Abstract
Cyclooxygenase (COX)-2 expression and release of prostaglandins (PGs) by macrophages are consistent features of lipopolysaccharide (LPS)-induced macrophage inflammation. The two major PGs, PGE2 and PGD2, are synthesized by the prostanoid isomerases, PGE synthases (PGES) and PGD synthases (PGDS), respectively. Since the expression profile and the individual role of these prostanoid isomerases-mediated inflammation in macrophages has not been defined, we examined the LPS-stimulated PGs production pattern and the expression profile of their synthases in the primary cultured mouse bone marrow derived macrophages (BMDM). Our data show that LPS induced both PGE2 and PGD2 production, which was evident by ∼8 hrs and remained at a similar ratio (∼1∶1) in the early phase (≤12 hrs) of LPS treatment. However, PGE2 production continued increase further in the late phase (16–24 hrs); whereas the production of PGD2 remained at a stable level from 12 to 24 hrs post-treatment. In response to LPS-treatment, the expression of both COX-2 and inducible nitric oxide synthase (iNOS) was detected within 2 to 4 hrs; whereas the increased expression of microsomal PGES (mPGES)-1 and a myeloid cell transcription factor PU.1 did not appear until later phase (≥12 hrs). In contrast, the expression of COX-1, hematopoietic-PGDS (H-PGDS), cytosolic-PGES (c-PGES), or mPGES-2 in BMDM was not affected by LPS treatment. Selective inhibition of mPGES-1 with either siRNA or isoform-selective inhibitor CAY10526, but not mPGES-2, c-PGES or PU.1, attenuated LPS-induced burst of PGE2 production indicating that mPGES-1 mediates LPS-induced PGE2 production in BMDM. Interestingly, selective inhibition of mPGES-1 was also associated with a decrease in LPS-induced iNOS expression. In summary, our data show that mPGES-1, but not mPGES-2 or c-PGES isomerase, mediates LPS-induced late-phase burst of PGE2 generation, and regulates LPS-induced iNOS expression in BMDM.
Introduction
The prostaglandins (PGs) are a group of biologically active lipid compounds that are derived enzymatically from arachidonic acid or other polyunsaturated fatty acids, and mediate a variety of important physiological and pathophysiological functions in vivo [1], [2]. Arachidonic acid is released from the cell membrane by phospholipase A2, and is converted to unstable intermediate products PGG2 and PGH2, the precursor of the series-2 prostanoids, by rate limiting COX enzymes including COX-1 and COX-2 [3], [4]. COX-1 is reported as the constitutively expressed isoform in most mammalian cells, and its role has been reported in tumorigenesis [5]. COX-2 is the inducible COX isoform, and is rarely expressed in quiescent cells. However, COX-2 is expressed in a wide range cell types (e.g., macrophages) following appropriate stimulation including growth factors, tumor promoters, hormones, and inflammatory agents [6]. COX contains two active sites: a COX site, where arachidonic acid is converted into PGG2; and a heme site with peroxidase activity, responsible for the reduction of PGG2 to PGH2 [3]. PGH2 serves as the common substrate for subsequent distal prostanoid isomerases including PGES and PGDS to generate various bioactive PGs in vivo [6], [7]. The production profiles of PGE2 and PGD2 in specific cells are dependent on both the stimulus and the cell type. Alteration of the production profiles of PGs in vivo (e.g., PGE2 vs. PGD2 ratio) is a critical determinant in the development of many diseases including cancer [8], atherosclerosis [9], arthritis [10], and pulmonary diseases [11], [12]. PGE2 and PGD2 are the two major PG isoforms involved in many inflammatory and pulmonary diseases including chronic obstructive pulmonary disease [13], bronchiectasis [14], and bronchial asthma [11]. PGD2 is reported to have strong proinflammatory and bronchoconstrictive action in human and animal models of asthma [15]; whereas PGE2 detected in bronchoalveolar lavage fluid of asthmatic patients appears to be bronchoprotective and anti-inflammatory [16]. Therefore, understanding the type, amount, source, and timing of the PGD2 or PGE2 production in the microenvironment of the diseased organs or tissues is critical for determining the precise roles of each PG isoform in the pathogenesis of many inflammatory and pulmonary diseases.
PGD2 is produced from PGH2 in vivo by one of the two PGDS isomerases including lipocalin-PGDS (L-PGDS) and H-PGDS [17], [18]; whereas PGE2 is produced from PGH2 by three PGES isomerases including mPGES-1, mPGES-2 and c-PGES [19]–[21]. mPGES-1 is a membrane-associated enzyme with glutathione-dependent activity, and its expression is highly inducible in response to inflammatory stimuli [22], [23]. mPGES-2 is also a membrane-associated enzyme, but doesn't require glutathione for its catalytic activity. The third PGES isomerase c-PGES is also a glutathione-dependent enzyme, and is expressed in the cytosol of a wide variety of tissues and cells [7]. H-PGDS is widely distributed in the peripheral tissues and is localized in the antigen-presenting cells, mast cells, and megakaryocytes [24]. Recently, we showed the critical role of H-PGDS isomerase in mediating LPS-induced PGD2 production in BMDM [25].
PU.1 is an ETS transcription factor expressed in a wide variety of hematopoietic cells including most of the myeloid cells [26]. The critical roles of PU.1 in macrophage maturation and inflammatory response to LPS had been reported by others and us [26], [27]. It also has been well documented that many inflammatory stimuli can induce iNOS expression in a variety of cells including macrophages in various inflammatory diseases [28].
We and others have previously reported that LPS-induced production of PGD2 and PGE2 in macrophages including RAW294.7 cells and BMDM can be precisely quantified by a highly sensitive and selective liquid chromatography–tandem mass spectrometry (LC–MS-MS) method [4], [29], [30]. The overall purpose of this study was to precisely characterize the production patterns and the signaling mechanisms of the inflammatory mediators PGs, and to define the complete expression profile of PGs biosynthesis-related enzymes including the involved PGES isomerases in the primary cultured BMDM, which is the most commonly used primary cell model for studying macrophage functions in the development of macrophage-related inflammatory diseases.
Materials and Methods
Materials
Lipopolysaccharide (LPS) and NS-398 were purchased from Sigma (St. Louis, MO). CAY10526, antibodies for COX-1, COX-2, c-PGES, mPGES-1, mPGES-2, H-PGDS were from Cayman Chemical, Inc. (Ann Arbor, MI). The PU.1 antibody was purchased from Cell Signaling (Beverly, MA). The anti-iNOS antibody was purchased from BD Biosciences (San Jose, CA). The anti-β-actin antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). All TaqMan gene expression assays for real-time RT-PCR studies were purchased from Life Technologies (Grand Island, NY). The ON-TARGET plus siRNA's for all 3 PGES isoforms and their control siRNA were purchased from Dharmacon RNAi Technologies (Thermo Scientific). The Amaxa mouse macrophage nuclefector kit was purchased from Lonza (Switzerland).
BMDM isolation and culture
Wild-type C57BL/6 mice were obtained from Harlan (Indianapolis, IN). All procedures and protocols using mice were approved by the animal care committee (ACC) of the University of Illinois at Chicago (UIC) and the Jesse Brown Veterans Affairs Medical Center. BMDM were isolated from adult C57BL/6 mice as we previously described [32]. Briefly, after mice were euthanized, bone marrow was flushed from the femurs. The cells were washed and resuspended in DMEM medium containing 10% endotoxin-free fetal bovine serum (FBS) and 10% (vol/vol) L929 cell-conditioned medium as a biological source of macrophage colony-stimulating factor. The medium was then replenished at Day 4 in culture, and the nonadherent cells were removed. The adherent bone marrow cells were split and plated at a density of 1×106/plate into P60 culture plates, and were used for the experiments after Day 7 in culture, corresponding to a mature macrophage phenotype.
siRNA transfection
Primary cultured BMDM were transfected with 25 nM either ON-TARGET plus control siRNA or siRNA for PGES isoforms including mPGES-1, mPGES-2 and c-PGES using the Amaxa mouse macrophage nuclefector kit and transfection protocol (Catalog # VPA-1009, Lonza, Switzerland). After 36 hrs, BMDM were stimulated by LPS (1 µg/ml) for 16 h.
The in vitro cell-free enzyme assay
The method is similar to our previous reports using recombinant mPGES-1 and COX-2 enzymes [30], [31]. Briefly, the induced COX-2 and mPGES-1 enzymes were immunoprecipitated (IP, 2 hrs at 4°C) separately from equal amount of BMDM cell lysates at either 8 hrs or 16 hrs of LPS treatment under the same experimental condition. The IP COX-2 or mPGES-1 enzyme concentration was determined using Western blotting, and equal amount of IP COX-2 or mPGES-1 enzyme from each time point was used to determine their enzyme activity of PGE2 production from their upstream substrates. COX-2 or mPGES-1 enzymes were incubated in 0.2 ml Tris•HCl buffer (pH 8.0 at 37°C) in vitro with their enzyme substrates arachidonic acid (for COX-2) or PGH2 (for mPGES-1), respectively. After 30 min incubation at 37°C, the reaction was stopped by adding 50 µl 2 M HCl, and the reaction end product PGE2 was extracted from each incubation mixture and PGE2 concentration was determined to compare their enzyme activities between 8 or 16 hrs. For COX-2 enzyme activity assay [31], the IP-COX-2 enzymes from BMDM were incubated with 5 µM arachidonic acid in the presence of 1 µM hematin (COX-2 co-factor), 10 µl recombinant human mPGES-1 (Cayman), and 2.5 mM GSH (mPGES-1 co-factor). For mPGES-1 enzyme activity assay [30], the IP-mPGES-1 enzymes from BMDM were incubated with 2 µM PGH2 in the presence of 2.5 mM GSH. PGE2 was then extracted from each of the above incubation mixture using 800 µL hexane/ethyl acetate (50∶50, v/v). The organic phase was removed, evaporated to dryness, and reconstituted in 100 µL methanol/water (50∶50, v/v) for analysis using LC-MS-MS (see details in the next section).
Mass spectrometry
After the LPS treatment, the BMDM culture medium was collected and immediately stored at −80°C. The concentration of PGE2 and PGD2 in the collected culture medium was quantified by liquid chromatography in conjunction with mass spectrometry (LC-MS-MS) as we previously described [4], [30]. Briefly, HPLC separations were carried out using a Shimadzu (Columbia, MD) Prominence HPLC system with a Waters (Milford, MA) XTerra MS C18 (2.1 mm×50 mm, 3.5 µm) analytical column and a 5-min isocratic mobile phase consisting of acetonitrile/aqueous 0.1% formic acid (37∶63, v/v) at a flow rate of 200 µl/min. The HPLC system was interfaced to a Thermo-Finnigan (San Jose, CA) TSQuantum triple quadruple mass spectrometer that was operated using negative ion electrospray. Isomeric PGD2 and PGE2 were measured using a SRM transition of m/z 351 to m/z 271, and the SRM transition of m/z 355 to m/z 275 was selected for the internal standards d4-PGE2 and d4-PGD2.
Western blot analysis
The method is similar to that we described previously [33]–[35]. Briefly, BMDM were lysed in lysis buffer (1% Triton X-100, 150 mM NaCl, 10 mM Tris, pH 7.4, 1 mM EDTA, 1 mM EGTA, 0.4 mM phenylmethylsulfonyl fluoride, 0.5% Nonidet P-40), and sonicated for 2 seconds to shear DNA. Soluble lysates were separated by microcentrifugation, and volume representing equal amount of proteins were boiled at 95°C for 5 min before being resolved by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were transferred to PVDF membrane (Amersham Pharmacia Biotech), blocked with 5% non-fat dry milk for 1 h at room temperature, and then incubated with primary antibody at 4°C overnight. Protein was detected with horseradish peroxidase conjugated secondary antibody (Santa Cruz) and SuperSignal chemiluminescent substrate solution (Pierce). The protein loading of each sample was verified by staining the membrane with 0.1% Ponceau S solution.
Quantitative real-time reverse transcription polymerase chain reaction (RT-PCR)
Real-time PCR was conducted using ABI Prism 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions as we previously reported [36]. The mRNA expression levels of targeted genes and an endogenous housekeeping gene encoding for β-actin as an internal control, were quantified using TaqMan gene expression assays (Life Technologies). Amplification of specific PCR product was detected using the TaqMan Fast Real-Time PCR Universal Master Mix (Life Technologies). The quantitative real-time PCR was performed in duplicate in a total reaction volume of 20 µl according to manufacturer's instruction. Blank and positive controls (calibrators) were run in parallel to determine amplification efficiency within each experiment. During the extension step, the ABI Prism 7900HT Sequence Detection System monitored PCR amplification in real time by quantitative analysis of the emitted fluorescence. Each run was completed with a melting curve analysis to confirm the specificity of amplification and lack of primer dimmer. Quantification was performed using the ΔΔCt method. The target amount of each mRNA sample was subsequently divided by the control gene amount (which was assigned a value of 1 arbitrary unit) to obtain a normalized target value.
Confocal microscopy
BMDM were cultured in Nunc Lab-Tek II 8-well chamber slide (Fisher Scientific, Pittsburgh, PA) at 37°C and treated with or without LPS for various periods of time. BMDM were washed with 1× PBS and fixed for 20 min in 4% paraformaldehyde in PBS on ice and permeablized by 0.1% Tween 20 at room temperature (RT) for 5 min. After incubation with primary antibodies, BMDM were washed and incubated with Alexa Fluro 488-labled secondary antibodies (Life Technologies) for one hour at RT. After washing, the chamber slide was mounted with cover slips using Vectashield mounting medium (Vector Laboratories, Burlingame, CA) containing 4′, 6′ diamidino-2-phenylindole (DAPI) to stain the nucleus. Confocal microscopy images were acquired from the Carl Zeiss LSM 510 laser scanning confocal microscope equipped with a 63× water-immersion objective. Beams of 488 nm from Ag/Kr laser and 361 nm from UV laser were used for excitation. Green and blue emissions were detected through LP505 and 420 filters, respectively. The two different fluorochromes were scanned sequentially by using multi-tracking function to avoid any bleed through between the two dyes.
Statistical analysis
All data are reported as mean ± SEM. Significance tests (student's t-test or ANOVA) were performed using the Prism program (GraphPAD Software). A p value<0.05 was considered to be significant.
Results
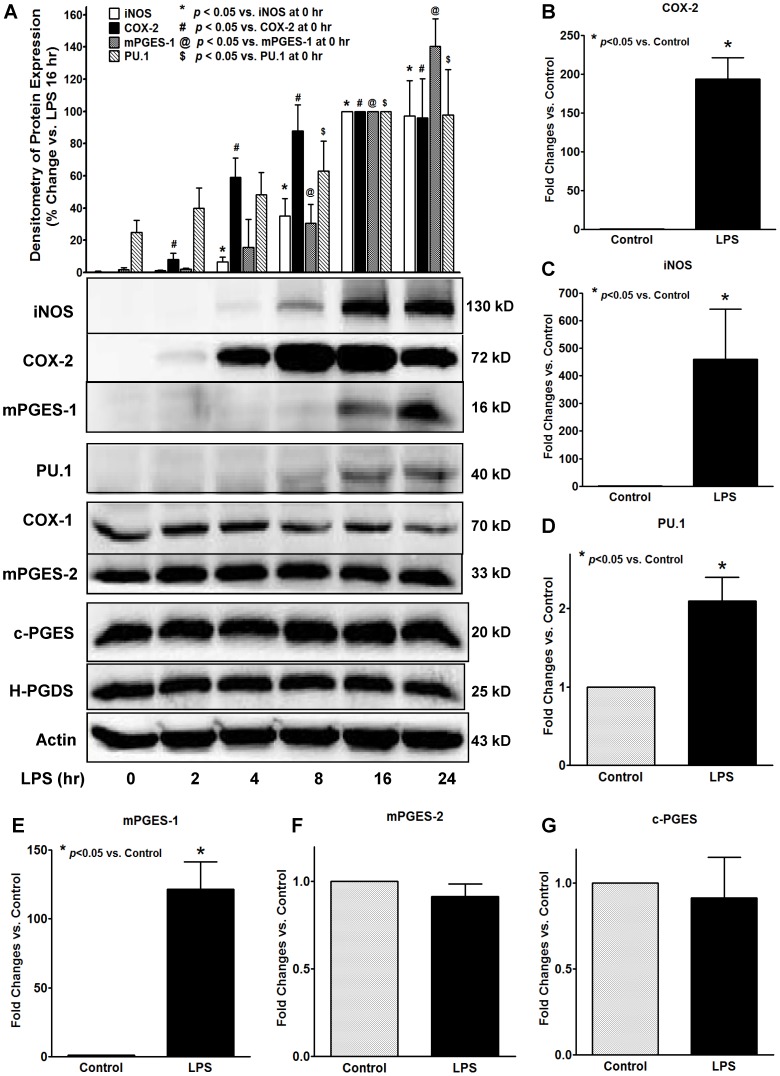
LPS stimulated expression of iNOS/COX-2/mPGES-1/PU.1 in BMDM
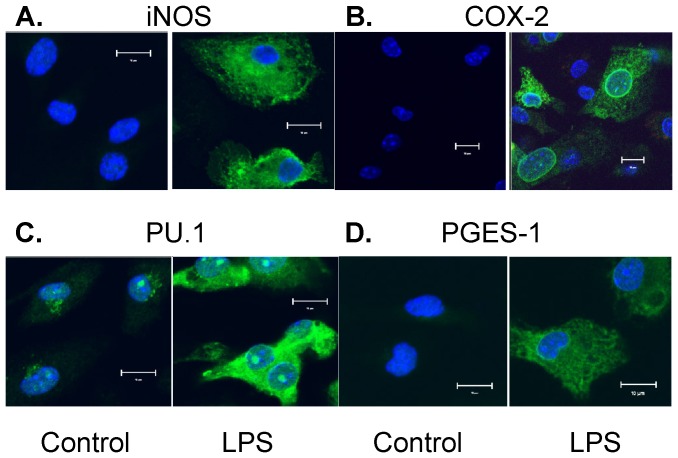
We first determined the protein expression profiles of a series of inflammation-related enzymes or transcription factor in primary cultured BMDM by Western blot assays, including enzymes related to macrophage inflammation (i.e., iNOS), PG biosynthesis (i.e., COX, PGES, and PGDS isoforms), and a transcription factor for macrophage maturation (i.e., PU.1). Since our previous studies had shown that LPS concentration-dependently (0.01 to 5 µg/ml) induced the production of both PGE2 and PGD2 in BMDM with a plateau level of PGs production at about 1 µg/ml [25], we used 1 µg/ml LPS for our current studies. Among the tested proteins, LPS significantly increased the protein expression of iNOS, COX-2, mPGES-1 and PU.1 in BMDM (Figures 1A, 2A–2D). Interestingly, the time-course results showed a distinct expression pattern of the above 4 inducible proteins in BMDM. LPS-induced protein expression of iNOS (∼4 hrs) and COX-2 (∼2 hrs) occurred earlier within 4 hrs of treatment; whereas the induction of the mPGES-1 and PU.1 protein expression did not appear until 12 hrs after LPS treatment (Figure 1A). In contrast, the other PGs synthases including COX-1, mPGES-2, c-PGES and H-PGDS were constitutively expressed in BMDM, and their protein expressions were not affected by LPS treatment (Figure 1A). The mRNA expression of iNOS, COX-2, mPGES-1 and PU.1 post-LPS treatment showed similar induction patterns (Figure 1B) as that seen for their protein expression (Figure 1A). Similarly, the protein expression profile and intra-cellular distribution of the above inducible proteins were also determined by immunostaining followed by confocol microscopy (Figure 2). LPS significantly increased the cytosolic protein staining for iNOS, COX-2, mPGES-1 and PU.1 (Figure 2), suggesting these are newly synthesized proteins in the cytosol in response to LPS treatment. In addition, we also found that LPS induced enhanced staining of both mPGES-1 and COX-2 in the perinuclear area of BMDM (Figure 2B and 2D), suggesting a potential protein-protein interaction of COX-2 and mPGES-1 enzymes working collectively in generation of PGE2. Unlike mPGES-1, although the expressions of the other two PGES isoforms mPGES-2 and c-PGES were detected in both LPS-treated and untreated BMDM, their protein and mRNA expressions were not induced or affected by LPS stimulation (Figures 1A, 1F, 1G) and they were diffusely expressed within the cytoplasmic compartment (data not shown).
Figure 1. LPS induces increased expression of iNOS/COX-2/mPGES-1/PU.1 in BMDM.
Primary cultured mouse BMDM was treated with or without 1 µg/ml LPS for various time points from 2 to 24 hrs. A. The protein expression of targeted proteins in BMDM cell lysates was detected by Western blot assays. Representative blots and the densitometry of iNOS/COX-2/mPGES-1/PU.1 protein expression are showed from 5 independent experiments. Protein expression of COX-2 and iNOS was detected within 2 to 4 hrs post-LPS treatment; whereas increased protein expression of mPGES-1 and PU.1 was detected after 8 hrs of LPS treatment. In contrast, the protein expression of COX-1, mPGES-2, c-PGES or H-PGDS was not affected by LPS treatment. The protein loading in each experiment was normalized by β-actin. B–G. The LPS-stimulated (16 hrs) mRNA expression of COX-2 (B), iNOS (C), PU.1 (D), mPGES-1 (E), mPGES-2 (F), or c-PGES (G) in BMDM was confirmed by real-time quantitative RT-PCR. The mRNA of each gene was normalized to the β-actin mRNA expression level in the same sample. The results shown are mean of at least 4 independent experiments for each gene, * p<0.05 vs. control.
Figure 2. Confocol fluorescent microscopy detects the expression and intracellular distribution of iNOS/COX-2/PU.1/mPGES-1 induced by LPS treatment.
Primary cultured mouse BMDM was treated with or without 1 µg/ml LPS for 16 hrs. The protein expression of iNOS (A), COX-2 (B), PU.1 (C), and mPGES-1 (D) was determined by immunostaining followed by confocol fluorescent microscopy. In each sample group, BMDM was stained with the green fluorescent-labeled antibody for the targeted protein and the blue fluorescent-labeled DAPI for nucleus; the overlay image of each targeted protein and nucleus are shown (magnification: ×200, scale bar: 10 µM). LPS stimulation significantly increased cytosolic expression of iNOS, COX-2, mPGES-1 and PU.1 in BMDM, suggesting newly synthesized protein expression in cytosol. PU.1 also showed strong nuclear staining (i.e., nuclear translocation) after LPS treatment; whereas both COX-2 and mPGES-1 showed similar enhanced perinuclear localization in LPS-treated groups. The results shown are representative images from 3 independent experiments.
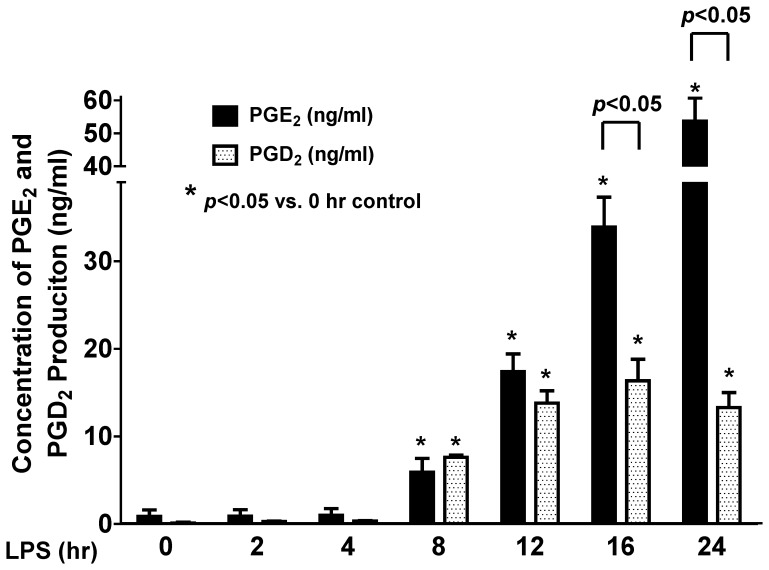
Patterns of LPS-stimulated PGE2 and PGD2 productions in BMDM
After LPS (1 µg/ml) treatment, the BMDM culture medium at each treatment time point was collected and frozen immediately at −80°C. The PGE2 and PGD2 concentration in the sample medium was quantified using LC-MS-MS method as we previously reported [4], [30]. LPS did not induce any detectable PGs production within the first 4 hrs of treatment, and the productions of both PGE2 and PGD2 were similar at 8 hrs (Figure 3). Although the production of PGE2 and PGD2 continued increase after 8 hrs, the ratio of PGE2 and PGD2 production was kept at ∼1∶1 between 8 and 12 hrs post-LPS treatment. However, the production of PGE2 significantly and continuously increased after 12 hrs; whereas the production of PGD2 stayed relatively stable from 12 to 24 hrs (Figure 3). These results indicated that although LPS-stimulated the production of both PGE2 and PGD2, their production patterns and signaling mechanisms are different in BMDM. The production of the purported pro-inflammatory PGD2 reached to a plateau level at about 12 hrs post-LPS treatment; whereas the production ability of the reported anti-inflammatory PGE2 still had not reached to its maximal level in BMDM.
Figure 3. LPS stimulates PGE2 and PGD2 production in BMDM.
Equal amount of BMDM was treated with or without 1 µg/ml LPS for various time points from 2 to 24 hrs. The BMDM culture medium was collected and the concentrations of PGE2 and PGD2 in the sample media were quantified using LC-MS-MS. LPS induced both PGE2 and PGD2 production at a similar lower level in the first 8 hrs of treatment. In contrast, the production of PGE2 in BMDM continuously increased after LPS treatment for 12 hrs; whereas the production of PGD2 kept at a relatively stable level after 12 hrs of LPS treatment (n = 5).
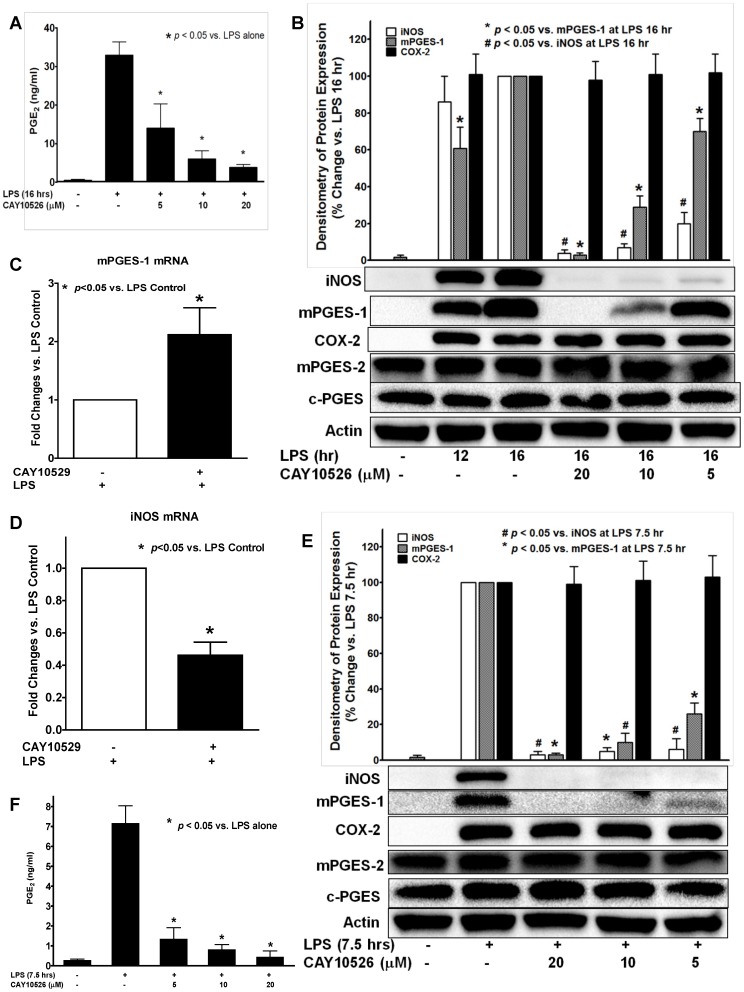
mPGES-1 selective inhibitor CAY10526 attenuated LPS-induced PGE2 production and iNOS expression in BMDM
Since the time course of the LPS-induced PGE2 production (Figure 3) highly correlated with the late-phase expression of mPGES-1 isomerase (Figure 1), we explored the potential role of the expression of mPGES-1 in the late-phase (16 hrs) burst of PGE2 production in BMDM. We found that the mPGES-1-selective inhibitor CAY10526 attenuated LPS-induced PGE2 production (Figure 4A) and protein expression of mPGES-1 in a concentration dependent manner, but did not alter the expression of other PGE2 synthesis-related enzymes including mPGES-2, c-PGES or COX-2 (Figure 4B), suggesting that mPGES-1 plays a critical role in the late-phase production of PGE2 in BMDM. Interestingly, we found that CAY10526 pretreatment significantly increased the mPGES-1 mRNA expression in BMDM as measured by real-time RT-PCR (Figure 4C). Previous reports have suggested that the inhibitory effect of CAY10526 on mPGES-1-synthesized PGE2 production was mediated via the selective inhibition of the mPGES-1 protein expression in RAW264 macrophages [37]. Our results confirm this finding in primary cultured BMDM and further indicate that this inhibitory effect of CAY10526 on mPGES-1 does not occur at its mRNA transcription level as these CAY10526-treated BMDM still could continue transcribe mPGES-1 mRNA without any inhibition in response to the LPS stimulation (Figure 4C). Instead, the inhibitory effect of CAY10526 possibly occurs at its mRNA translation step (leading to decreased expression of newly-synthesized mPGES-1 protein), or at the post-translational level (i.e., increasing mPGES-1 protein degradation). The increase of LPS-induced mPGES-1 mRNA transcription in the CAY10526-treated group (Figure 4C) suggested a novel positive-feedback mechanism in BMDM to overcome the loss of mPGES-1 enzyme activity and its protein expression (i.e., decreased translation of mPGES-1 mRNA into protein, or increased mPGES-1 protein degradation) due to the selective inhibition of CAY10526 on mPGES-1. Taken together, these data showed that the CAY10526 inhibited late-phase PGE2 production in BMDM was mediated via the selective inhibition of LPS-induced expression of mPGES-1, but not PGES-2, c-PGES, or COX-2. Surprisingly, we observed that CAY10526 pretreatment also attenuated LPS-induced expression of iNOS both at protein (Figure 4B) and mRNA (Figure 4D) levels in BMDM, but had no inhibitory effect on the other PGE2 synthesis-related enzymes including COX-2, mPGES-2 or c-PGES (Figure 4B). This result suggests that the mPGES-1 expression or mPGES-1-mediated PGE2 production contributes to the further induction of late-phase iNOS expression in BMDM. This finding was subsequently confirmed by our data using mPGES-1 siRNA (Figure 5B). In comparison, we also tested the effect of mPGES-1 inhibition on early phase (<8 hrs) LPS-induced iNOS expression in BMDM. In order to ensure the sufficient and detectable mPGES-1 expression and PGE2 production by LPS treatment, we chose 7.5 hrs as the early phase time point. Like in the late phase of LPS treatment (16 hrs), the mPGES-1-selective inhibitor CAY10526 also concentration-dependently inhibited both mPGES-1 and iNOS expression (Figure 4E), as well as the LPS-induced PGE2 production in BMDM (Figure 4F), suggesting the critical role of mPGES-1 in both earlier and late phase LPS-induced PGE2 production in BMDM.
Figure 4. mPGES-1-selective inhibitor CAY10526 concentration-dependently attenuated LPS-induced PGE2 production in BMDM.
BMDM was pretreated with mPGES-1-selective inhibitor CAY10526 at 5, 10, or 20 µM for 0.5 hr prior to LPS (1 µg/ml) treatment for either 16 hrs or 7.5 hrs. CAY10526 concentration-dependently inhibited the LPS-induced PGE2 production at 16 hrs (A) and 7.5 hrs (F) as measured by LC-MS-MS assay. Results are the mean of at least 3 independent experiments.. Western blot assays showed CAY10526 concentration-dependently attenuated the LPS-induced mPGES-1 and iNOS protein expression at either 16 hrs (B) or 7.5 hrs (E) in BMDM, but had no inhibitory effect on the protein expression of mPGES-2, c-PGES or COX-2. Representative blots and the densitometry of iNOS/mPGES-1/COX-2 protein expression were showed from at least 3 independent experiments. C–D. Real time RT-PCR results showed the efforts of CAY10526 pretreatment (10 µM, 0.5 hr prior to LPS treatment) significantly attenuated the LPS-induced (16 hrs) iNOS mRNA expression in BMDM (D); whereas LPS-induced (16 hrs) mPGES-1 mRNA expression in BMDM was significantly increased after CAY10526 treatment (C), confirming that CAY10526 selectively inhibited the translation step of mPGES-1 mRNA into proteins. Results shown were mean from 4 independent experiments.
Figure 5. Selective siRNA inhibition of mPGES-1, but not mPGES-2 or c-PGES, attenuated LPS-induced late-phase PGE2 production.
BMDM were transfected with siRNA's for mPGES-1, mPGES-2, c-PGES mRNA, or a control siRNA for 36 hrs, and were then treated with 1 µg/ml LPS for 16 hrs. A. The PGE2 level in culture medium was determined by LS-MC-MC. mPGES-1 siRNA significantly attenuated LPS-induced PGE2 production at 16 hrs in BMDM compared to that of BMDM transfected with the control siRNA. In contrast, the siRNA for either mPGES-2 or c-PGES did not affect the LPS-induced PGE2 production in BMDM compared to the control siRNA group. B. Western blot results showed that mPGES-1 siRNA not only selectively inhibit the protein expression of mPGES-1, but also that of iNOS. C. In contrast, transfection of BMDM with siRNA's for either mPGES-2 or c-PGES selectively attenuated the expression of its targeted protein expression accordingly, but had no inhibitory effect on LPS-induced expression of iNOS, COX-2, or mPGES-1. D. Real-time RT-PCR result confirmed that mPGES-1 siRNA significantly attenuated the LPS-induced mRNA expression of iNOS in BMDM at 16 hrs. E–G. Real-time RT-PCR results showed that siRNA's for mPGES-1 (E), mPGES-2 (F), or c-PGES (G) specifically inhibited the mRNA expression of its targeted PGES isoform compared to the control siRNA group, but did not affect the mRNA expression of the other two PGES isoforms in BMDM.
Selective siRNA inhibition of mPGES-1, but not mPGES-2 or c-PGES isomerase, prevented LPS-induced late-phase PGE2 production
To more precisely show a role for the various PGES isomerases in LPS-induced late-phase PGE2 production in BMDM, we used siRNA transfection method to test the effect of each of the three PGES isomerases on LPS-induced PGE2 production. siRNA's for mPGES-1, mPGES-2, c-PGES mRNA, and a control siRNA were used to transfect the BMDM prior to LPS treatment. mPGES-1 siRNA selectively inhibited LPS-induced expression of both mPGES-1 (Figures 5B & 5E) and iNOS (Figures 5B & 5D), and significantly attenuated the late-phase PGE2 production (Figure 5A). In contrast, siRNA inhibition of either mPGES-2 (Figures 5C & 5F) or c-PGES (Figures 5C & 5G) isomerase did not affect the PGE2 production compared to that of the BMDM transfected with the control siRNA (Figure 5A). These data indicated that mPGES-1 isomerase, but not mPGES-2 or c-PGES, mediated LPS-induced late-phase PGE2 production in BMDM. Furthermore, these siRNA results also confirmed our above finding with CAY10526 that mPGES-1 regulates LPS-induced iNOS expression in BMDM, as siRNA for mPGES-1, but not for mPGES-2 or c-PGES, significantly attenuated LPS-induced iNOS expression (Figures 5B & 5C). This finding is consistent with our above observation using the mPGES-1 selective inhibitor CAY10526 in BMDM.
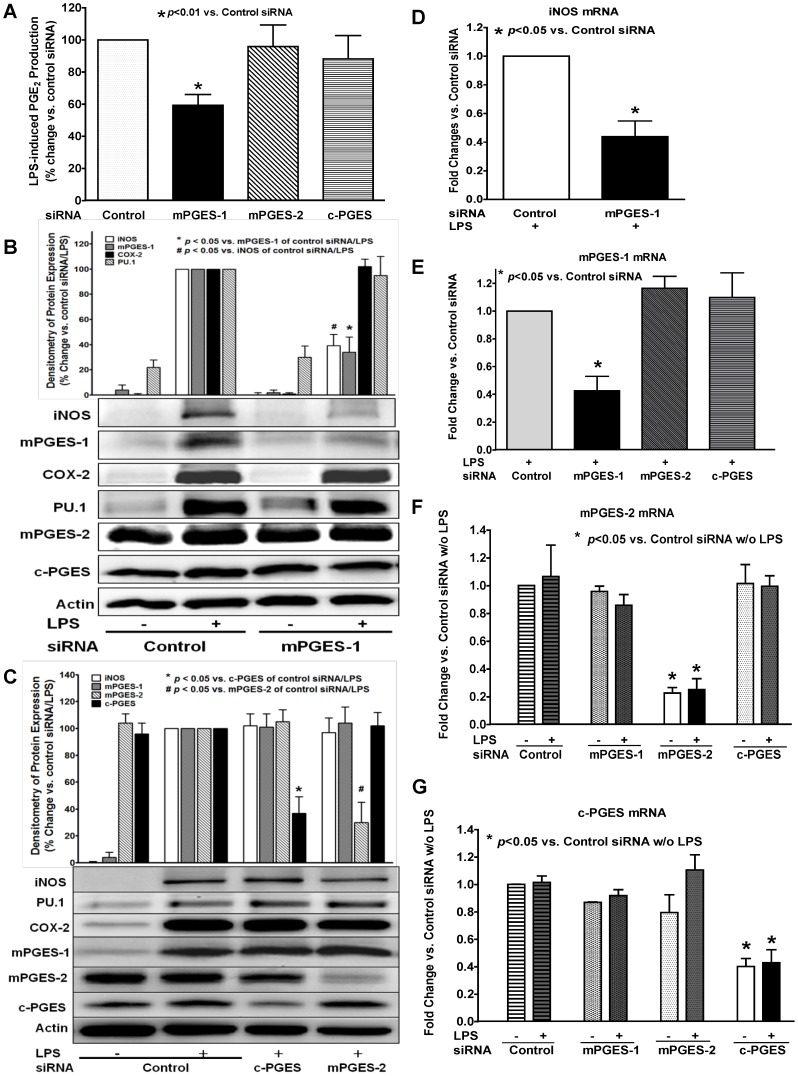
Selective siRNA inhibition of PU.1 in BMDM did not affect LPS-induced mPGES-1 or late-phase PGE2 production
We and others had previously reported the potential important roles of the ETS family transcriptional factor PU.1 in macrophage maturation and inflammatory response to LPS stimulation [26], [27]. In the current studies, we found LPS significantly induced PU.1 protein expression at late phase (∼16 hrs) similar to that seen in mPGES-1 expression. Thus, we explored the potential link between PU.1 expression and late-phase PGE2 production in response to LPS treatment using siRNA for PU.1 mRNA. We found that although PU.1 siRNA transfection significantly prevented LPS-induced expression of PU.1 at both protein (Figure 6A) and mRNA (Figure 6B) levels, it showed no inhibitory effect on either PGE2 production (Figure 6C) or the expression of any of the three PGES isomerases (Figure 6A). Conversely, we found that siRNA selective inhibition of any of the above three PGES isomerases also did not affect the LPS-induced PU.1 expression in BMDM (Figure 5B–5C). Therefore, our data suggested that although the LPS-induced PU.1 induction in BMDM appeared with a similar time-course (late-phase expression) to those of mPGES-1 expression (Figure 1) and PGE2 production burst, PU.1 expression may be parallel to or independent of the mPGES-1 expression signaling pathway, and thus may not directly contribute to the late-phase PGE2 production induced by LPS.
Figure 6. Selective inhibition of PU.1 didn't affect LPS-induced mPGES-1 expression or late-phase PGE2 production in BMDM.
BMDM was transfected with either PU.1 siRNA or a control siRNA for 36 hrs, and were then treated with or without 1 µg/ml LPS for 16 hrs. The concentration of PGE2 and PGD2 in culture medium was then determined by LS-MC-MC. The protein and mRNA expression in BMDM was determined by Western blot assay or real-time RT-PCR, respectively. A. PU.1 siRNA significantly prevented LPS-induced protein expression of PU.1 in BMDM, but had no inhibitory effect on the protein expression of iNOS, mPGES-1, mPGES-2, or c-PGES (representative blots and the densitometry of iNOS/mPGES-1/PU.1 protein expression were showed from 3 independent experiments). B. Real-time RT-PCR result confirmed that PU.1 siRNA significantly attenuated the PU.1 mRNA expression with or without LPS (16 hrs) treatment in BMDM. C. LPS-induced (16 hrs) PGE2 and PGD2 production in BMDM was not affected by PU.1 siRNA (n = 3).
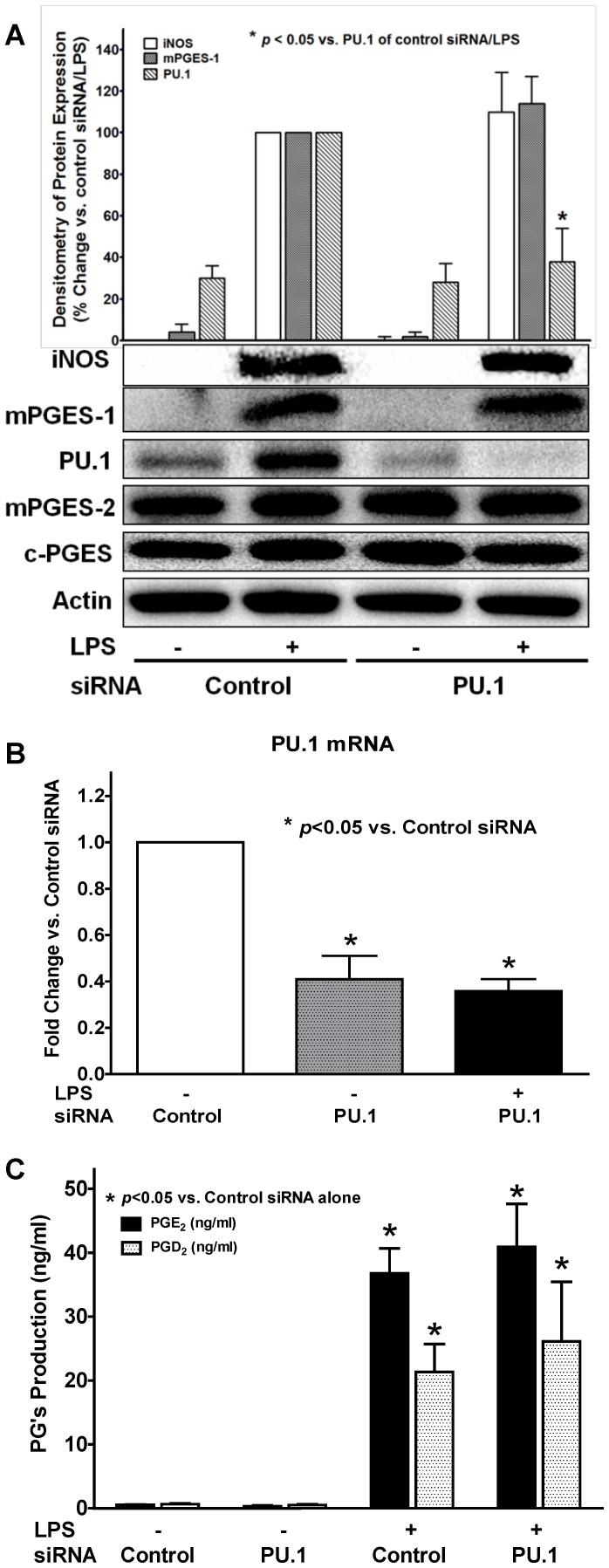
Selective inhibition of COX-2 completely inhibited PGs production, but did not affect LPS-induced mPGES-1 expression
It has been well documented in literature that LPS-induced PGs production in macrophages is mediated via selective expression and activation of the upstream PGs synthase COX-2 [6]. In order to test the potential role of COX-2 in LPS-induced mPGES-1 expression and late-phase PGE2 production, BMDM were pretreated with a COX-2-selective inhibitor NS-398 prior to LPS treatment. NS-398 pretreatment completely prevented LPS-induced both PGD2 and PGE2 production (Figure 7A) in BMDM via inhibition of COX-2 enzyme activity as previously reported [38], [39] and also partial inhibition of its protein expression (Figure 7B), but showed no inhibitory effect on LPS-induced expressions of the three PGES isomerases or COX-1 (Figure 7B) in BMDM. These data suggested that LPS-induced mPGES-1 expression is not directly triggered by or dependent on the expression and activation of its immediate upstream PG synthesis enzyme COX-2. In addition, we found that NS-398 pretreatment not only prevented LPS-induced PGE2 production but also attenuated the iNOS expression in BMDM, which was consistent with our findings that inhibition of late-phase PGE2 production using mPGES-1 siRNA or CAY10526 could attenuate LPS-induced iNOS expression in BMDM.
Figure 7. Selective inhibition of COX-2 abolished LPS-induced PGs production, but didn't affect LPS-induced mPGES-1 expression.
BMDM were pretreated with COX-2-selective inhibitor NS-398 (20 µM, 0.5 hr) prior to the treatment of 1 µg/ml LPS for 16 hrs, the PGE2 and PGD2 levels in culture medium were measured by LS-MC-MC. A. NS-398 completely prevented LPS-induced both PGE2 and PGD2 production (n = 3). B. NS-398 also partially inhibited LPS-induced protein expression of COX-2 and iNOS, but had no inhibitory effect on the expression of COX-1/PU.1/mPGES-1/mPGES-2/c-PGES by Western blot assay. The representative blots and the densitometry of iNOS/COX-2/mPGES-1/PU.1 protein expression were from 3 independent experiments.
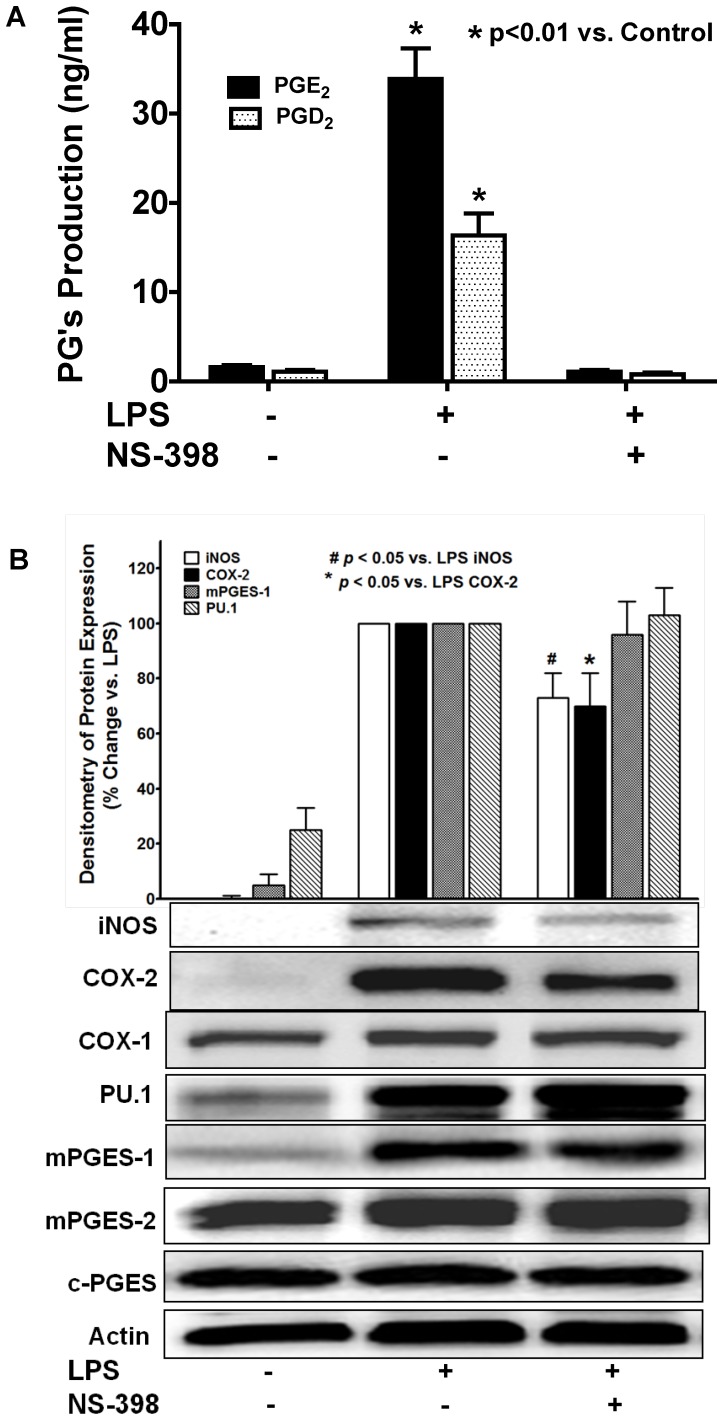
The enzyme activities of expressed COX-2 and mPGES-1 between early and late phase of LPS treatment were not significantly different
Although our results clearly demonstrated the significant difference of mPGES-1 protein expression in BMDM between early and late phase of LPS treatment (Figure 1), which also correlated with the changes of PGE2 production (Figure 3) at different time points, it is also possible that the enzyme activities of mPGES-1 or COX-2 changed in response to LPS at different time points. In order to explore this possibility, we compared the COX-2 and mPGES-1enzyme activities between 8 and 16 hrs of LPS treatment using equal amount of IP COX-2 or mPGES-1 enzymes. The enzyme activities were determined by in vitro enzyme assay measuring enzyme-mediated PGE2 production. Our results showed (Figure 8) that there is no significant difference in either COX-2 or mPGES-1 enzyme activity of PGE2 production in vitro between 8 and 16 hrs of LPS treatment, suggesting the difference of PGE2 production between 8 and 16 hrs of LPS treatment was not due to the changes of COX-2 or mPGES-1 enzyme activities, but more likely because of the difference in enzyme expression levels (i.e. mPGES-1) and/or other upstream factors such as substrate availability (e.g., arachidonic acid released by PLA2) at early or late phase of LPS treatment.
Figure 8. The enzyme activities of expressed COX-2 and mPGES-1 enzymes between early and late phase of LPS treatment were not different.
The induced COX-2 and mPGES-1 enzymes were immunoprecipitated (IP, 2 hrs at 4°C) separately from equal amount of BMDM lysates at either 8 or 16 hrs of LPS treatment under the same experimental condition. The IP COX-2 or mPGES-1 enzyme concentrations were determined, and equal amount of IP COX-2 or mPGES-1 enzymes from each time point was used to determine their enzyme activity of PGE2 production from their substrates (either arachidonic acid or PGH2) in vitro. No significant difference in either COX-2 or mPGES-1 enzyme activity of PGE2 production was detected between 8 and 16 hrs of LPS treatment (n = 3).
Discussion
PGE2 and PGD2 are the two major PGs in the human body with distinct biological functions in different tissues or cells in vivo. Macrophages are a major producing source of PGs and play an important role in the development of in many pulmonary and inflammatory diseases in vivo. BMDM are the most commonly used primary cell model to study the functions of macrophages in disease states [27], [32], [40]. However, the complete expression profile of PGs synthesis-related enzymes and the precise time courses and ratio of PGE2 and PGD2 production in response to LPS stimulation has not been previously characterized in BMDM. In the current study, we investigated the expression profile of PGs synthases and other inflammation-related proteins in BMDM; we also studied the LPS-induced PGs production patterns and the potential signaling mechanisms of the PGE2 production in BMDM.
We found that the LPS stimulation significantly increased the expression of inducible COX-2 and iNOS proteins similar to what we and others showed in macrophage cell line RAW294.7 [4]. The expression of these two proteins occurs relatively earlier at about 2 to 4 hrs post-LPS treatment, suggesting the direct cellular responses of synthesis of these two proteins in response to LPS stimulation. In contrast, unlike in RAW294.6 macrophages [4], the expression of prostanoid isomerase mPGES-1 and a transcription factor PU.1 in BMDM showed a distinct time course and only appeared to be inducible at a much later time (8–16 hrs) after LPS stimulation. The LPS-induced expression of the above four inducible proteins in BMDM at both protein and mRNA levels were confirmed by confocol microscopy and real-time RT-PCR, respectively. This is an interesting finding suggesting that mPGES-1 (late-phase expression) may have distinct activation mechanisms from COX-2 (early-phase expression) in response to LPS stimulation. In addition, we found that the increase in expression of mPGES-1 directly correlated with the significantly increased production of PGE2 at late phase post-LPS treatment in BMDM. Inhibition of mPGES-1 expression or its function by either mPGES-1-specific siRNA or mPGES-1 selective inhibitor CAY10526 prevented the LPS-induced production of PGE2, indicating a causal relationship between mPGES-1 expression and the burst of PGE2 production. In contrast, inhibition of the other two PGES isoforms including mPGES-2 and c-PGES did not affect the LPS-induced late-phase PGE2 production. These results clearly show a role for mPGES-1 in mediating the LPS-induced late-phase PGE2 production in BMDM.
Although mPGES-1 has been reported as an inducible isomerase previously [22], [23], we also found that it is constitutively expressed in RAW264.7 macrophages and not inducible by LPS treatment [4]. Therefore, our finding showed that the expression pattern and response of mPGES-1 enzyme in primary cultured BMDM are different from those in the commonly used macrophage cell line like RAW 264.7, which may explain the relatively lower production level of PGE2 vs. PGD2 production in our previous report using RAW264.7 cells [4], [36]. Our results are consistent with previous reports that the c-PGES and mPGES-2 are mainly constitutively expressed enzymes. c-PGES was reported to use PGH2 produced by COX-1 to generate PGE2 [21]; whereas mPGES-1 uses COX-2-dervived PGH2 as its preferred substrate [19]. mPGES-2 can use PGH2 generated from both sources [20]. In our studies, the mPGES-1 showed an enhanced perinuclear expression post-LPS treatment in addition to the elevated cytosolic expression using confocol microscopy, which is consistent with the COX-2 expression pattern in BMDM (Figure 2B and 2D), suggesting that the expression of these two sequential PG synthases are physically in close proximity to each other after LPS treatment. Since we found that the COX-2 protein is already highly expressed at about 4 hrs post-LPS treatment, thus BMDM should be able to generate PGH2 that accordingly serve as substrate for the available cellular PGES isomerases (i.e., mPGES-2 and c-PGES) at the time to produce PGE2 in BMDM. However, although both mPGES-2 and c-PGES isoforms are consistently present in the cytosol of BMDM, we did not detect any significant PGs production until 8 hrs post-LPS treatment. The results show that the expression of COX-2 alone with the presence of mPGES-2 and c-PGES isomerases may not be sufficient in biosynthesis of PGE2 in BMDM. Among the three PGES isomerases, mPGES-1 is reported to be the most efficient enzyme in catalyzing PGH2 to PGE2 (310 mM−1 s−1) [41] vs. mPGES-2 (65 mM−1 s−1) [42] or c-PGES (∼57 mM−1 s−1) [21], which is consistent with our finding of the significant increase of PGE2 production at late phase as soon as the mPGES-1 starts to appear. However, our results don't completely exclude a possible minor role for mPGES-2 or c-PGES in mediating the basal or low level of PGE2 production at the earlier phase (≤12 hrs) of LPS treatment in BMDM. In fact, since COX-2 expression already appears at 2∼4 hrs post-LPS treatment but the mPGES-1 expression was not detectable until about 8 hrs, it is likely that either mPGES-2 or c-PGES may contribute to the earlier phase basal or low level of PGE2 production in BMDM, as they could use the PGH2 generated by either the constitutively expressed COX-1 or the early expressed COX-2 enzymes to synthesize PGE2. Moreover, we recently reported that a portion of the COX-generated PGH2 (∼21%) can spontaneously covert to PGE2 or PGD2 in cell-free enzymatic assays in vitro without the presence of any PGDS or PGES enzymes in the reaction system [30]. We confirmed that neither COX-1 nor COX-2 has the PGES or PGDS isomerase activity in vitro, and thus this portion of spontaneous conversion of PGH2 to PGE2 or PGD2 in cell-free system is independent of COX-1 or COX-2 enzyme activity in vitro. It is possible that in addition to the potential roles of the mPGES-2 or c-PGES in LPS-induced early phase of PGE2 production, the COX-2 synthesized PGH2 might spontaneously convert to PGE2 in BMDM prior to the LPS-induced expression of mPGES-1. Nevertheless, our current studies focus on the late-phase burst of PGE2 production in BMDM, which generates incomparable large amount of the reported anti-inflammatory PGE2 and thus may potentially have more impact on its downstream signaling pathways or cellular functions in macrophages than that generated from the early phase.
Since COX-2 is the major rate-limiting upstream enzyme in PGE2 synthesis in response to LPS treatment in macrophages [6], we thus investigated the potential role of COX-2 in LPS-induced mPGES-1 expression and late-phase PGE2 production, we found that inhibition of COX-2 enzyme by its selective inhibitor NS-398 completely prevented the LPS-induced both PGE2 and PGD2 production, but did not have any effects on the LPS-induced mPGES-1 expression, suggesting that the expression of mPGES-1 is independent of COX-2 expression and the generation of PGs. Therefore, the signaling pathway of LPS-induced mPGES-1 expression in BMDM is not necessarily downstream from the COX-2 expression, but rather parallel to the signaling pathway leading to COX-2 expression, although both these two enzymes work coordinately in biosynthesis of PGE2 in response to LPS stimulation. In addition, our in vitro enzyme activity assays showed that the enzyme activities of the induced COX-2 and mPGES-1 enzymes were not different between early and late phase of LPS treatment in BMDM, suggesting the significant increase of PGE2 in late phase is most likely due to the LPS-induced increase of mPGES-1 protein expression in BMDM, but not increase of enzyme activities.
The role of PU.1 in macrophage maturation and LPS-stimulated inflammatory response has been previously reported by others and us [26], [27]. In the current studies, we found similar induction time-courses of PU.1 and mPGES-1 protein expression in response to LPS treatment (i.e., late-phase expression); we thus studied the potential relationship between PU.1 expression and late-phase PGE2 production and the potential regulatory role of PU.1 in mPGES-1 expression using siRNA method. We found that selective inhibition of LPS-induced PU.1 expression had no inhibitory effect on either PGE2 production or the protein expression of any of the three PGES isomerases. Conversely, selective inhibition of PGES isomerases by their siRNA's also did not affect the LPS-induced PU.1 expression in BMDM. Therefore, our results suggested that although LPS stimulated PU.1 expression in BMDM with a similar time-course to those of mPGES-1 expression and the burst of PGE2 production, PU.1 expression is required for neither mPGES-1 expression nor PGE2 production, and thus may not directly contribute to the LPS-induced late-phase mPGES-1 expression and PGE2 production.
Another interesting and potentially important finding of our studies is the effect of mPGES-1 expression on the LPS-induced iNOS expression in BMDM. Our data clearly showed that inhibition of mPGES-1 expression using either mPGES-1 siRNA or its selective inhibitor CAY10526 not only attenuated LPS-induced PGE2 production, but also prevented the iNOS expression at both protein and mRNA levels. Conversely, the expression of iNOS is strongly enhanced when the mPGES-1 expression starts to appear (Figure 1, 4B, 4E). These results were also confirmed by selective inhibition of COX-2 using NS-398, which inhibited both PGs production and the LPS-induced iNOS expression. To our knowledge, this is the first report showing the potential regulatory role of mPGES-1 in the expression of iNOS in macrophages. The expression of iNOS in phagocytes is considered as a hallmark for inflammatory response after pathogen exposure, and many inflammatory stimuli could lead to the expression of iNOS in macrophages [28]. PGE2 is a potent immunomodulator in inflammation, its actions in phagocytes could lead to either immunosuppressive conditions possibly via the activation of its EP2 and EP4 receptors that results in the production of cAMP [43], [44], or immunostimulatory effects via the activation of its EP3 receptor that results in decreased production of cAMP [45], [46]. We found that the burst of PGE2 production in BMDM is mediated via mPGES-1, and the inhibition of either mPGES-1 or COX-2 prevented PGE2 production and iNOS expression in BMDM. These data suggested that the PGE2 generated from mPGES-1 may contribute to and mediate the LPS-induced iNOS expression in BMDM. However, the precise signaling mechanism how mPGES-1 expression regulates LPS-induced iNOS expression still awaits further investigation.
In summary, our studies first determined the precise expression profile of PGs synthesis-related enzymes including the prostanoid isomerases and COX, and the production patterns of PGE2 and PGD2 in BMDM. LPS induces expression of iNOS, COX-2, mPGES-1 and PU.1 in BMDM with distinct expression time courses: The protein expression of COX-2 and iNOS appears within early phase (2–4 hrs) after LPS treatment; whereas LPS-induced mPGES-1 and PU.1 expression appears at a much later phase (after 8 hrs). LPS stimulates both PGD2 and PGE2 production in BMDM at a similar level between 8–12 hrs post-LPS treatment, but triggers significant burst of PGE2 production at later time (16–24 hrs). This late-phase burst of PGE2 production in BMDM is mediated via LPS-induced mPGES-1 expression, but not mPGES-2 or c-PGES isomerase, nor changes of enzyme activities of mPGES-1 or COX-2. Although COX-2 expression directly regulates LPS-induced production of PGE2 and PGD2, LPS-induced mPGES-1 expression in BMDM is independent of COX-2 or PU.1 expression. In addition, mPGES-1 expression directly regulates the LPS-induced iNOS expression likely via its production of PGE2 as its downstream mediator, because inhibition of mPGES-1 expression or PGE2 production prevented LPS-induced iNOS expression in BMDM. Our studies showed the unique late expression pattern of mPGES-1 in primary cultured macrophages for the first time, which mediates the burst of late-phase PGE2 production and regulates the iNOS expression. This late-phase burst of PGE2 production shifts the balance of the production of PGD2 and PGE2 in BMDM, and alters the ratio of the purported proinflammatory PGD2 and the anti-inflammatory PGE2 in macrophages, which may indicate an important in vivo functional adjustment in host defense in order to better manage the cellular response to bacterial infection.
Funding Statement
Supported in part by National Institutes of Health (NIH, http://www.nih.gov/) grants 1R01 HL083218 (LX), 3R01 HL083218-01A2S1 (LX), and 5R01 HL075557 (JWC); Merit Review Grant 1I01BX000108 (JWC) from Jesse Brown VA Medical Center (http://www.va.gov/); Campus Research Board Grant S06-118 (LX) and Faculty Scholarship Support Grants (LX) from the University of Illinois at Chicago(http://www.uic.edu/uic/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Funk CD (2001) Prostaglandins and leukotrienes: advances in eicosanoid biology. Science 294: 1871–1875. [DOI] [PubMed] [Google Scholar]
- 2. Hata AN, Breyer RM (2004) Pharmacology and signaling of prostaglandin receptors: multiple roles in inflammation and immune modulation. Pharmacol Ther 103: 147–166. [DOI] [PubMed] [Google Scholar]
- 3. Marnett LJ, Rowlinson SW, Goodwin DC, Kalgutkar AS, Lanzo CA (1999) Arachidonic acid oxygenation by COX-1 and COX-2: Mechanisms of catalysis and inhibition. J Biol Chem 274: 22903–22906. [DOI] [PubMed] [Google Scholar]
- 4. Cao H, Xiao L, Park G, Wang X, Azim AC, et al. (2008) An improved LC-MS/MS method for the quantification of prostaglandins E2 and D2 production in biological fluids. Anal Biochem 372: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sales KJ, Katz AA, Howard B, Soeters RP, Millar RP, et al. (2002) Cyclooxygenase-1 is up-regulated in cervical carcinomas. Cancer Res 62: 424–432. [PMC free article] [PubMed] [Google Scholar]
- 6. Simmons DL, Botting RM, Hla T (2004) Cyclooxygenase isozymes: The biology of prostaglandin synthesis and inhibition. Pharmacol Rev 56: 387–437. [DOI] [PubMed] [Google Scholar]
- 7. Samuelsson B, Morgenstern R, Jakobsson PJ (2007) Membrane prostaglandin E synthase-1: A novel therapeutic target. Pharmacol Rev 59: 207–224. [DOI] [PubMed] [Google Scholar]
- 8. Badawi AF, Eldeen MB, Liu Y, Ross EA, Badr MZ (2004) Inhibition of rat mammary gland carcinogenesis by simultaneous targeting of cyclooxygenase-2 and peroxisome proliferator-activated receptor γ. Cancer Res 64: 1181–1189. [DOI] [PubMed] [Google Scholar]
- 9. Burleigh ME, Babaev VR, Oates JA, Harris RC, Gautam S, et al. (2002) Cyclooxygenase-2 promotes early atherosclerotic lesion formation in LDL receptor-deficient mice. Circulation 105: 1816–1823. [DOI] [PubMed] [Google Scholar]
- 10. Li X, Afif H, Cheng S, Martel-Pelletier J, Pelletier JP, et al. (2005) Expression and regulation of microsomal prostaglandin E synthase-1 in human osteoarthritic cartilage and chondrocytes. J Rheumatol 32: 887–895. [PubMed] [Google Scholar]
- 11. Yoshimura T, Yoshikawa M, Otori N, Haruna S, Moriyama H (2008) Correlation between the prostaglandin D2/E2 ratio in nasal polyps and the recalcitrant pathophysiology of chronic rhinosinusitis associated with bronchial asthma. Allergol Int 57: 429–436. [DOI] [PubMed] [Google Scholar]
- 12. Scher JU, Pillinger MH (2009) The anti-inflammatory effects of prostaglandins. J Investig Med 57: 703–708. [DOI] [PubMed] [Google Scholar]
- 13. Montuschi P, Toni GC, Paredi P, Pantelidis P, du Bois RM, et al. (1998) 8-Isoprostane as a biomarker of oxidative stress in interstitial lung diseases. Am J Respir Crit Care Med 158: 1524–1527. [DOI] [PubMed] [Google Scholar]
- 14. Peebles RS Jr, Hashimoto K, Morrow JD, Dworski R, Collins RD, et al. (2002) Selective cyclooxygenase-1 and -2 inhibitors each increase allergic inflammation and airway hyperresponsiveness in mice. Am J Respir Crit Care Med 165: 1154–1160. [DOI] [PubMed] [Google Scholar]
- 15. Wenzel SE, Westcott JY, Smith HR, Larsen GL (1989) Spectrum of prostanoid release after bronchoalveolar allergen challenge in atopic asthmatics and in control groups: An alteration in the ratio of bronchoconstrictive to bronchoprotective mediators. Am Rev Respir Dis 139: 450–457. [DOI] [PubMed] [Google Scholar]
- 16. Sastre B, Fernandez-Nieto M, Molla R, Lopez E, Lahoz C, et al. (2008) Increased prostaglandin E2 levels in the airway of patients with eosinophilic bronchitis. Allergy 63: 58–66. [DOI] [PubMed] [Google Scholar]
- 17. Urade Y, Fujimoto N, Hayaishi O (1985) Purification and characterization of rat brain prostaglandin D synthetase. J Biol Chem 260: 12410–12415. [PubMed] [Google Scholar]
- 18. Kanaoka Y, Ago H, Inagaki E, Nanayama T, Miyano M, et al. (1997) Cloning and crystal structure of hematopoietic prostaglandin D synthase. Cell 90: 1085–1095. [DOI] [PubMed] [Google Scholar]
- 19. Jakobsson PJ, Thoren S, Morgenstern R, Samuelsson B (1999) Identification of human prostaglandin E synthase: A microsomal, glutathione-dependent, inducible enzyme, constituting a potential novel drug target. Proc Natl Acad Sci USA 96: 7220–7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Murakami M, Nakashima K, Kamei D, Masuda S, Ishikawa Y, et al. (2003) Cellular prostaglandin E2 production by membrane-bound prostaglandin E synthase-2 via both cyclooxygenases-1 and -2. J Biol Chem 278: 37937–37947. [DOI] [PubMed] [Google Scholar]
- 21. Tanioka T, Nakatani Y, Semmyo N, Murakami M, Kudo I (2000) Molecular identification of cytosolic prostaglandin E2 synthase that is functionally coupled with cyclooxygenase-1 in immediate prostaglandin E2 biosynthesis. J Biol Chem 275: 32775–32782. [DOI] [PubMed] [Google Scholar]
- 22. Mancini JA, Blood K, Guay J, Gordon R, Claveau D, et al. (2001) Cloning, expression, and up-regulation of inducible rat prostaglandin E synthase during lipopolysaccharide-induced pyresis and adjuvant-induced arthritis. J Biol Chem 276: 4469–4475. [DOI] [PubMed] [Google Scholar]
- 23. Murakami M, Naraba H, Tanioka T, Semmyo N, Nakatani Y, et al. (2000) Regulation of prostaglandin E2 biosynthesis by inducible membrane-associated prostaglandin E2 synthase that acts in concert with cyclooxygenase-2. J Biol Chem 275: 32783–32792. [DOI] [PubMed] [Google Scholar]
- 24. Herlong JL, Scott TR (2006) Positioning prostanoids of the D and J series in the immunopathogenic scheme. Immunol Lett 102: 121–131. [DOI] [PubMed] [Google Scholar]
- 25. Thomas S, Zhao G, Yu R, Deng J, Christman JW, et al. (2011) Critical role of reactive oxygen species in differential regulation of lipopolysaccharide-induced prostaglandins production in mouse bone marrow-derived macrophages. Am J Respir Crit Care Med 183: A1076. [Google Scholar]
- 26. Dahl R, Simon MC (2003) The importance of PU.1 concentration in hematopoietic lineage commitment and maturation. Blood Cells Mol Dis 31: 229–233. [DOI] [PubMed] [Google Scholar]
- 27. Karpurapu M, Wang X, Deng J, Park H, Xiao L, et al. (2011) Functional PU.1 in macrophages has a pivotal role in NFγB activation and neutrophilic lung inflammation during endotoxemia. Blood 118: 5255–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jorens PG, Matthys KE, Bult H (1995) Modulation of nitric oxide synthase activity in macrophages. Mediators Inflamm 4: 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Raetz CR, Garrett TA, Reynolds CM, Shaw WA, Moore JD, et al. (2006) Kdo2-lipid A of escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J Lipid Res 47: 1097–1111. [DOI] [PubMed] [Google Scholar]
- 30. Yu R, Xiao L, Zhao G, Christman JW, van Breemen RB (2011) Competitive enzymatic interactions determine the relative amounts of prostaglandins E2 and D2 . J Pharmacol Exp Ther 339: 716–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cao H, Yu R, Tao Y, Nikolic D, van Breemen RB (2011) Measurement of cyclooxygenase inhibition using liquid chromatography-tandem mass spectrometry. J Pharm Biomed Anal 54: 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Deng J, Wang X, Qian F, Vogel S, Xiao L, et al. (2012) Protective role of reactive oxygen species in endotoxin-induced lung inflammation through modulation of IL-10 expression. J Immunol 188: 5734–5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xiao L, Pimental DR, Amin JK, Singh K, Sawyer DB, et al. (2001) MEK1/2-ERK1/2 mediates α1-adrenergic receptor-stimulated hypertrophy in adult rat ventricular myocytes. J Mol Cell Cardiol 33: 779–787. [DOI] [PubMed] [Google Scholar]
- 34. Xiao L, Pimentel DR, Wang J, Singh K, Colucci WS, et al. (2002) Role of reactive oxygen species and NAD(P)H oxidase in α1-adrenoceptor signaling in adult rat cardiac myocytes. Am J Physiol Cell Physiol 282: C926–C934. [DOI] [PubMed] [Google Scholar]
- 35. Xiao L, Zhao Q, Du Y, Yuan C, Solaro RJ, et al. (2007) PKCε increases phosphorylation of the cardiac myosin binding protein C at serine 302 both in vitro and in vivo. Biochemistry 46: 7054–7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ornatowska M, Azim AC, Wang X, Christman JW, Xiao L, et al. (2007) Functional genomics of silencing TREM-1 on TLR4 signaling in macrophages. Am J Physiol Lung Cell Mol Physiol 293: L1377–L1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guerrero MD, Aquino M, Bruno I, Terencio MC, Paya M, et al. (2007) Synthesis and pharmacological evaluation of a selected library of new potential anti-inflammatory agents bearing the gamma-hydroxybutenolide scaffold: a new class of inhibitors of prostanoid production through the selective modulation of microsomal prostaglandin E synthase-1 expression. J Med Chem 50: 2176–2184. [DOI] [PubMed] [Google Scholar]
- 38. Futaki N, Yoshikawa K, Hamasaka Y, Arai I, Higuchi S, et al. (1993) NS-398, a novel non-steroidal anti-inflammatory drug with potent analgesic and antipyretic effects, which causes minimal stomach lesions. Gen Pharmacol 24: 105–110. [DOI] [PubMed] [Google Scholar]
- 39. Barnett J, Chow J, Ives D, Chiou M, Mackenzie R, et al. (1994) Purification, characterization and selective inhibition of human prostaglandin G/H synthase 1 and 2 expressed in the baculovirus system. Biochim Biophys Acta 1209: 130–139. [DOI] [PubMed] [Google Scholar]
- 40. Thoren S, Weinander R, Saha S, Jegerschold C, Pettersson PL, et al. (2003) Human microsomal prostaglandin E synthase-1: purification, functional characterization, and projection structure determination. J Biol Chem 278: 22199–22209. [DOI] [PubMed] [Google Scholar]
- 41. Hume DA, Allan W, Fabrus B, Weidemann MJ, Hapel AJ, et al. (1987) Regulation of proliferation of bone marrow-derived macrophages. Lymphokine Res 6: 127–139. [PubMed] [Google Scholar]
- 42. Tanikawa N, Ohmiya Y, Ohkubo H, Hashimoto K, Kangawa K, et al. (2002) Identification and characterization of a novel type of membrane-associated prostaglandin E synthase. Biochem Biophys Res Commun 291: 884–889. [DOI] [PubMed] [Google Scholar]
- 43. Takayama K, Sukhova GK, Chin MT, Libby P (2006) A novel prostaglandin E receptor 4-associated protein participates in antiinflammatory signaling. Circ Res 98: 499–504. [DOI] [PubMed] [Google Scholar]
- 44. Medeiros AI, Serezani CH, Lee SP, Peters-Golden M (2009) Efferocytosis impairs pulmonary macrophage and lung antibacterial function via PGE2/EP2 signaling. J Exp Med 206: 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Breyer RM, Bagdassarian CK, Myers SA, Breyer MD (2001) Prostanoid receptors: subtypes and signaling. Annu Rev Pharmacol Toxicol 41: 661–690. [DOI] [PubMed] [Google Scholar]
- 46. Goulet JL, Pace AJ, Key ML, Byrum RS, Nguyen M, et al. (2004) E-Prostanoid-3 receptors mediate the proinflammatory actions of prostaglandin E2 in acute cutaneous inflammation. J Immunol 173: 1321–1326. [DOI] [PubMed] [Google Scholar]