). We note here that ubiquitous expression of Cre in the Rosa26.CreERT mice including hets and Cre-alone, caused weight loss at days 5-7, which is presumably associated with toxic effects of Cre expression [34]. The number of whole bone marrow cells and lineage negative Sca1- c-Kit+ myeloid progenitor (MP) cells was diminished at day 4 (Figure S3A and B), as previously reported [35]. In contrast, the number of LSK cells was not noticeably decreased by Cre expression (Figure S3A and B). Thus, although Cre exerted significant effects on bone marrow cells, this system might still be suitable for examining the effects of gene deletions in HSCs. Nonetheless, to exclude any effects of Cre toxicity, we used Cre-expressing controls, Cre-alone or hets, in all experiments.
Figure 1. TAK1, TAB1 and TAB2 in bone marrow cells.
(A) Expression of TAK1, TAB1 and TAB2 proteins. Whole cell extracts of the BMN cells (BM), splenocytes (Spleen) and thymocytes (Thymus) were analyzed by SDS-PAGE and Western blotted (WB) using the indicated antibodies. The amount of β-actin is shown as a loading control. (B) Expression of Tak1, Tab1 and Tab2 mRNA. Total RNA was isolated from BMN cells (BM), splenocytes (Spleen) and thymocytes (Thymus), and analyzed by qPCR. Expression level of each gene was normalized to that of Actb and shown as the relative value to BM. Data are presented as mean ± S.D. of three independent experiments. (C) Expression of Tak1, Tab1 and Tab2 mRNA. The cells in the LT-HSC (CD34− Flt3− LSK), ST-HSC (CD34+ Flt3− LSK), MPP (CD34+ Flt3+ LSK), MP (Lineage− c-Kit+ Sca-1−) or Lineage+ fractions from wild type mouse bone marrow were sorted by FACS, and total RNA was prepared. The relative amounts of Tak1, Tab1, Tab2 and Actb mRNA were determined by qPCR. Expression level of each gene was normalized to that of Actb and shown as the relative value to LT-HSC. Data are presented as mean ± S.D. of four independent experiments.
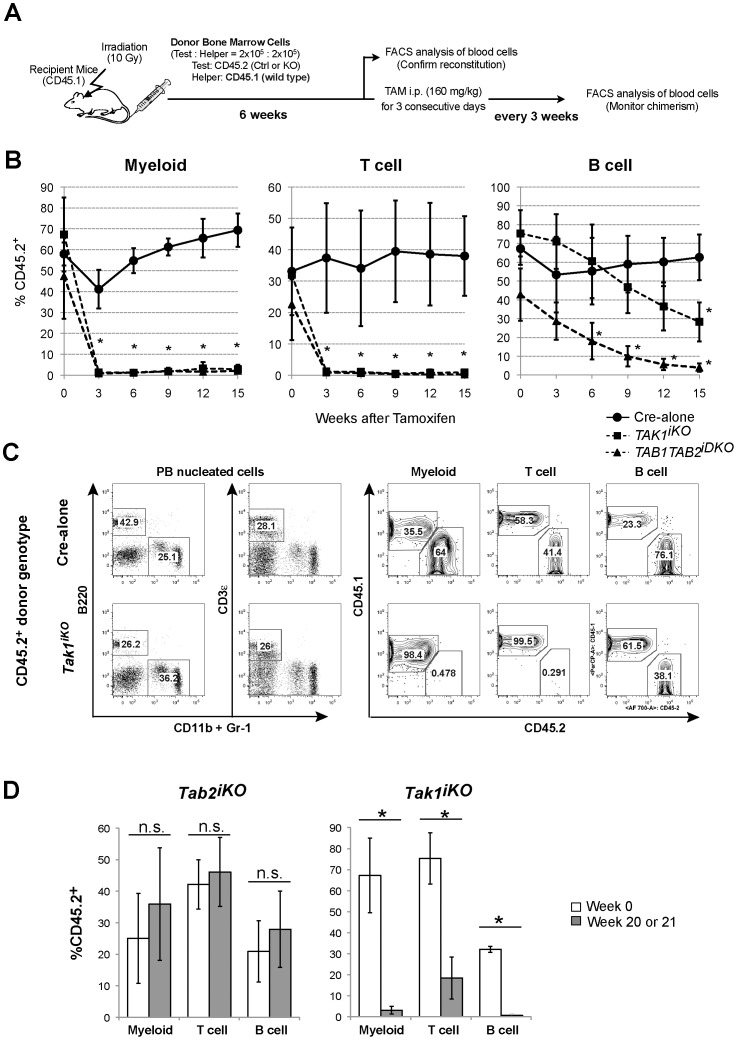
To examine whether TAK1, TAB1 and TAB2 are involved in HSC function in vivo, we determined long-term chimerism in the competitive transplantation assay. Tak1iKO, Tab1iKO, Tab2iKO, Tab1Tab2iDKO, hets, Cre-alone and no-Cre control bone marrow cells, which express CD45.2, were mixed with CD45.1+ wild type competitors at a 1∶1 ratio (and 5∶1 ratio in some experiments as indicated in the figure legends) and were transplanted into lethally irradiated CD45.1+ recipients. After engraftment of CD45.2+ hematopoietic cells was confirmed, tamoxifen was injected, and myeloid, T and B cells in peripheral blood were analyzed (see a schematic drawing in Figure 2A). To monitor the reconstitution potential of HSCs, chimerism was monitored every three weeks for a 15-week period. Bone marrow from control including Cre alone and hets exhibited the same trend of chimerism; a transient decrease in myeloid and B-cell chimerism of Cre-alone and hets cells was observed around three weeks post tamoxifen injection (Figure 2B). However; chimerism was restored to the original levels by 6–12-weeks post-tamoxifen injection. Thus, Cre expression alone reduces myeloid cells and/or their progenitors as shown in Figure S3, but does not have a major impact on HSC function and has no impact on LT-HSC function.
Figure 2. Competitive reconstitution assay.
(A) Schematic representation of competitive transplantation. 2×105 BMN cells from Cre-alone, Tak1iKO or Tab1Tab2iDKO mice (CD45.2+) were transplanted into lethally irradiated recipients (CD45.1) together with 2×105 competitor wild type BMN cells (CD45.1+). At six weeks post-transplantation (designated as Week 0), the chimerism of myeloid, T and B cell populations in the recipients’ peripheral blood (PB) was analyzed, then the recipients were i.p. injected with tamoxifen at 160 mg/kg body weight for three consecutive days. (B) The chimerism of PB cell populations was monitored every three weeks, and shown as mean ± S.D. (*p<0.05). [Cre-alone (solid line, circles, n = 3), Tak1iKO (dashed line, squares, n = 3) or Tab1Tab2iDKO (dashed line, triangles, n = 4)]. (C) Representative flow cytometry data of blood cell chimerism. PB cells were collected 15 weeks after tamoxifen injection. Myeloid cells (CD11b+ or Gr-1+), B cells (B220+) and T cells (CD3ε+) in PB mononuclear cells were analyzed for the expression of CD45.1 or CD45.2. (D) Competitive reconstitution assay for Tab2iKO and Tak1iKO mice. Donor-derived chimerism of PBs was analyzed. The percentage of Tab2iKO or Tak1iKO PB myeloid, T and B cells (CD45.2+) in total PB myeloid, T and B cells (CD45.1+ and CD45.2+) before (Week 0, open bars) and 20 weeks or 21 weeks, respectively, after tamoxifen injection is shown (gray bars). Data are presented as mean ± S.D. (*p<0.05, n.s. not significant, n = 3).
In contrast, deletion of Tak1 sharply diminished the myeloid and T-cell populations and gradually reduced the B-cell population (Figures 2B-D). Since myeloid cells have a short life-span, this sharp drop of cell number is likely to be the result of MP and/or HSC impairment due to Tak1 deletion. In contrast to myeloid cells, T cells are known to live for several weeks to months. Therefore, the common lymphoid progenitor (CLP) and/or HSC impairment by Tak1 deletion cannot account for the sharp drop of the peripheral T-cell population. It has previously been reported that TAK1 is essential for T-cell survival using a T cell-specific Tak1 deletion [36]. Therefore, the depletion of Tak1-deficient T cells in the peripheral blood is likely due to reduced survival. B-cell chimerism was gradually but constantly decreased after tamoxifen injection. This observation suggests that B cells do not require TAK1 for their survival, which is consistent with the findings of an earlier study [30]. This gradual decline indicates that HSC function is impaired by deletion of Tak1. Collectively, these results suggest that TAK1 is essential for repopulating potential of LT-HSC, and in addition to that, TAK1 is important for T-cell survival in the peripheral blood.
While Tak1 deletion donor derived cells were almost completely depleted at 21 weeks, Tab2 single deletion donor-derived cells exhibited repopulating ability for more than 20 weeks post-tamoxifen treatment, (Figure 2D and Figure S4A). Tamoxifen-induced gene deletion was confirmed by immunoblotting of splenocytes from Tab2iKO bone marrow chimera mice at 21 weeks post-tamoxifen injection (Figure S4B). TAB2 protein was not detectable in the CD45.2+ population even after 21 weeks, indicating that TAB2 is largely dispensable for HSC function. Tab1 single deletion donor bone marrow cells also exhibited ability to repopulate, although these modified cells were slightly less efficient compared to “no-Cre” controls (Figure S4A). Thus, either TAB1 or TAB2 is not essential for HSC function. Since we have recently found that TAB1 and TAB2 are functionally redundant in the activation of TAK1 in the epidermis and the intestinal epithelium [22], we further generated Rosa26.CreERT Tab1flox/flox Tab2flox/flox (referred to as Tab1Tab2iDKO) mice and performed the competitive transplantation assay. In contrast to Tab1 or Tab2 single deletion, double deletion of Tab1 and Tab2 generated the identical chimerism pattern produced by Tak1 deletion (Figure 2B). Thus, TAB1 and TAB2 redundantly function to activate TAK1 and together play an indispensable role in the reconstitution activity of HSC. TAB1 is important for stress-induced activation of TAK1 [21], while TAB2 functions as an adaptor for cytokine receptor signaling pathways [15], [19]. Bone marrow cells are constantly exposed to stressors such as reactive oxygen species [8] and cytokines such as TNF [37]. Thus, it is likely that both stressors and cytokines activate TAK1 through TAB1 and TAB2 in bone marrow under normal conditions. Because stressors and cytokines potentially damage HSCs, we assume that stressor- and cytokine-induced activation of TAK1 is required for HSC maintenance.
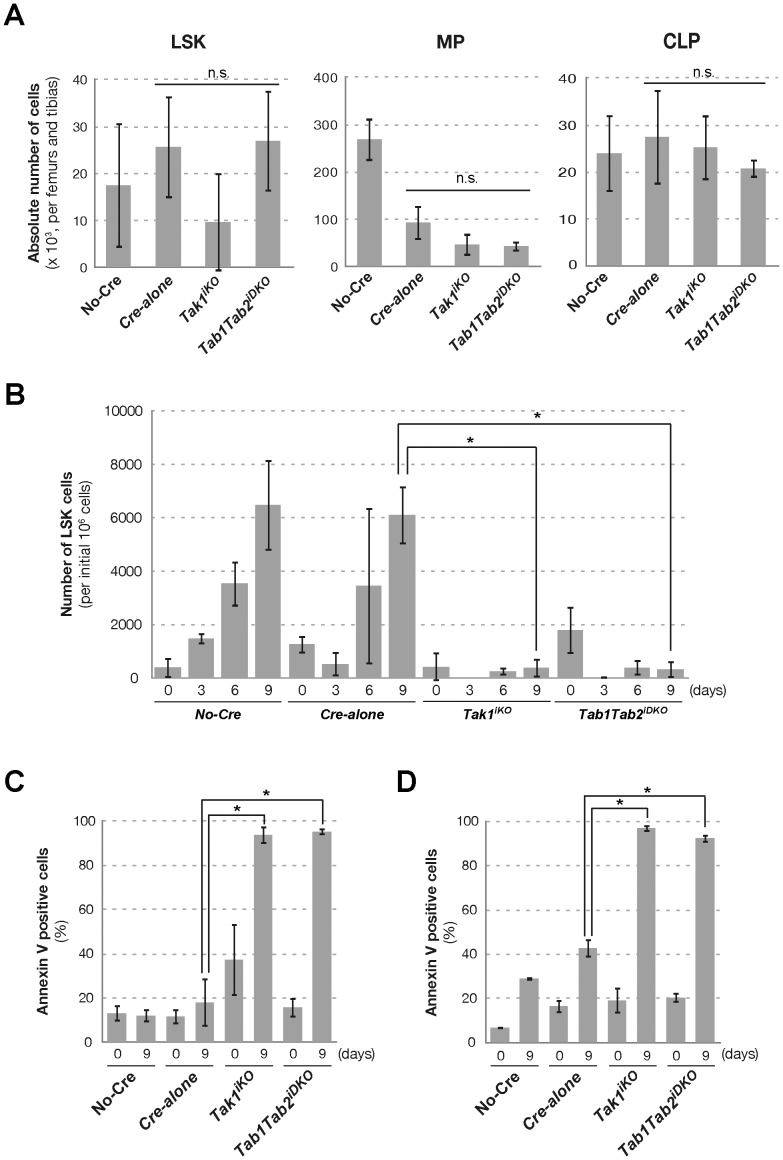
TAK1 is Important for LSK Expansion
Reconstitution potential of HSCs was abolished by Tak1 or Tab1 Tab2 double deletion as shown in Figure 2B. This suggests that ablation of TAK1 signaling impairs either ability of differentiation, expansion or survival of HSCs. If only the capacity for differentiation is impaired, then Tak1iKO and Tab1Tab2iDKO HSCs should still be able to expand in the in vitro expansion assay. Accordingly, we treated Tak1iKO and Tab1Tab2iDKO and control mice with tamoxifen in vivo for three consecutive days, and at the next day, bone marrow cells were isolated. At day 4, LSK cells were found to be largely viable, and LSK population number was not significantly reduced in Tak1iKO and Tab1Tab2iDKO (Figure 3A). MP, CLPs, B and T cells and granulocytes were not reduced by Tak1 or Tab1 Tab2 double deficiency at this point (Figure 3A and Figure S5). To test the capacity for expansion, the isolated bone marrow cells were plated in StemSpan serum-free medium containing the growth factors SCF, TPO, IGF-2, FGF-1 and Angptl3 (STIFA medium) [33] (Figure 3B). As expected, the number of LSK cells was increased in control no-Cre and Cre-alone bone marrow cells, while Tak1iKO and Tab1Tab2iDKO LSK cells failed to expand. Annexin V-binding, which is associated with apoptosis, was greatly increased at day 9 of the expansion assay in both the Tak1iKO and Tab1Tab2iDKO LSK fractions (Figure 3C). Differentiated lineage positive Tak1iKO and Tab1Tab2iDKO cells were also found to be largely annexin V binding positive at day 9 (Figure 3D). These results indicate that deficiency of Tak1 impairs LSK expansion due to increased apoptosis, raising the possibility that TAK1 signaling may be required for survival of HSCs.
Figure 3. In vitro expansion of LSK population.
(A) The mice with indicated genotypes were i.p. injected with tamoxifen (160 mg/kg) for three consecutive days and sacrificed at day 4. The total LSK, MP and CLP cell numbers in the femurs and tibias from each mouse were determined. Data are presented as mean ± S.D. (n.s. not significant, n = 3) (B) Whole BMN cells isolated at day 4 described in (A) were cultured in STIFA medium. At the time of plating, 2.5×106 cells/well, 2×106 cells/well and 1×106 cells/well were plated for 3-, 6- and 9-day culture, respectively. Cells were harvested and analyzed by flow cytometry for lineage, Sca-1 and c-Kit surface markers. The absolute number of the LSK population in the harvested cells is shown as per 1×106 initial BMN cells. Data are presented as mean ± S.D. (*p<0.05, n = 3) (C, D) Annexin V-binding assay of LSK (C) and lineage positive cells (D) after nine days of cell expansion in STIFA medium. Data are presented as mean ± S.D. (*p<0.05, n = 3).
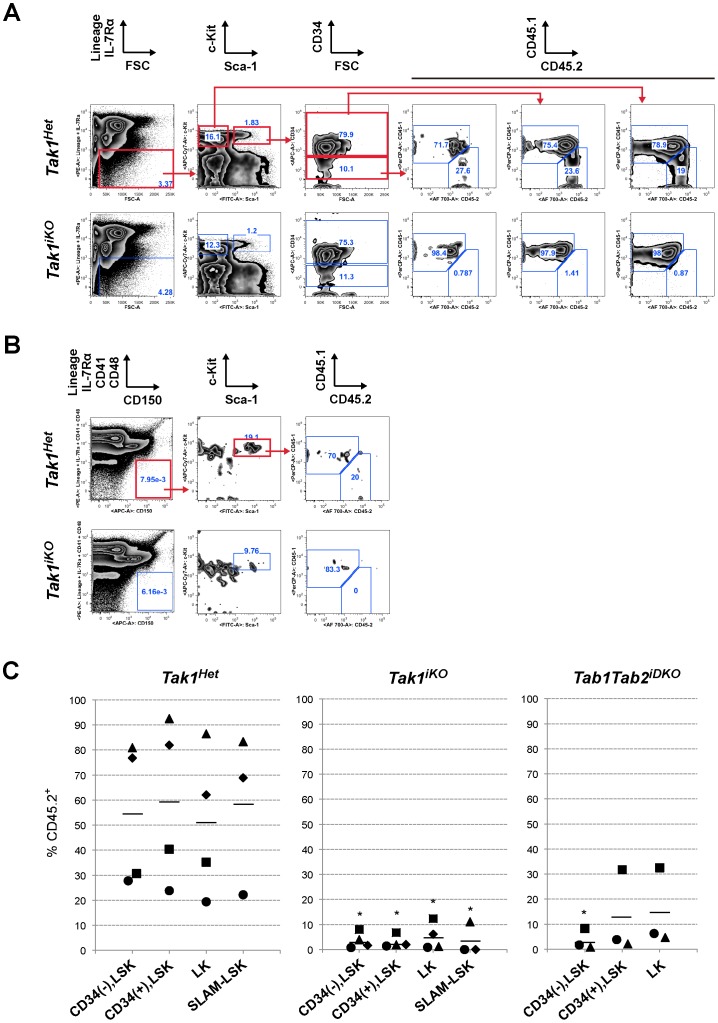
Ablation of TAK1 Signaling Depletes Long-term HSCs
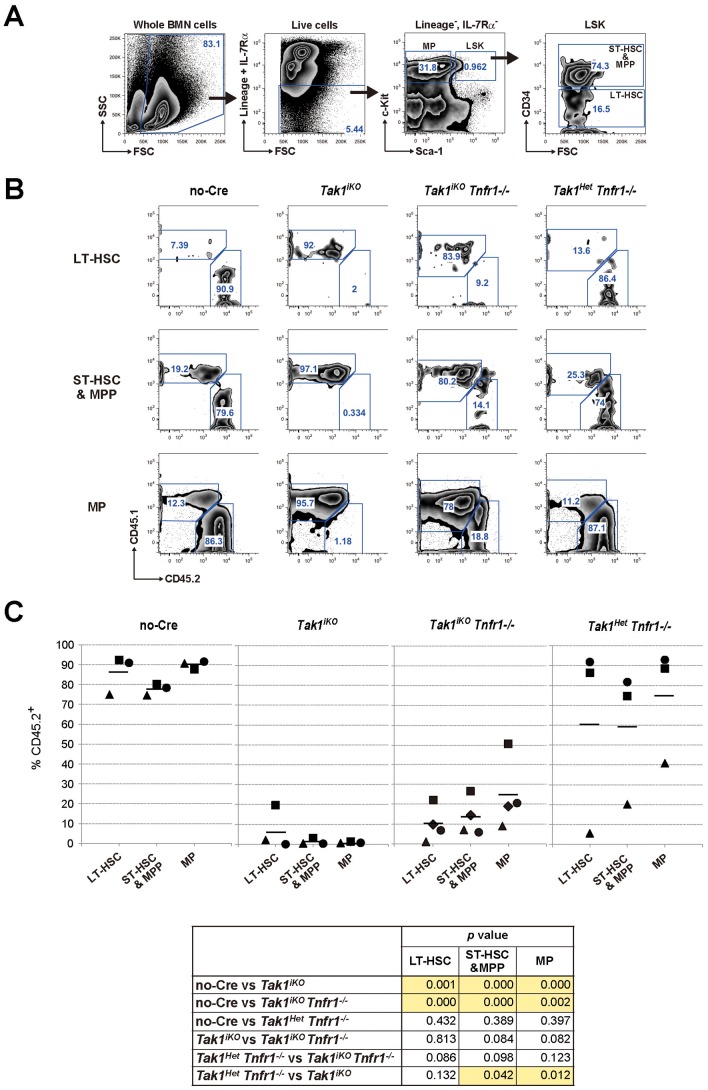
Since LT-HSCs have been reported to double approximately every 36 days (activated HSCs) or 145 days (dormant HSCs) in mice [38], and the number of LT-HSCs is less than 0.001% of bone marrow cells, the in vitro expansion assay could not permit the determination of whether the ablation of TAK1 signaling caused the failure of expansion or death of LT-HSCs. To further assess TAK1 regulation of LT-HSCs, we determined the populations of LT-HSCs at a relatively early time point after gene deletion in the competitive transplantation assay. If the TAK1 complex regulates self-renewal but not survival of LT-HSCs, then Tak1iKO LT-HSCs should still be present at three weeks post-gene deletion. Therefore, we analyzed bone marrow cells three weeks after tamoxifen injection in the competitively transplanted Tak1iKO bone marrow chimeric mice. While control Tak1Het bone marrow cells reconstituted CD45.2+ LT-HSC and ST-HSC/MPP populations with a reasonable level of chimerism of 30–90%, only small numbers of Tak1iKO CD45.2+ LT-HSC and ST-HSC/MPP cell numbers were present in in the competitively transplanted Tak1iKO bone marrow chimeric mice (Figures 4A, gating strategy, and 4C). Additionally, LSK population with SLAM family receptor CD150+, which is another phenotypic marker of LT-HSCs [5], was also diminished in the Tak1iKO CD45.2+ population (Figures 4B, gating strategy, and 4C). Similar to Tak1iKO donor-derived LSKs, both of the Tab1Tab2iDKO donor-derived LT-HSCs were also eliminated within three weeks post-tamoxifen injection (Figure 4C). In aggregate, these data suggest that TAB1/TAB2-dependent TAK1 signaling is required for survival of LT-HSCs.
Figure 4. HSC chimerism.
(A) The chimerism of LT-HSC, ST-HSC/MPP and MP in the recipient mice BM in competitive transplantation assay. The recipient mice (CD45.1+) were lethally irradiated and transplanted with a mixture of 1×106 test (Tak1Het and Tak1iKO, CD45.2+) and 2×105 competitor wild type (CD45.1+) BMN cells. Three weeks after tamoxifen injection, these recipients were analyzed for the chimerism of LT-HSC (CD34(–), LSK), ST-LSK/MPP (CD34(+) LSK) and MP (LK). Gating strategy and one result for each of the Tak1Het and Tak1iKO test BMN transplanted animals are shown. (B) Gating strategy for SLAM-LSK and one result for each of the Tak1Het and Tak1iKO test BMN transplanted animals are shown. (C) The chimerism of LT-HSC (CD34(–), LSK), ST-LSK/MPP (CD34(+) LSK), MP (LK) (n = 4) and SLAM-LSK (n = 3) in the recipient mice BM. The chimerism in each recipient is plotted, and the bars represent the average. * p<0.05 [Tak1Het versus Tak1iKO or Tab1Tab2iDKO].
Tnfr1 Deficiency Partially Rescues the Failure of Tak1-deficient HSC Function
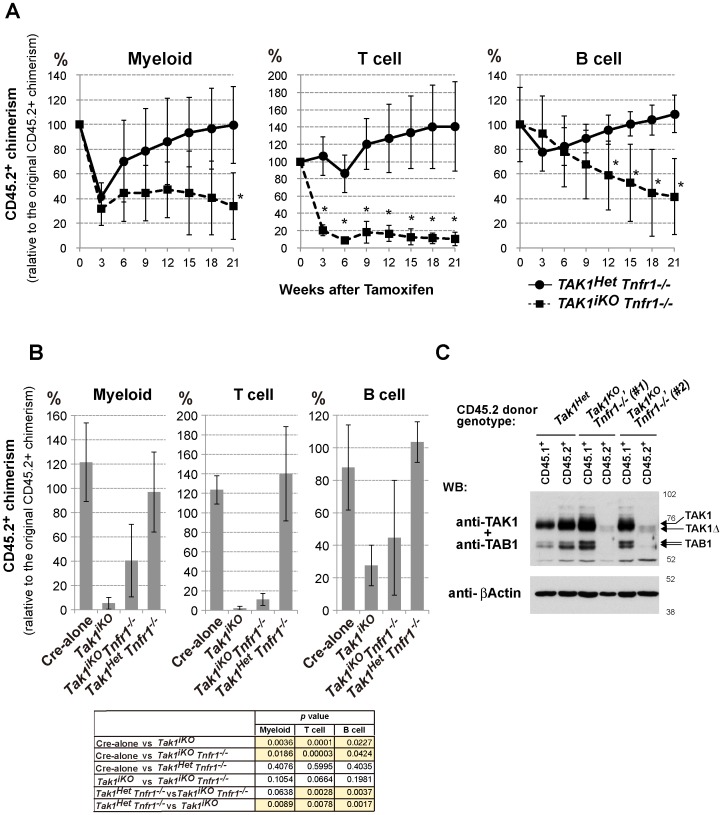
TNF receptor deficiency is reported to partially restore the reconstitution ability of Tak1-deficient bone marrow cells [27]. Tak1-deficiency is known to sensitize cells to TNF-induced cell death without altering TNF production in epithelial tissues [25], [26]. Therefore, Tak1-deficient HSCs may be killed by TNF which is expressed at a constant level in the bone marrow. Since the earlier study in Tak1-deficient bone marrow cells focused on relatively short-term reconstitution assays and did not provide detailed analyses for the capacity of Tak1 and TNF receptor double deficient HSCs for reconstitution, the contribution of TNF signaling to the inability of Tak1-deficient HSCs to reconstitute the hematopoietic system is not fully defined. Thus, we next investigated the reconstitution potential of Tak1iKO bone marrow cells in a Tnfr1−/− background over both a short and long term. The short-term reconstitution reflects ST-HSC function, while the long-term reconstitution requires LT-HSC function. Since TNF signaling is not known to profoundly impact on HSC function [37], we compared Tak1iKO Tnfr1−/− with control Tak1Het Tnfr1−/− and also with Tak1iKO cells. The Tak1iKO Tnfr1−/− and control bone marrow cells were subjected to a competitive transplantation assay using the same procedure as described in Figure 2A. The reconstitution from donor-derived cells was analyzed for 18 weeks after tamoxifen injection (Figure 5A). The level of reconstitution from Cre-positive control (Tak1Het Tnfr1−/−) donor cell was transiently reduced at three weeks (myeloid and B cells) and at six weeks (T cells) post-tamoxifen injection, which is associated with Cre toxicity, as described above. The transient reduction in control test donor cells was restored by six to nine weeks, and the controls exhibited stable reconstitution thereafter. In contrast, Tak1iKO Tnfr1−/− T cells were significantly reduced within three weeks and remained at a low level, and Tak1iKO Tnfr1−/− myeloid cells were also reduced, but without statistical significance until 21 weeks post-treatment (Figure 5A). The level of Tak1iKO Tnfr1−/− B-cell reconstitution slowly but persistently declined over 18 weeks, which resembles the Tak1iKO phenotype shown in Figure 2B. Thus, Tak1 deficiency still impaired HSC function even in the absence of TNFR1 signaling. However, the reconstitution level of Tak1iKO test donor at 18 weeks was found to be lower than that of Tak1iKO Tnfr1−/− (Figure 5B), suggesting that Tnfr1 deficiency might partially rescue the ST-HSC function in Tak1iKO. To confirm the gene deletion, the TAK1 protein in splenocytes from two independent Tak1iKO Tnfr1−/− donor mice (#1 and #2) and control mice was analyzed at 15 weeks (Figure 5C and Figure S6). Test donor- and competitor-derived splenocytes were separated by FACS, and immunoblotting of TAK1 and TAB1 was performed. In both Tak1iKO Tnfr1−/− donor transplanted mice, we could collect live test donor-derived splenocytes as shown in Figure S6, although #2 exhibited a lower test donor-derived cell number. The Tak1iKO Tnfr1−/− donor-derived splenocytes exhibited almost no expression of TAK1 (Figure 5C). TAB1 was also found to be greatly reduced in Tak1iKO Tnfr1−/− splenocytes (Figure 5C), as in other Tak1-deficient cell types [22]. Thus, TAK1 is not absolutely required for at least the short-term reconstitution of the hematopoietic system if the Tnfr1 gene is deleted. These results suggest that Tak1iKO Tnfr1−/− ST-HSCs are at least in part functionally intact and can reconstitute the hematopoietic system in the short term.
Figure 5. Ablation of TNF signaling partially restores the reconstitution potential of Tak1-deficient HSCs.
(A) Competitive reconstitution assay. 2×105 BMN cells from control Tak1Het Tnfr1−/− (n = 3) or Tak1iKO Tnfr1−/− mice (n = 4) (CD45.2+) were transplanted into lethally irradiated recipients (CD45.1+) together with 2×105 competitor wild type BMN cells (CD45.1+). At six weeks post transplantation (designated as Week 0), the chimerism of myeloid, T and B cells in the recipients’ PB was analyzed, and then the recipients were i.p. injected with tamoxifen at 160 mg/kg body weight for three consecutive days. The chimerism of PB cells was monitored every three weeks, and is shown as the mean ± S.D. (*p<0.05) (B) Competitive reconstitution assay of Cre-alone (n = 3) and Tak1iKO (n = 3) was also performed as described above, and compared with Tak1Het Tnfr1−/− (n = 3) and Tak1iKO Tnfr1−/− (n = 4) at 18 weeks post-tamoxifen injection. The table shown below the graphs indicates statistical significance for the indicated comparisons. P values of less than 0.05 are highlighted. (C) Expression of TAK1 and TAB1 proteins in the donor-derived splenocytes. Whole spleen cells from control Tak1Het and two independent Tak1iKO Tnfr1−/− transplanted mice (#1 and #2) at 15 weeks post-tamoxifen injection were sorted into the CD45.1+ or CD45.2+ population. Total cell lysates from the sorted splenocytes from control Tak1Het, and Tak1iKO Tnfr1−/− #1 and #2 were analyzed by SDS-PAGE and Western blotted with anti-TAK1, TAB1 or anti-β-actin antibodies. The positions of molecular weight markers are shown on the right. The arrows indicate the bands corresponding to endogenous TAK1 and TAB1, and truncated TAK1 (TAK1Δ) resulting from Cre-mediated recombination.
TAK1 is Required for HSC Maintenance Independently of TNF-induced Cell Death
Tak1iKO Tnfr1−/− bone marrow cells could reconstitute the hematopoietic system to some extent for at least 18 weeks. However, all Tak1iKO Tnfr1−/− derived myeloid, T- and B-cell chimerism levels were low and gradually declined, while control chimerism was not altered for a long time (Figure 5A). This phenomenon might indicate impaired LT-HSC function in Tak1iKO Tnfr1−/− bone marrow cells. To investigate this possibility, we analyzed bone marrow cells from competitively transplanted mice more than 22 weeks post Tak1 gene deletion (Figure 6). We phenotypically determined LT-HSC, ST-HSC/MPP and myeloid progenitor (MP) (Figure 6A, gating strategy). Controls including those receiving no-Cre and Tak1Het Tnfr1−/− test donor cells exhibited a chimerism level of 70–90% (Figure 6B-C) except one animal receiving Tak1Het Tnfr1−/− cells. Tak1iKO ST-HSC/MPP and MP cells were completely depleted as expected, and Tak1iKO Tnfr1−/− ST-HSC/MPP and MP cells were also reduced (Figure 6B-C). However, the level of Tak1iKO Tnfr1−/− MP cells was higher than Tak1iKO MP cells, suggesting that Tnfr1 deficiency might have partially rescued MPs. In contrast, Tak1iKO Tnfr1−/− LT-HSC and ST-HSC/MPP cells were still significantly reduced compared to the controls. Thus, TAK1 is required for the maintenance of the HSC pool, which is independent of the prevention of TNF-induced cell death.
Figure 6. Impaired LT-HSC function by Tak1 deficiency cannot be rescued by ablation of TNF signaling.
(A) Gating strategy for LT-HSC, ST-HSC/MPP and MP in the competitive reconstitution assay. (B) Representative results of chimerism analysis of No-Cre, Tak1iKO, Tak1iKO Tnfr1−/−, and Tak1Het Tnfr1−/− test donor transplanted mice are shown. (C) The chimerism of LT-HSC, ST-HSC/MPP and MP in the recipient mouse BM. Lethally irradiated recipient mice (CD45.1+) were transplanted with a mixture of 5×105 test (controls, No-Cre and Tak1Het Tnfr1−/−; and Tak1 deficient, Tak1iKO and Tak1iKO Tnfr1−/−, CD45.2+) and 5×105 competitor wild type (CD45.1+) BMN cells. The recipients were i.p. injected with tamoxifen at 160 mg/kg body weight for three consecutive days starting at six weeks post transplantation. Twenty-two weeks or more after tamoxifen injection, recipients were analyzed for the chimerism of LT-HSC (CD34−, LSK), ST-HSC/MPP (CD34+, LSK) and MP (LK) in BMN cells. The chimerism in each recipient is plotted, and the bars represent the average. The table shown below the graphs indicates statistical significance for the indicated comparisons. P values of less than 0.05 are highlighted. Mice were i.p. injected with tamoxifen at 160 mg/kg body for three consecutive days (Tak1iKO, Tab1iKO, Tab2iKO and Tab1Tab2iDKO) or untreated (Tak1FF, Tak1FF Cre or Tab1Tab2FF) and sacrificed on day 4. Genomic DNA isolated from BMN cells was analyzed by qPCR using primers designed to detect a portion of the genome flanked by two loxP sites to determine the relative copy number of intact Tak1, Tab1 or Tab2 genome. Data are presented as mean ± S.D. of three independent experiments. Mice with the indicated genotype were i.p. injected with tamoxifen (160 mg/kg) for three consecutive days, and splenocytes were collected at Day 14 (left panels) or Day 4 (right panels). Whole cell extracts were prepared and analyzed by Western blot using the indicated antibodies. Asterisks indicate non-specific bands. Mice with the indicated genotype were i.p. injected with tamoxifen (160 mg/kg body weight) for three consecutive days, and BMN cells were collected at Day 4. (A) The number of total BMN cells of femurs and tibias were counted on hemacytometer. The cell number in MP or LSK population was determined by FACS analysis. Data are presented as mean ± S.D. (n = 3) (*p<0.05) (B) Representative FACS plots for Sca-1 vs c-Kit in lineage-negative population of BMN cells are shown. The gates for MP and LSK and percentages of each population are indicated. (A) 2×105 BMN cells from Tab1FF, Tab1iKO, Tab2FF, or Tab2iKO mice (CD45.2+) were transplanted into lethally irradiated recipients (CD45.1+) together with 2×105 competitor wild type BMN cells (CD45.1+). At six weeks post transplantation, the chimerism of myeloid, T and B cells in the recipients’ PB was analyzed, then the recipients were i.p. injected with tamoxifen (160 mg/kg body weight) for three consecutive days. The chimerism of PB cells was monitored every three weeks. In each experiment, a CD45.1/CD45.2 mixture of BMN cells was transplanted into two recipients, and the average of their blood cell chimerism is shown. (B) Splenocytes from control No-Cre or Tab2iKO transplanted mice (#1 and #2) at 21 weeks post-tamoxifen injection were sorted into the CD45.1+ or CD45.2+ population. Total cell lysates from the sorted splenocytes were analyzed by SDS-PAGE and Western blotted with indicated antibodies. The positions of molecular weight markers are shown on the right. The asterisks indicate the non-specific bands. Mice with the indicated genotype (the same animals used for the in vitro LSK expansion assay in Figure 3) were i.p. injected with tamoxifen (160 mg/kg body weight) for three consecutive days, and BMN cells were collected at Day 4. BMN cells were analyzed by FACS to determine the number of B cell (B220+), T cell (CD3ε+) and granulocyte (CD11b+ and Gr-1+) per femurs and tibias. Data are presented as mean ± S.D (n = 3).
Our results demonstrate that TAK1 protects the hematopoietic system through two distinct mechanisms: one is by the prevention of TNF-induced cell death, which is important for the maintenance of ST-HSCs, and the second is via a TNF-independent process that contributes to LT-HSC maintenance. Undoubtedly, our understanding of HSC homeostasis will be enhanced by further study defining the TNF-independent mechanism through which TAK1 signaling regulates the LT-HSC compartment.
Supporting Information
Gene deletions of Tak1 , Tab1 and Tab2 in BMN cells. Mice were i.p. injected with tamoxifen at 160 mg/kg body for three consecutive days (Tak1iKO, Tab1iKO, Tab2iKO and Tab1Tab2iDKO) or untreated (Tak1FF, Tak1FF Cre or Tab1Tab2FF) and sacrificed on day 4. Genomic DNA isolated from BMN cells was analyzed by qPCR using primers designed to detect a portion of the genome flanked by two loxP sites to determine the relative copy number of intact Tak1, Tab1 or Tab2 genome. Data are presented as mean ± S.D. of three independent experiments.
(TIF)
Deletion of TAB1 and TAB2 in splenocytes. Mice with the indicated genotype were i.p. injected with tamoxifen (160 mg/kg) for three consecutive days, and splenocytes were collected at Day 14 (left panels) or Day 4 (right panels). Whole cell extracts were prepared and analyzed by Western blot using the indicated antibodies. Asterisks indicate non-specific bands.
(TIF)
Effect of CreERT activation on hematopoietic cells. Mice with the indicated genotype were i.p. injected with tamoxifen (160 mg/kg body weight) for three consecutive days, and BMN cells were collected at Day 4. (A) The number of total BMN cells of femurs and tibias were counted on hemacytometer. The cell number in MP or LSK population was determined by FACS analysis. Data are presented as mean ± S.D. (n = 3) (*p < 0.05) (B) Representative FACS plots for Sca-1 vs c-Kit in lineage-negative population of BMN cells are shown. The gates for MP and LSK and percentages of each population are indicated.
(TIF)
Either Tab1 or Tab2 is not essential for HSC function. (A) 2×105 BMN cells from Tab1FF, Tab1iKO, Tab2FF, or Tab2iKO mice (CD45.2+) were transplanted into lethally irradiated recipients (CD45.1+) together with 2×105 competitor wild type BMN cells (CD45.1+). At six weeks post transplantation, the chimerism of myeloid, T and B cells in the recipients’ PB was analyzed, then the recipients were i.p. injected with tamoxifen (160 mg/kg body weight) for three consecutive days. The chimerism of PB cells was monitored every three weeks. In each experiment, a CD45.1/CD45.2 mixture of BMN cells was transplanted into two recipients, and the average of their blood cell chimerism is shown. (B) Splenocytes from control No-Cre or Tab2iKO transplanted mice (#1 and #2) at 21 weeks post-tamoxifen injection were sorted into the CD45.1+ or CD45.2+ population. Total cell lysates from the sorted splenocytes were analyzed by SDS-PAGE and Western blotted with indicated antibodies. The positions of molecular weight markers are shown on the right. The asterisks indicate the non-specific bands.
(TIF)
Deletion of Tak1 or Tab1/Tab2 does not impact the number of B and T cell and granulocyte. Mice with the indicated genotype (the same animals used for the in vitro LSK expansion assay in Figure 3) were i.p. injected with tamoxifen (160 mg/kg body weight) for three consecutive days, and BMN cells were collected at Day 4. BMN cells were analyzed by FACS to determine the number of B cell (B220+), T cell (CD3ε+) and granulocyte (CD11b+ and Gr-1+) per femurs and tibias. Data are presented as mean ± S.D (n = 3)
(TIF)
Cell sorting strategy and efficiency. Presort gates were set around splenocytes, then CD45.1+CD45.2- and CD45.1-CD45.2+ cells were sorted. The sorting yielded 93 to 99% purity of target populations. Whole cell extracts were prepared from the sorted cells and subjected to Western blot analysis shown in Figure 5C.
(TIF)
Acknowledgments
We thank S. Akira for Tak1-floxed and Tab2-floxed mice, and S. Elliott, L. Hester, and J. Dow for support.
Funding Statement
This work was supported by “Keio Kanrinmaru Project”, the Promotion of Environmental Improvement for Independence of Young Researchers under the Special Coordination Funds for Promoting Science and Technology (JST, Japan) (to GT), and National Institutes of Health Grant GM068812 (to JN-T). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Orkin SH, Zon LI (2008) Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132: 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moore KA, Lemischka IR (2006) Stem Cells and Their Niches. Science 311: 1880–1885. [DOI] [PubMed] [Google Scholar]
- 3. Adams GB, Scadden DT (2006) The hematopoietic stem cell in its place. Nat Immunol 7: 333–337. [DOI] [PubMed] [Google Scholar]
- 4. Christensen JL, Weissman IL (2001) Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc Natl Acad Sci U S A 98: 14541–14546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, et al. (2005) SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121: 1109–1121. [DOI] [PubMed] [Google Scholar]
- 6. Suda T, Takubo K, Semenza GL (2011) Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 9: 298–310. [DOI] [PubMed] [Google Scholar]
- 7. Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, et al. (2007) FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128: 325–339. [DOI] [PubMed] [Google Scholar]
- 8. Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, et al. (2004) Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature 431: 997–1002. [DOI] [PubMed] [Google Scholar]
- 9. Liu J, Cao L, Chen J, Song S, Lee IH, et al. (2009) Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature 459: 387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nakada D, Saunders TL, Morrison SJ (2010) Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature 468: 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gurumurthy S, Xie SZ, Alagesan B, Kim J, Yusuf RZ, et al. (2010) The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature 468: 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gan B, Hu J, Jiang S, Liu Y, Sahin E, et al. (2010) Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature 468: 701–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ono K, Ohtomo T, Sato S, Sugamata Y, Suzuki M, et al. (2001) An evolutionarily conserved motif in the TAB1 C-terminal region is necessary for interaction with and activation of TAK1 MAPKKK. J Biol Chem 276: 24396–24400. [DOI] [PubMed] [Google Scholar]
- 14. Scholz R, Sidler CL, Thali RF, Winssinger N, Cheung PC, et al. (2010) Autoactivation of transforming growth factor β-activated kinase 1 is a sequential bimolecular process. J Biol Chem 285: 25753–25766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kanayama A, Seth RB, Sun L, Ea CK, Hong M, et al. (2004) TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell 15: 535–548. [DOI] [PubMed] [Google Scholar]
- 16. Jin G, Klika A, Callahan M, Faga B, Danzig J, et al. (2004) Identification of a human NF-κB-activating protein, TAB3. Proc Natl Acad Sci U S A 101: 2028–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheung PC, Nebreda AR, Cohen P (2004) TAB3, a new binding partner of the protein kinase TAK1. Biochem J 378: 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ishitani T, Takaesu G, Ninomiya-Tsuji J, Shibuya H, Gaynor RB, et al. (2003) Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO J 22: 6277–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen ZJ, Bhoj V, Seth RB (2006) Ubiquitin, TAK1 and IKK: is there a connection? Cell Death Differ 13: 687–692. [DOI] [PubMed] [Google Scholar]
- 20. Sanjo H, Takeda K, Tsujimura T, Ninomiya-Tsuji J, Matsumoto K, et al. (2003) TAB2 is essential for prevention of apoptosis in fetal liver but not for interleukin-1 signaling. Mol Cell Biol 23: 1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Inagaki M, Omori E, Kim JY, Komatsu Y, Scott G, et al. (2008) TAK1-binding Protein 1, TAB1, Mediates Osmotic Stress-induced TAK1 Activation but Is Dispensable for TAK1-mediated Cytokine Signaling. J Biol Chem 283: 33080–33086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Omori E, Inagaki M, Mishina Y, Matsumoto K, Ninomiya-Tsuji J (2012) Epithelial transforming growth factor beta-activated kinase 1 (TAK1) is activated through two independent mechanisms and regulates reactive oxygen species. Proc Natl Acad Sci U S A 109: 3365–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kajino-Sakamoto R, Omori E, Nighot PK, Blikslager AT, Matsumoto K, et al. (2010) TGF-β-activated kinase 1 signaling maintains intestinal integrity by preventing accumulation of reactive oxygen species in the intestinal epithelium. J Immunol 185: 4729–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Omori E, Morioka S, Matsumoto K, Ninomiya-Tsuji J (2008) TAK1 regulates reactive oxygen species and cell death in keratinocytes, which Is essential for skin integrity. J Biol Chem 283: 26161–26168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Omori E, Matsumoto K, Sanjo H, Sato S, Akira S, et al. (2006) TAK1 is a master regulator of epidermal homeostasis involving skin inflammation and apoptosis. J Biol Chem 281: 19610–19617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kajino-Sakamoto R, Inagaki M, Lippert E, Akira S, Robine S, et al. (2008) Enterocyte-derived TAK1 signaling prevents epithelium apoptosis and the development of ileitis and colitis. J Immunol 181: 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xiao Y, Li H, Zhang J, Volk A, Zhang S, et al. (2011) TNF-alpha/Fas-RIP-1-induced cell death signaling separates murine hematopoietic stem cells/progenitors into 2 distinct populations. Blood 118: 6057–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tang M, Wei X, Guo Y, Breslin P, Zhang S, et al. (2008) TAK1 is required for the survival of hematopoietic cells and hepatocytes in mice. J Exp Med 205: 1611–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Inagaki M, Komatsu Y, Scott G, Yamada G, Ray M, et al. (2008) Generation of a conditional mutant allele for Tab1 in mouse. Genesis 46: 431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sato S, Sanjo H, Takeda K, Ninomiya-Tsuji J, Yamamoto M, et al. (2005) Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol 6: 1087–1095. [DOI] [PubMed] [Google Scholar]
- 31. Badea TC, Wang Y, Nathans J (2003) A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci 23: 2314–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pfeffer K, Matsuyama T, Kundig TM, Wakeham A, Kishihara K, et al. (1993) Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell 73: 457–467. [DOI] [PubMed] [Google Scholar]
- 33. Zhang CC, Kaba M, Ge G, Xie K, Tong W, et al. (2006) Angiopoietin-like proteins stimulate ex vivo expansion of hematopoietic stem cells. Nat Med 12: 240–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schmidt-Supprian M, Rajewsky K (2007) Vagaries of conditional gene targeting. Nat Immunol 8: 665–668. [DOI] [PubMed] [Google Scholar]
- 35. Higashi AY, Ikawa T, Muramatsu M, Economides AN, Niwa A, et al. (2009) Direct hematological toxicity and illegitimate chromosomal recombination caused by the systemic activation of CreERT2. J Immunol 182: 5633–5640. [DOI] [PubMed] [Google Scholar]
- 36. Wan YY, Chi H, Xie M, Schneider MD, Flavell RA (2006) The kinase TAK1 integrates antigen and cytokine receptor signaling for T cell development, survival and function. Nat Immunol 7: 851–858. [DOI] [PubMed] [Google Scholar]
- 37. Pronk CJ, Veiby OP, Bryder D, Jacobsen SE (2011) Tumor necrosis factor restricts hematopoietic stem cell activity in mice: involvement of two distinct receptors. J Exp Med 208: 1563–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, et al. (2008) Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135: 1118–1129. 208: 1563–1570. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene deletions of Tak1 , Tab1 and Tab2 in BMN cells. Mice were i.p. injected with tamoxifen at 160 mg/kg body for three consecutive days (Tak1iKO, Tab1iKO, Tab2iKO and Tab1Tab2iDKO) or untreated (Tak1FF, Tak1FF Cre or Tab1Tab2FF) and sacrificed on day 4. Genomic DNA isolated from BMN cells was analyzed by qPCR using primers designed to detect a portion of the genome flanked by two loxP sites to determine the relative copy number of intact Tak1, Tab1 or Tab2 genome. Data are presented as mean ± S.D. of three independent experiments.
(TIF)
Deletion of TAB1 and TAB2 in splenocytes. Mice with the indicated genotype were i.p. injected with tamoxifen (160 mg/kg) for three consecutive days, and splenocytes were collected at Day 14 (left panels) or Day 4 (right panels). Whole cell extracts were prepared and analyzed by Western blot using the indicated antibodies. Asterisks indicate non-specific bands.
(TIF)
Effect of CreERT activation on hematopoietic cells. Mice with the indicated genotype were i.p. injected with tamoxifen (160 mg/kg body weight) for three consecutive days, and BMN cells were collected at Day 4. (A) The number of total BMN cells of femurs and tibias were counted on hemacytometer. The cell number in MP or LSK population was determined by FACS analysis. Data are presented as mean ± S.D. (n = 3) (*p < 0.05) (B) Representative FACS plots for Sca-1 vs c-Kit in lineage-negative population of BMN cells are shown. The gates for MP and LSK and percentages of each population are indicated.
(TIF)
Either Tab1 or Tab2 is not essential for HSC function. (A) 2×105 BMN cells from Tab1FF, Tab1iKO, Tab2FF, or Tab2iKO mice (CD45.2+) were transplanted into lethally irradiated recipients (CD45.1+) together with 2×105 competitor wild type BMN cells (CD45.1+). At six weeks post transplantation, the chimerism of myeloid, T and B cells in the recipients’ PB was analyzed, then the recipients were i.p. injected with tamoxifen (160 mg/kg body weight) for three consecutive days. The chimerism of PB cells was monitored every three weeks. In each experiment, a CD45.1/CD45.2 mixture of BMN cells was transplanted into two recipients, and the average of their blood cell chimerism is shown. (B) Splenocytes from control No-Cre or Tab2iKO transplanted mice (#1 and #2) at 21 weeks post-tamoxifen injection were sorted into the CD45.1+ or CD45.2+ population. Total cell lysates from the sorted splenocytes were analyzed by SDS-PAGE and Western blotted with indicated antibodies. The positions of molecular weight markers are shown on the right. The asterisks indicate the non-specific bands.
(TIF)
Deletion of Tak1 or Tab1/Tab2 does not impact the number of B and T cell and granulocyte. Mice with the indicated genotype (the same animals used for the in vitro LSK expansion assay in Figure 3) were i.p. injected with tamoxifen (160 mg/kg body weight) for three consecutive days, and BMN cells were collected at Day 4. BMN cells were analyzed by FACS to determine the number of B cell (B220+), T cell (CD3ε+) and granulocyte (CD11b+ and Gr-1+) per femurs and tibias. Data are presented as mean ± S.D (n = 3)
(TIF)
Cell sorting strategy and efficiency. Presort gates were set around splenocytes, then CD45.1+CD45.2- and CD45.1-CD45.2+ cells were sorted. The sorting yielded 93 to 99% purity of target populations. Whole cell extracts were prepared from the sorted cells and subjected to Western blot analysis shown in Figure 5C.
(TIF)