Abstract
Somatic Nucleophosmin (NPM1) mutation frequently occurs in acute myeloid leukemia (AML), but its role in leukemogenesis remains unclear. This study reports the first “conventional” knock-in mouse model of Npm1 mutation, which was achieved by inserting TCTG after nucleotide c.857 (c.854_857dupTCTG) to mimic human mutation without any “humanized” sequence. The resultant mutant peptide differed slightly different from that in humans but exhibited cytoplasmic pulling force. Homozygous (Npm1c+/c+) mice showed embryonic lethality before day E8.5, wheras heterozygous (Npm1wt/c+) mice appeared healthy at birth and were fertile. Approximately 36% of Npm1wt/c+ mice developed myeloproliferative disease (MPD) with extramedullary hematopoiesis. Those Npm1wt/c+ mice that did not develop MPD nevertheless gradually developed monocytosis and showed increased numbers of marrow myeloid precursors. This second group of Npm1wt/c+ mice also showed compromised cobblestone area formation, suggesting pathology in the hematopoietic niche. Microarray experiments and bioinformatic analysis on mice myeloid precursor cells and 227 human samples revealed the expression of CXCR4/CXCL12-related genes was significantly suppressed in mutant cells from both mice and humans. Thus, our mouse model demonstrated that Npm1 mutation can result in MPD, but is insufficient for leukemogenesis. Perturbation of hematopoietic niche in mutant hematopoietic stem cells (implied by underrepresentation of CXCR4/CXCL12-related genes) may be important in the pathogenesis of NPM1 mutations.
Introduction
NPM1 mutation in acute myeloid leukemia (AML) was first identified by the aberrant cytoplasmic localization of NPM1 protein, which is normally located in the nucleoli of non-mitotic cells [1]. The mutation consists of a tetra-nucleotide insertion near the C terminal end of the coding sequence of NPM1. The most frequent form of mutation is duplication of TCTG (type A, c.860_863dupTCTG), which results in alteration of the peptide sequence from DLWQWRKSL* to DLCL AVEEVSLRK* [1],[2]. The insertion is near the stop codon of the gene, but nonetheless disrupts 2 tryptophan residues that are critical for the localization of wild-type NPM1 protein in nucleoli [3]. Moreover, the mutant peptide contains a nuclear export signal (NES), causing the mutant NPM1 protein to shift into cytoplasm [4]. Some less frequent mutant forms involve insertion of 4 nucleotides between coding sequence position 863 and 864 or further 3′ ends; invariably, these mutations disrupt at least 1 tryptophan residue [2]. In vitro study has shown that an increased number of disrupted tryptophan residues is associated with incremental “traction” power of mutant NPM1 protein toward the cytoplasm [5].
The NPM1 protein is ubiquitously expressed in mammalian cells. The protein shuttles between nuclei and cytoplasm, with pleiotropic functions including ribosomal biogenesis [6], [7], centrosome duplication [8], and regulation of p53 and ARF proteins [9], [10]. Constitutional knock-out of Npm1 in mice results in embryonic lethality [11]. Heterozygous Npm1 knock-out produced phenotypes mimicking human myelodysplastic syndrome or malignancies of myeloid or lymphoid cells [12]. These findings suggest a haploinsufficient tumor suppressor role of Npm1 in vivo. However, the phenotypes of Npm1 haploinsufficiency of differ from those seen in AML patients with NPM1 mutation. Lymphoid malignancies and cytogenetic changes are associated with the phenotypes of Npm1 haploinsufficiency, whereas these 2 phenomena are rarely seen in human AML patients with NPM1 mutation [12]. This finding, together with the fairly fixed pattern of mutation in the NPM1 gene, suggests that the mutation is likely a gain-of-function one rather than simple haploinsufficiency.
Although the clinical and biological features of AML patients with NPM1 mutations have been well characterized [1], [13]–[18], the pathophysiological roles of NPM1 mutation remain to be defined. In mice bearing transgenic human NPM1 mutation, a number of animals developed myeloproliferation [19], indicating a pathological role of mutant NPM1 protein in myeloid disorders. Similarly, zebrafish bearing enforced human NPM1 mutant expression showed an expansion of hematopoietic cells [20]. Furthermore, a mouse model of knock-in of “humanized” NPM1 mutation (which resulted in a peptide sequence identical to the human mutation) showed expansion of the myeloid pool but shrinkage of B cells. In addition, AML phenotypes were evident in a percentage of aging mice [21]. All these animal models suggest that mutant NPM1 causes hematopoietic expansion, but is insufficeint for leukemogenesis. The pathogenetic role of NPM1 mutation in AML awaits further clarification.
This study was the firstto document a mouse Npm1 mutant knock-in model. In a previous study using a humanized NPM1 mutant knock-in model [21], human mutation nucleotide sequence was directly used to replace the wild-type mouse sequence. By contrast, we inserted TCTG after coding sequence nucleotide 857 (c.854_857dupTCTG) to mimic human NPM1 mutation. The minor difference in nucleotide sequences between humans and mice meant that a disruption in the reading frame would result in discrepancy of peptide sequences. The mutation of our model caused a shift of wild-type C terminal peptide sequence WQWRKSL* (amino acid 286–292) to LCLAVEEISLRKGFKQFEIFCLHFCNS* (amino acid 285 to 311), a pattern that differed slightly from the change observed in human NPM1 mutation, LCLAVEEVSLRK*. Similar to the humanized NPM1 mutant knock-in model, our homozygous Npm1 mutant knock-in mice showed embryonic lethality. About a third of the heterozygous mutant mice (Npm1wt/c+) in our study developed fatal myeloproliferative disease (MPD) but not AML. The Npm1wt/c+ mice without MPD nevertheless developed monocytosis in the peripheral blood, and increased numbers of myeloid precursors in the marrow, as they aged. Furthermore, hematopoietic stem-progenitor cells (HSPC) from the Npm1wt/c+ mice exhibited defective formation of the cobblestone areas. Microarray experiments and gene set enrichment analysis (GSEA) conducted on the cells from the mice and 227 human samples showed that expression of CXCR4/CXCL12-related genes was suppressed in the mutant cells, compared with NPM1 wild-type cells. This finding implied a perturbation of niche functions related to mouse and human NPM1 mutation.
Materials and Methods
Generation of Npm1-TCTG knock-in mouse model
The targeting vector was generated by retrieving whole Npm1 gene from bMQ-282-D14 BAC clone (LifeSciences, Nottingham, UK) and subcloning into a pL253 vector; this was followed by engineering with insertion of loxP-neo-loxP cassette and replacement of the wild- type sequence with TCTG insertion after coding sequence nucleotide 857 (c.854_857dupTCTG) (Figure 1A). The targeting vectors were electroporated into cultured embryonic stem (ES) cells from 129 mouse strain. The correctly targeted ES clones were identifieded by Southern blot analysis, followed by excision of the loxP-neo-loxP cassette and implantation into foster mothers. The mice containing germ-line transmission of the TCTG-inserted Npm1 mutant were confirmed by genotyping with PCR and sequencing of single TA clones. The PCR genotyping was performed using the following primers: forward: 5′ -GGCAACACT GGCCATAAAGT-3′; reverse: 5′ -GGTGGAGTTCCATCCTTGAA-3′. All animal protocols were approved by the Institutional Animal Care and Use Committee.
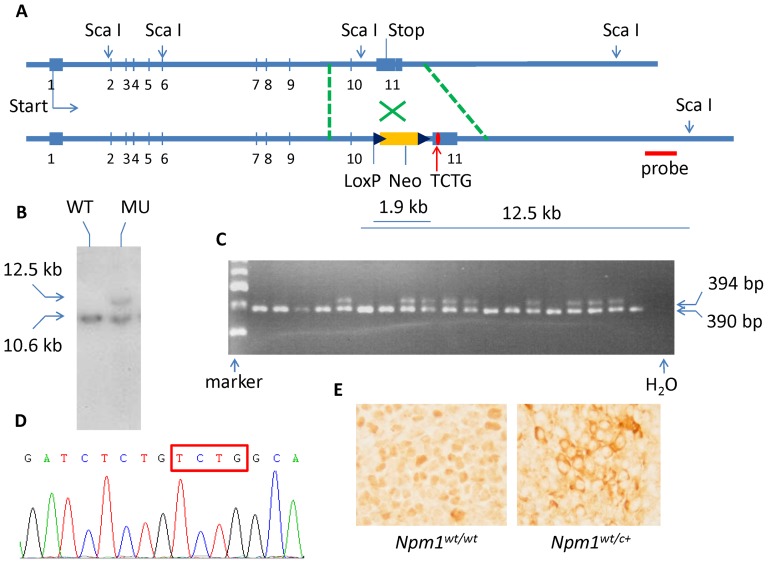
Figure 1. Generation of Npm1wt/c+ mutation knock-in mice.
(A) Mouse knock-in mutation construct map with the sites of ScaI restriction sites and fragment length. The loxP-neo-loxP cassette was removed before injection into foster mothers. (B) The knock-in was confirmed by Southern blot using a probe (as shown in panel A) which hybridized DNA digested with ScaI. (C) The mutant mice were confirmed by genotyping of the tail vein DNA. The upper band (394 bp) and lower band (390 bp) represented mutant and wild-type PCR products, respectively. Please note no homozygous mutant mice were born. (D) The mutant Npm1 sequence was confirmed in DNA level by direct sequencing of the individual TA clones from PCR products of mutant mice. The box indicates the mutated nucleotides. (E) The immunohistochemistry of Npm1 protein in mononuclear cells obtained from mouse spleen cells. The wild-type protein is located in the nuclei, whereas the mutated Npm1 shows aberrant cytoplasmic localization.
Blood and Tissue Collection
Peripheral blood was obtained from the submandibular vein in mice at various time points up to 24 months. Blood was collected into EDTA for hematologic analysis or heparinized tubes for flow cytometry. Mouse femurs and spleens were prepared as described [22]. Briefly, sterile bone marrow (BM) was obtained by flushing the femurs and tibias with D-PBS supplemented with 2% heat-inactivated FBS using a 25-gauge needle and a 3-ml disposable syringe. Cell suspensions from the spleen tissues were made by grinding the tissues between frosted ends of microscope slides, followed by cell counting. The average number of cells in representative samples of blood, spleen, or BM under flow cytometry was used to calculate the absolute number of positive cells. All the organs and tissues were obtained postmortem under sterile conditions.
Flow cytometery and sorting
Flow cytometric analysis of BM cells, blood, and splenocytes was performed with the following monoclonal antibodies conjugated to phycoerythrin, fluorescein, or PE-Cy5. For blood lineage analysis, we used rat anti-mouse Gr-1, F4/80, CD11b, CD3e, B220, Ter119, or NK1.1 antibodies (eBioscience, CA, USA). The hematopoietic stem-progenitor cells (HSPC), MPP (multipotent progenitor), CMP (common myeloid progenitor), GMP (granulocyte-macrophage progenitor), monocytes, and granulocytes were represented by c-kit+/sca-1+/Lin−, F4/80-/c-kit+, F4/80+/c-kit+, F4/80+/Gr-1+, F4/80+/Gr-1−, and F4/80−/Gr-1+, respectively. For marrow HSC and progenitor analyses, we used the CELLection magnetic dynabead separation kit to deplete the lineage-positive cells, and c-kit, sca-1, CD34, F4/80, or Gr-1 antibodies for staining. All data were collected and analyzed using a CyFlow instument (Partec, Germany). For collection of myeloid progenitors, a group of c-kit+/sca-1− cells were sorted using a FACSArial sorter (BD Bioscience, CA, USA).
Generation of GFP-Npm1 vectors
Wild-type and mutant mouse Npm1- and human NPM1-coding sequences were fused in frame with GFP and expressed in pcDNA3.1 (Life Technologies, Grand Island, NY, USA). This was followed by forced expression in 293T-HEK cells to identify and compare the cellular localization of mutant nucleophosmin proteins from mouse and human tissues.
Histology and immunohistochemistry
Tissue samples were fixed in PBS-buffered paraformaldehyde (4%) and dehydrated by gradual sucrose solution. Thereafter, they were embedded in Tissue-Tek O.C.T. compound medium for cryostat section. Tissues were cryosectioned at a thickness of 5 to 7 µm and used for histology or immunohistochemistry. Cryostat sections were stained with hematoxylin and eosin for histopathologic examination. For immunohistochemistry staining for Npm1 protein, we used a rabbit primary antibody (Cat. No. 3542, Cell Signaling Technology, Danvers, MA, USA).
Hematopoietic colony-Forming Unit Assay
BM or spleen cells were plated in M3434 methylcellulose medium (Stem Cell Technologies, Canada) as described previously [23]. All cultures were repeated in triplicate, and the colonies were scored at Days 6 to 9 and Days 12 to 15 of culture. Colony types were determined by in situ observation using an inverted microscope.
Cobblestone-area formation assay
To evaluate the hematopoietic differentiation and the contact behavior of hematopoietic stem and progenitor cells with marrow stromal cells, we seeded 5×104 hematopoietic cells from Npm1wt/c+ or wild-type mice on a confluent MS-5 BM stromal cell layer in 6-well plates. Cobblestone formation was determined after 5–6 days by counting the phase-dark cobblestone areas of at least 5 cells, using an inverted microscope. In all the experimental groups, we counted the number of cobblestone colonies as the average of 5 independent areas, each measuring 1 mm2 areas, using an inverted microscope.
Microarray labeling and hybridization
Gene expression profiling was performed for both human and mouse tissues. We used Human HT-12 v4.0 BeadChip (Illumina, San Diego, CA, USA) for RNA samples obtained from mononuclear cells of BM from human samples, and we used Mouse Ref-8 BeadChip (Illumina, San Diego, CA, USA) for RNA samples from sorted mouse c-kit+/sca-1− cells (with myeloid progenitors including MPP, CMP, and GMP). Because of limitation of the cell number, we pooled RNA from 3 mice each of either wild-type or non-MPD mutant mice for one single pair of microarray experiment. High quality RNA weighing 500 ng was converted to double-stranded cDNA, followed by an amplification step (in vitro transcription, Ambion, Austin, TX, USA) to generate biotin labeled cRNA (Ambion). Subsequently, 1.5 µg cRNA was hybridized to BeadChips at 58°C. After 14–20 hours hybridization, the BeadChips were stained by Cy3 and were then washed. The intensities of the bead fluorescence were detected by the Illumina BeadArray Reader, and the results were analyzed and quantile-normalized using GenomeStudio v2010.1 software. This study has been approved by the Institutional Review Board (IRB) of the National Taiwan University Hospital. Written informed consents were obtained from the human participants who provided tissue samples, in accordance with the regulations of the IRB.
Gene set enrichment analysis
We used GSEA software v2.07 (www.broadinstitute.org/gsea/index.jsp) to study specific groups of genes. We were interested in 22 gene sets related to HSPC functions, which we selected from the database of Ingenuity Pathway Analysis (IPA, Ingenuity Systems, www.ingenuity.com) (Table S1). The method determines whether predefined sets of genes are enriched in the observed gene expression profile. Genes are ranked according to the fold-change, or significant differences in expression, observed among phenotypes in the whole-genome expression data. Given a predefined gene set, an enrichment score (ES) is calculated to measure the overrepresentation of members of that gene set appearing at the extremes (top or bottom) of the ranked gene list. The ES is then evaluated for significance using phenotype-based or gene-based permutation tests. The ES, together with the permutation P value, indicates the degree to which the defined gene set is enriched in the gene expression data. Parameters and settings used in the analyses are shown in Table S2. The gene sets include (i) 2 sets of validated up-stream regulators and down-stream targets of CXCR4 and CXCL12 respectively, and (ii) 20 groups of genes involved in cellular functions in HSPCs, of which (i) and (ii) were exported from the database of IPA. Table S1 shows the complete contents of these 22 gene sets.
Statistical analysis
All data are presented as means ± S.E.M., and the differences between groups were assessed using the Student t test or one-way ANOVA with a significance level of P<0.05.
Results
In vivo expression of conventional Npm1wt/c+ mutation in mice
To study the role of Npm1 mutation in hematopoiesis and leukemogenesis in vivo, we inserted TCTG after nucleotide c.857 of murine Npm1 coding sequence (c.854_857dupTCTG), because this pattern mimics human NPM1 mutation but without any “humanized” sequence (Figure S1). This mutation causes a shift of peptide sequence from WQWRKSL* (amino acid 286–292) to LCLAVEEISLRKGFKQFEIFCLHFCNS* (amino acid 285 to 311), a pattern that differs slightly from the change in human NPM1 mutation. Nonetheless it also harbors a canonical NES, [LIVFM]–x(2,3)–[LIVFM]–x(2,3)–[LIVFM]–x–[LIVFM], and is predicted to generate an NES according to NES predictor (NetNES) [24]. We first examined the localization of this putative mutant Npm1 protein by forced expression of the mutant protein that was fused with GFP protein in 293T-HEK cells. The localization of the mutant protein in cytoplasm, compared with the restricted localization of wild-type Npm1 protein in the nucleoli, was similar to human condition (Figure S2). The process of generating heterozygous mutant mice (Npm1wt/c+) is shown in Figure 1A. Npm1wt/c+ genotype in our mice was confirmed by Southern blot and sequencing (Figure 1B–D). The subcellular distribution of Npm1 in blood cells from heterozygous mice was then confirmed by immunohistochemistry stain, which showed that the wild type protein was mainly located in the nuclei, whereas the mutated Npm1 protein had appeared in cytoplasm in spleen sections (Figure 1E). Homozygous Npm1 (Npm1c+/c+) mutation resulted in embryonic lethality before E8.5 day, while Npm1wt/c+ mice survived and were fertile.
Npm1wt/c+ mice showed myeloproliferative features
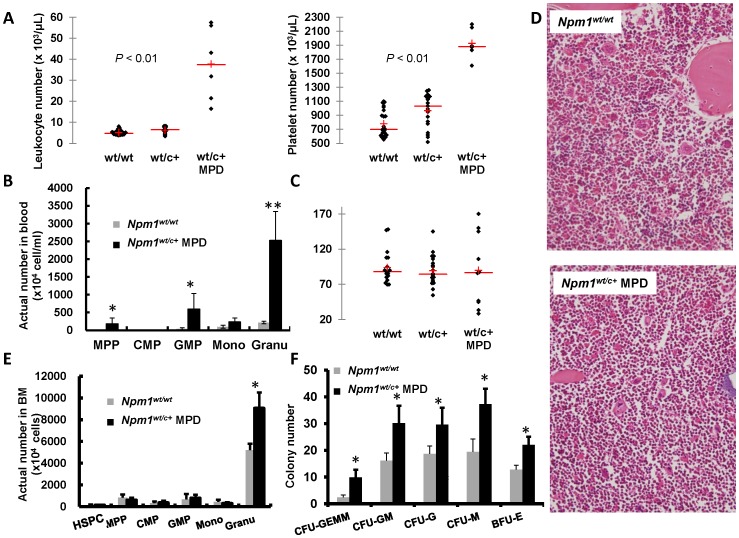
Data from mice aged between 2 to 24 months were analyzed. Eleven mice among 30 (36%) Npm1wt/c+ mice developed either leukocytosis (n = 5), thrombocytosis (n = 5), or both (n = 1); this pattern mimicked MPD in humans (Table 1). The 11 Npm1wt/c+ MPD mice displayed significantly higher platelet counts or white blood cell counts (P<0.01) compared with the age-matched wild-type or Npm1wt/c+ mice without MPD (Figure 2A). The red cell volumes, red blood cell count, hemoglobin, and mean platelet volume did not differ between the groups (data not shown). In Npm1wt/c+ mice with MPD, leukocytosis appeared at various ages, whereas thrombocytosis occurred only late in life (Table 1). Several mice displaying myeloproliferative features were fatally affected, but others were not moribund at the time of sacrifice. Flow cytometric analysis of blood cells showed a significant increase in the number of mature granulcoytes in Npm1wt/c+ mice with MPD (Figure 2B). Specifically, a group of hematopoietic progenitors including multipotent progenitors (MPP) (F4/80−/c-kit+) and granulocyte-macrophage progenitors (GMP) (F4/80+/Gr-1+), were detected in blood taken from the mice (Figure 2B). Although the total bone marrow (BM) cell count and cellularity in histopathology section did not differ significantly between the groups (Figure 2C and 2D), a significant higher count of mature granulocytes in BM was observed in mutant mice having myeloproliferative features (Figure 2E). Also, marrow cells from these mutant mice showed an increase in myeloid and erythroid colony formation (Figure 2F).
Table 1. The hematopoietic changes in white blood cell count, platelet count, and hemoglobulin of peripheral blood in wild type, Npm1wt/c+ mice without signs of myeloproliferative disease (MPD), and Npm1wt/c+ mice with sign of MPD.
| n | WBC (106/mL) | PLT (106/mL) | Hb (g/dL) | Age of MPDa | |
| Npm1wt/wt | 24b | 5.15±0.22d | 805.42±42.84 | 13.26±0.44 | |
| Npm1wt/c+ | 19b | 5.91±0.31 | 967.52±49.14 | 12.92±0.29 | |
| Npm1wt/c+ MPD | 11 | 23.95±6.06 | 1518.27±153.77 | 11.55±0.683 | |
| numberc | #5 | 5.2e | 1610 | 13.5 | 22 mon. |
| #7 | 10 | 2153 | 11 | 22 mon. | |
| #9 | 21.45 | 1295 | 14.4 | 24 mon. | |
| #18 | 56 | 1048 | 12.8 | 22 mon. | |
| #17 | 7.5 | 1891 | 11 | 22 mon. | |
| #24 | 43.1 | 1061 | 9.9 | 6 mon. | |
| #36 | 4.3 | 2200 | 12 | 22 mon. | |
| #54 | 10 | 1831 | 8.5 | 22 mon. | |
| #57 | 16.5 | 1874 | 13.8 | 24 mon. | |
| #97 | 57.5 | 1017 | 8.2 | 9 mon. | |
| #66 | 31.9 | 721 | 12 | 12 mon. |
Age of MPD: the age when MPD developed in Npm1-mutated mice.
The leukocytes (WBC), platelets (PLT), and hemoglobulin (Hb) count of peripheral blood from wild type and mutant mice without MPD were collected at 12 months.
number: the code representing each Npm1wt/c+ mouse with sign of MPD.
Data is represented as mean ± SE.
Data is represented as individual value of each Npm1wt/c+ mouse with MPD.
Figure 2. Hematopoietic changes in peripheral blood and BM in Npm1wt/c+ mice with myeloproliferative features.
(A) Monthly blood counts in peripheral blood were monitored in 24 Npm1wt/wt and 30 Npm1wt/c+ mice, respectively. Five, five, and one Npm1wt/c+ mice had abnormal increase of leukocytes, platelets or both in blood, respectively. (B) The Npm1wt/c+ mice with myeloproliferative features showed higher numbers of MPP (F4/80−/c-kit+), GMP (F4/80+/Gr-1+) progenitor cells, and granulocytes, compared with wild-type mice; n = 3 in each group. (C) No differences of total BM cells among the mice whether they are Npm1-wild type or Npm1-mutant with myeloproliferative features or without (n = 20 for wild-type; n = 20 for Npm1wt/c+ without MPD; n = 10 for Npm1wt/c+ with MPD). (D) Histologic sections of BM from wild-type and MPD mutant mice did not show obvious difference in cellularity (H & E staining, 400×). However, the mature granulocyte number is significantly higher for mutant mice with myeloproliferation ; n = 3 each for wild-type and Npm1wt/c+ with MPD (E). In addition, the colony forming ability is significantly elevated in those mutant mice with myeloproliferation; n = 4 each for wild-type and Npm1wt/c+ with MPD (F). * means P<0.05; ** means P<0.01. MPP, multipotent progenitor; CMP, common myeloid progenitor; GMP, granulocyte-macrophage progenitor; CFU-GEMM, CFU-granulocyte, erythroid, macrophage, megakaryocyte; CFU-GM, CFU-granulocyte macrophage; CFU-G, CFU-granulocyte; CFU-M, CFU-macrophage, and BFU-E, burst-forming unit-erythroid.
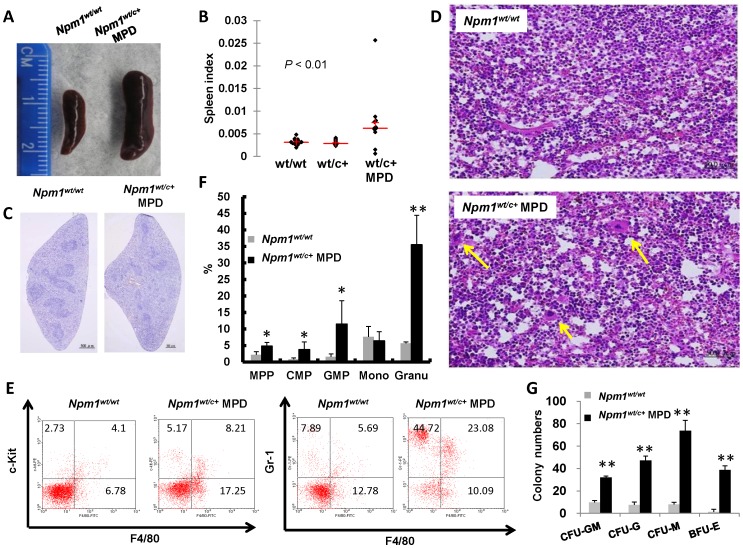
The Npm1wt/c+ mice with MPD exhibited splenomegaly and higher spleen-weight index compared with wild-type or non-MPD mutant mice (Figure 3A–B). Histomorphological examination of the Npm1wt/c+ mouse spleens showed effacement of follicle structure, and extramedullary hematopoiesis with megakaryocyte infiltration (Figure 3C–D). Flow cytometric analysis and colony formation assay confirmed the existence of hematopoietic precursor cells and panmyelosis in the spleens (Figure 3E–G).
Figure 3. Hematopoietic changes of spleen in Npm1wt/c+ mice with myeloproliferation.
(A, B) Mutated Npm1wt/c+ mice with signs of myeloproliferation had splenomegaly and higher spleen weight index (spleen weight/body weight). n = 20 for wild-type; n = 20 for Npm1wt/c+ without MPD; n = 10 for Npm1wt/c+ with MPD). (C, D) Spleen sections from wild-type and MPD mutant mice stained with hematoxylin demonstrated changes in the follicle structure (C), and infiltration with megakaryocytes (arrows), indicating extramedullary hematopoiesis (D). (E) Npm1wt/c+ mice with signs of myeloproliferation had more CMP (c-kit+/F4/80+, right upper quadrant of the left panel), GMP (Gr-1+/F4/80+, right upper quadrant of the right panel), and granulocytes (Gr-1+/F4/80−, left upper quadrant of the right panel), as shown by flow cytometry. (F, G) Npm1wt/c+ mice with signs of myeloproliferation had extramedullary hematopoiesis as shown by increased immature and mature myeloid cells (n = 3 each; * P≤0.05) and hematopoietic colony formation in spleen cells (n = 3).
Npm1wt/c+ mice without MPD revealed myeloid expansion
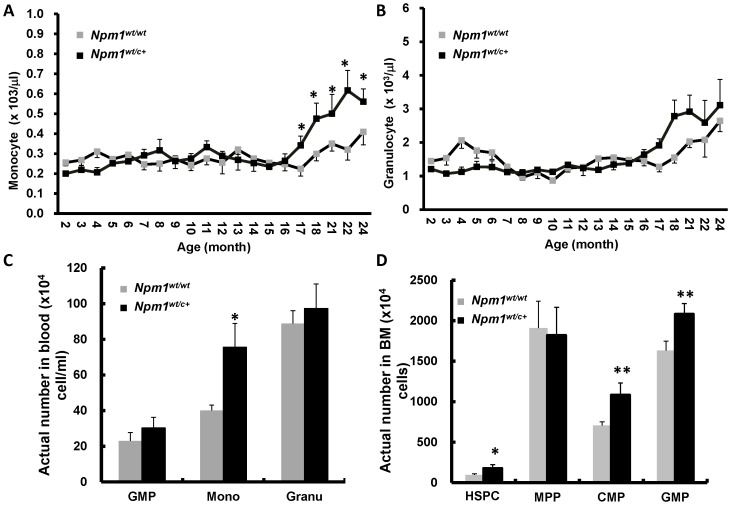
Most Npm1wt/c+ mice obtained normal hemograms and appeared clinically disease-free throughout their lives. However, these mice developed a delayed-onset increase of monocytes (but not granulocytes, lymphocytes or platelets) in their peripheral blood after the age of 17 to 18 months (Figure 4A–B, and data not shown). Flow cytometric analysis of blood cells confirmed an increased proportion of mature monocytes (F4/80+/Gr-1−) in 18-month-old Npm1wt/c+ mice without MPD (Figure 4C). These BM of these mice also exhibited expansion of the myeloid compartment, with an increased number of HSPC (c-kit+/sca-1+), as well as progenitors (CMP, c-kit+/F4/80+ and GMP, F4/80+/Gr-1+) (Figure 4D).
Figure 4. The hematologic changes of peripheral blood and BM in Npm1wt/c+ mice without signs of myeloproliferation.
Monthly blood collection from wild-type and Npm1wt/c+ mice without signs of myeloproliferation showed increase in the compartment of monocytes (A) but not granulocytes (B), lymphocytes, or platelet (data not shown). Each data point was the average of at least 10 mice. (C) There were significant more monocytes (F4/80+/Gr-1−) in non-myeloproliferative Npm1wt/c+ mice compared to wild-type mice at age of 18 months (n = 7 each). (D) At age of 12 months (n = 7 each), quantification of marrow cells of HSPC (c-kit+/sca-1+), MPP (F4/80−/c-kit+), CMP (F4/80+/c-kit+), GMP (F4/80+/Gr-1+), monocytes (F4/80+/Gr-1−), and granulocytes (F4/80−/Gr-1+) by flow cytometry revealed significant increase in marrow stem cell and progenitor cell compartments of HSPC, CMP, and GMP, but not MPP. Data are expressed as mean ± SE. * means P<0.05; ** P≤0.01.
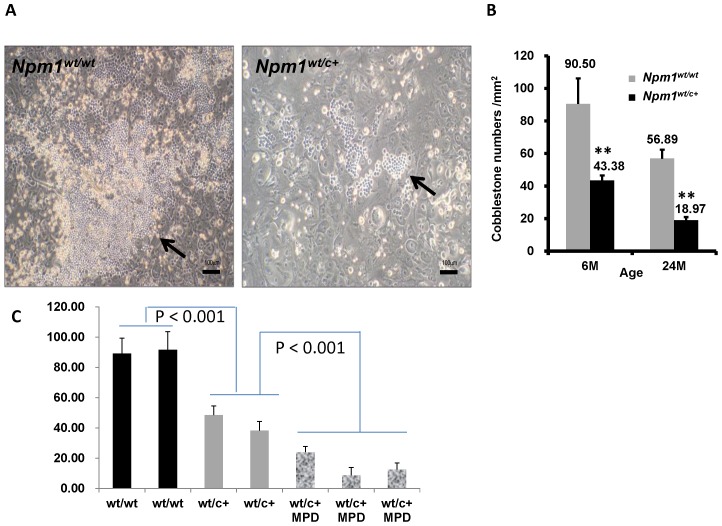
Compromised cobblestone area formation in BM cells of Npm1wt/c+ mice
The majority of mutant mice did not develop clinically evident MPD. However, the gradual development of monocytosis and increased numbers of myeloid precursors in these mice might imply the presence of a pathological hematopoietic niche in BM. The extramedullary hematopoiesis observed in the MPD mice suggests the same possibility. We thus examined the effect of Npm1wt/c+ mutant cells on the interaction of HSPCs and marrow stromal cells by ex vivo cobblestone area formation assay. We collected 5×104 hematopoietic cells from Npm1wt/c+ mice without MPD, and from wild-type mice of various ages; the collected cells were placed on the MS5 stromal cell layers for cobblestone-like colony formation. The mutant cells exhibited severely compromised cobblestone area formation compared with wild-type cells (Figure 5A). In 6-month-old Npm1wt/c+ mice without MPD, a 2-fold decrease in cobblestone area numbers was noted, and by 24 months the decrease was 3-fold compared with wild-type mice (Figure 5B). These findings imply abnormalities in the contact signal pathways of mutant HSPC in the context of the microenvironment. The same defect was seen and even more evident in mutant mice with MPD (Fig. 5C).
Figure 5. Age-dependent decline of cobblestone area formation in Npm1wt/c+ mice.
(A) The cobblestone area formation (arrows) is obviously inhibited in Npm1wt/c+ HSPC. (B) The cobblestone area numbers are also much lower in mutant mice. The difference seems more augmented as mice became older (n = 7 each). (C) The mutant mice with MPD exhibited even more severe compromised cobblestone area formation when compared with mutant but non-MPD mice (P<0.001, one bar represents triplicate experiments from one mouse).
Correlation between Npm1 mutation and expression of CXCL12/CXCR4 pathway genes
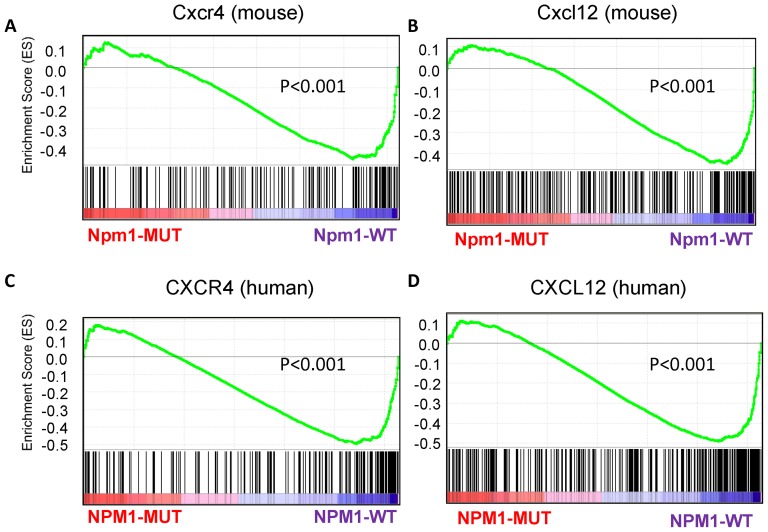
To explore the mechanisms underlying the defective cobble stone area formation, we compared the gene expression profiles of sorted BM myeloid progenitors (c-kit+/sca-1−/F4/80−) between wild-type and Npm1wt/c+ mice without MPD. We conducted GSEA to explore pathways that might be responsible for defective cobblestone area formation in Npm1wt/c+ HSPC. We compared the degree of enrichment across 22 gene sets between Npm1wt/c+ and wild-type expression profiles; these gene sets were related to HSPC functions, and were selected from the IPA (Table S1). Among the gene sets, only pathways related to Cxcl12 and Cxcr4 were significantly downregulated in Npm1wt/c+ mouse (both P-value<0.001, normalized) (Table S3 and Figure 6A–B for enrichment plots). These pathways both imply interaction between HSPCs and the marrow niche.
Figure 6. Enrichment plots from gene set enrichment analysis (GSEA).
The enrichment plots contain profiles of the running enrichment scores (ES) and positions of gene set members on the rank ordered list in GSEA. (A) Cxcr4 and (B) Cxcl12 interaction signatures on the mouse data. (C) CXCR4 and (D) CXCL12 interaction signatures on the human data.
Concordant suppresion of CXCL12 and CXCR4 pathway signatures in NPM1-mutated AML patients
To investigate whether differential enrichment of CXCL12/CXCR4 pathway-related genes occurs in human AML, we performed genome-wide expression microarray analysis on BM mononuclear cells. The sample cells were collected from a cohort of 197 AML patients who exhibited either 147 NPM1-wild-type or 50 NPM1-mutated, and 30 normal BM transplantation donors. The GSEA showed obvious suppression of CXCL12 and CXCR4 pathway genes in NPM1-mutated patients compared to NPM1-wild AML and normal controls (normalized P-values of both <0.001, Table S3; enrichment plots, Figure 6C–D). The results of the GSEA are shown in Table S4. In addition to measuring the degree of enrichment, GSEA identified a leading edge subset in each gene set, comprising core gene members that contributed the most to the enrichment scores. The leading-edge members represent genes that might participate in biological functions governed by that specific gene set in the observed data (Table S5).
Discussion
The earliest report of NPM1 mutation in AML was published 2005 [1], but to date the molecular mechanisms underlying the pathogenesis are not yet well understood. Apart from this study, 3 other animal models (including transgenic mouse and zebrafish models [19], [20] and a “humanized” knock-in mouse model [21]) have been generated and analyzed. All of these studies have used human mutation sequences, but ours was the first to use an “endogenous” mouse sequence, which we named “conventional” mouse model. The mutated peptide sequence in our mutant mice differed from that in humans because of a discrepancy in the nucleotide sequence of the gene in mice versus humans. Nonetheless, we showed that cellular localizations of mutated and wild-type Npm1 proteins were different. However, the phenotypes of our mouse model were not identical to those present in the previous 2 mouse studies. In the transgenic mouse model, mice developed non-fatal myeloproliferation in the BM and spleen, but did not show leukocytosis in peripheral blood [19]. In addition, 30% of the “humanized” knock-in mice showed expansion of mature myeloid cells and late-onset AML [21]. By contrast, in our “conventional” mouse model, approximately one third of the mutant mice showed myeloproliferation at various ages, but no AML was seen throughout their lifetimes. The phenotypic discrepancy between the mice in our study and those in the transgenic model can be explained by their use of exogenous promoter to drive human NPM1 mutant gene in the presence of full-dose wild-type mouse Npm1 protein. The differences of the phenotypes between our mice and the “humanized” knock-in model are likely related to different mutation sequences. Although both mutant proteins can be localized in cytoplasm, the strength of cytoplasmic retention strength may differ in the 2 proteins, which would result in different nucleus-to-cytoplasm ratios for Npm1 protein. Alternatively, the extra amino acids observed after the NES in our study might have led to unexpected consequences in cells. Nevertheless, all the models suggest a role of mutant nucleophosmin protein in aberrant myelopoiesis. Overall, all the models suggest that NPM1 mutation alone does not provide sufficient conditions for AML formation. Other pathological abnormalities are necessary to induce full-blown leukemia.
Although in human, MPD is usually associated with increased marrow cellularity, splenomegaly, and peripheral blood cytosis, there is occasional inconsistency among these organs/tissues in mice. For example, in a report, the MPD was shown in BM and spleen but not PB [19]. In another study where a myeloproliferative syndrome was induced in mice, elevated myeloid cells were seen in spleen and blood, but BM showed paradoxically reduced cellularity [25]. We did not see difference in the BM cellularity or precursor cell number among the wild type, non-MPD mutant, and MPD mutant mice. It is likely that the origin of MPD may arise from extramedullary organs, such as spleen, related to extramedullary hematopoiesis.
Is the defective cobblestone area formation, which reflects the repopulating ability of HSPC, in Npm1-mutated mice truly microenvironment-dependent or cell-intrinsic? To address this question, we have to clarify two points: first, did the cells used in the cobblestone assays contain the same number of progenitor cells (which are supposed to be the source of cobblestone colonies) between wild-type and mutant mice? To answer this point, we seeded equal number of Lin- progenitor cells on murine stromal cells and found that the colony numbers in wild-type cells were still significantly greater than mutant cells (data not shown). Secondly, do the progenitor cells from wild-type or mutant mice grow at the same speed without stromal cells? We noted that the CFU-GEMM and CFU-GM numbers were not different between wild-type mice and non-MPD mutant mice at 12 months of age (data not shown). Therefore, it seems that the proliferation of the progenitor cells from these 2 groups of mice differ only when stromal cells are added for co-culture. Therefore, these results suggest that the decreased cobblestone colony number in mutant cells is likely related to microenvironment rather than intrinsic cell factors. However, further investigations are warranted to get more definitive answers.
To investigate the possible mechanisms underlying this phenomenon, we employed GSEA tool, which is a powerful computational tool for interpreting whole-genome gene expression data at the level of gene sets [26], [27]. The main advantage of GSEA over the traditional strategies of examining differentially expressed genes is the capacity to detect overall trends and differences throughout the entire network. In addition, threshold-free analysis enables the identification of gene-set level enrichment when the underlying changes in individual genes are modest and subtle. Studied gene sets can include built-in gene sets of the Molecular Signature Database [27], including functionally defined groups (e.g., Gene Ontology terms), canonical pathways (e.g., BioCarta and KEGG), and signatures of chemical perturbations. Alternatively, user-defined sets of genes that share common features of interest may be studied. The prowess and flexibility of GSEA have resulted in this software being widely used in genomic data analyses [28]–[32].
The results of our study showed that expression of CXCR4/CXCL12-related genes was significantly downregulated in Npm1wt/c+ mouse BM precursor cells and in human AML samples displaying the NPM1 mutation. These genes are critical in maintaining the, HSPC pool in the BM niche [33]. Downregulated expression thus suggests an aberration in the hematopoietic niche, which might be related to abnormal interaction between these cells and stromal cells. This type of microenvironment perturbation has been implicated in human diseases such as primary myelofibrosis [34], [35]. The extramedullary hematopoiesis and myeloid expansion observed in our mutant mice mimicked the clinical features of idiopathic myelofibrosis and other myeloproliferative neoplasms. These diseases might be related to a pathological hematopoietic niche and enhanced mobilization of the HSPC to alternative niches, such as liver and spleen [34], [35]. This niche pathology worsened as our mutant mice aged. In Npm1wt/c+ mice without MPD, the gradual development of monocytosis and increased numbers of myeloid precursors with aging, combined with defective cobblestone area formation, may be early signs of MPD. As-yet-unidentified genetic hits might be required to accelerate the development of clinically evident MPD for these mice.
The aging-related decline of cobblestone area formation coincides with the fact that NPM1-mutation AML is more prevalent among older than younger patients [1], [18]. We surmise that additional hits of genetic abnormalities (which are more likely to occur in older people) interact with the age-dependent perturbation of the hematopoietic niche to result in clinical NPM1-mutated AML among older patients. High CXCR4 expression in AML cells is an independent unfavorable prognostic factor [36]. A recent study showed that a CXCR4 antagonist, plerixafor, increased the sensitivity of leukemia cells to chemotherapy [37]. Previous reports have documented a similar increase in certain patients' responsiveness to chemotherapy [1], [13]. The possibility that defective CXCR4/CXCL12 interaction in AML patients bearing an NPM1 mutation might contribute to this increased responsiveness deserves further investigation.
In conclusion, this study presented a “conventional” knock-in model of Npm1 mutation. The main phenotypes were myeloproliferative neoplasms with extramedullary hematopoiesis, which occurred in approximately 30% of the mutant mice. Other non-myeloproliferative Npm1wt/c+ mice showed delayed onset of monocytosis in the peripheral blood and increased numbers of BM myeloid precursors as the animal aged. The results of ex vivo cobblestone assay implied a perturbation of the hematopoietic niche in mutant mice from as early as 6 months of age. This finding might be related to defective CXCR4/CXCL12 pathways. This hypothesis was supported by genome-expression data from a large cohort of human samples. The findings of our study provide further evidence that NPM1 mutation alone is insufficient to cause AML formation. The relatively suppressed expression of CXCR4/CXCL12 pathway genes, observed in our mutant mice and NPM1-mutated AML patients, might be critical in the pathogenesis of NPM1 mutation.
Supporting Information
The alignment of nucleotide and protein sequences of wild-type and mutant NPM1 of human and mice. CDS, coding sequence.
(TIF)
The cytoplasmic localization of mutated nucleophosmin protein from human and mice. Upper panel: GFP fused at the N terminal end of human wild-type NPM1 (left) localizes mainly in the nucleoli as punctate pattern. In contrast, GFP fused with human mutant NPM1 protein mainly resides in cytoplasm, although some appears in the nucleoli. Lower panel: GFP fused at the N terminal end of wild-type mouse Npm1 protein (left) and expected mutant Npm1 protein (right) exhibit similar patterns as the human condition.
(TIF)
Updated defined gene sets for GSEA.
(XLS)
Parameters and settings of GSEA in this Study.
(DOCX)
Summary of mouse and human gene expression data in gene set enrichment analysis.
(DOCX)
Complete summary of GSEA.
(XLS)
Leading-edge genes from GSEA.
(XLSX)
Acknowledgments
We thank the technical services provided by the “Transgenic Mouse Model Core Facility of the National Core Facility Program for Biotechnology, National Science Council” and the “Gene Knockout Mouse Core Laboratory of National Taiwan University Center of Genomic Medicine”.
Funding Statement
This work was supported by grants NSC 98-2314-B-002-033-MY3, NSC 98-2314-B-030-003-MY3, NSC 100-2325-B-002-032, NSC 100-2325-B-002-033, and NSC 101-2325-B-002-029 from National Science Council (Taiwan), NHRI-EX97-9731BI from National Health Research Institute, and DOH101-TD-C-111-001 from Department of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, et al. (2005) Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med 352: 254–266. [DOI] [PubMed] [Google Scholar]
- 2. Falini B, Nicoletti I, Martelli MF, Mecucci C (2007) Acute myeloid leukemia carrying cytoplasmic/mutated nucleophosmin (NPMc+ AML): biologic and clinical features. Blood 109: 874–885. [DOI] [PubMed] [Google Scholar]
- 3. Nishimura Y, Ohkubo T, Furuichi Y, Umekawa H (2002) Tryptophans 286 and 288 in the C-terminal region of protein B23.1 are important for its nucleolar localization. Biosci Biotechnol Biochem 66: 2239–2242. [DOI] [PubMed] [Google Scholar]
- 4. Falini B, Bolli N, Shan J, Martelli MP, Liso A, et al. (2006) Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood 107: 4514–4523. [DOI] [PubMed] [Google Scholar]
- 5. Bolli N, Nicoletti I, De Marco MF, Bigerna B, Pucciarini A, et al. (2007) Born to be exported: COOH-terminal nuclear export signals of different strength ensure cytoplasmic accumulation of nucleophosmin leukemic mutants. Cancer Res 67: 6230–6237. [DOI] [PubMed] [Google Scholar]
- 6. Savkur RS, Olson MO (1998) Preferential cleavage in pre-ribosomal RNA byprotein B23 endoribonuclease. Nucleic Acids Res 26: 4508–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hingorani K, Szebeni A, Olson MO (2000) Mapping the functional domains of nucleolar protein B23. J Biol Chem 275: 24451–24457. [DOI] [PubMed] [Google Scholar]
- 8. Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, et al. (2000) Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103: 127–140. [DOI] [PubMed] [Google Scholar]
- 9. Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG (2002) Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol 4: 529–533. [DOI] [PubMed] [Google Scholar]
- 10. Kurki S, Peltonen K, Latonen L, Kiviharju TM, Ojala PM, et al. (2004) Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell 5: 465–475. [DOI] [PubMed] [Google Scholar]
- 11. Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, et al. (2005) Role of nucleophosmin in embryonic development and tumorigenesis. Nature 437: 147–153. [DOI] [PubMed] [Google Scholar]
- 12. Sportoletti P, Grisendi S, Majid SM, Cheng K, Clohessy JG, et al. (2008) Npm1 is a haploinsufficient suppressor of myeloid and lymphoid malignancies in the mouse. Blood 111: 3859–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Verhaak RG, Goudswaard CS, van Putten W, Bijl MA, Sanders MA, et al. (2005) Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood 106: 3747–3754. [DOI] [PubMed] [Google Scholar]
- 14. Schnittger S, Schoch C, Kern W, Mecucci C, Tschulik C, et al. (2005) Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood 106: 3733–3739. [DOI] [PubMed] [Google Scholar]
- 15. Dohner K, Schlenk RF, Habdank M, Scholl C, Rucker FG, et al. (2005) Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood 106: 3740–3746. [DOI] [PubMed] [Google Scholar]
- 16. Boissel N, Renneville A, Biggio V, Philippe N, Thomas X, et al. (2005) Prevalence, clinical profile, and prognosis of NPM mutations in AML with normal karyotype. Blood 106: 3618–3620. [DOI] [PubMed] [Google Scholar]
- 17. Suzuki T, Kiyoi H, Ozeki K, Tomita A, Yamaji S, et al. (2005) Clinical characteristics and prognostic implications of NPM1 mutations in acute myeloid leukemia. Blood 106: 2854–2861. [DOI] [PubMed] [Google Scholar]
- 18. Chou WC, Tang JL, Lin LI, Yao M, Tsay W, et al. (2006) Nucleophosmin mutations in de novo acute myeloid leukemia: the age-dependent incidences and the stability during disease evolution. Cancer Res 66: 3310–3316. [DOI] [PubMed] [Google Scholar]
- 19. Cheng K, Sportoletti P, Ito K, Clohessy JG, Teruya-Feldstein J, et al. (2010) The cytoplasmic NPM mutant induces myeloproliferation in a transgenic mouse model. Blood 115: 3341–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bolli N, Payne EM, Grabher C, Lee JS, Johnston AB, et al. (2010) Expression of the cytoplasmic NPM1 mutant (NPMc+) causes the expansion of hematopoietic cells in zebrafish. Blood 115: 3329–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vassiliou GS, Cooper JL, Rad R, Li J, Rice S, et al. (2011) Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat Genet 43: 470–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chou SH, Chawla A, Lee TH, Zhou Y, Busch MP, et al. (2001) Increased engraftment and GVHD after in utero transplantation of MHC-mismatched bone marrow cells and CD80low, CD86(-) dendritic cells in a fetal mouse model. Transplantation 72: 1768–1776. [DOI] [PubMed] [Google Scholar]
- 23. Nakahata T, Ogawa M (1982) Identification in culture of a class of hemopoietic colony-forming units with extensive capability to self-renew and generate multipotential hemopoietic colonies. Proc Natl Acad Sci U S A 79: 3843–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, et al. (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel 17: 527–536. [DOI] [PubMed] [Google Scholar]
- 25. Johnson GR, Gonda TJ, Metcalf D, Hariharan IK, Cory S (1989) A lethal myeloproliferative syndrome in mice transplanted with bone marrow cells infected with a retrovirus expressing granulocyte-macrophage colony stimulating factor. EMBO J 8: 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, et al. (2003) PGC-1[alpha]-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273. [DOI] [PubMed] [Google Scholar]
- 27. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. (2005) Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bourquin J-P, Subramanian A, Langebrake C, Reinhardt D, Bernard O, et al. (2006) Identification of distinct molecular phenotypes in acute megakaryoblastic leukemia by gene expression profiling. Proceedings of the National Academy of Sciences of the United States of America 103: 3339–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Leval L, Rickman DS, Thielen C, Reynies Ad, Huang Y-L, et al. (2007) The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood 109: 4952–4963. [DOI] [PubMed] [Google Scholar]
- 30. Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, et al. (2008) Gene Expression in Fixed Tissues and Outcome in Hepatocellular Carcinoma. New England Journal of Medicine 359: 1995–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oron AP, Jiang Z, Gentleman R (2008) Gene set enrichment analysis using linear models and diagnostics. Bioinformatics 24: 2586–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alles MC, Gardiner-Garden M, Nott DJ, Wang Y, Foekens JA, et al. (2009) Meta-Analysis and Gene Set Enrichment Relative to ER Status Reveal Elevated Activity of MYC and E2F in the “Basal” Breast Cancer Subgroup. PLoS One 4: e4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sugiyama T, Kohara H, Noda M, Nagasawa T (2006) Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 25: 977–988. [DOI] [PubMed] [Google Scholar]
- 34. Tefferi A (2005) Pathogenesis of myelofibrosis with myeloid metaplasia. J Clin Oncol 23: 8520–8530. [DOI] [PubMed] [Google Scholar]
- 35. Lataillade JJ, Pierre-Louis O, Hasselbalch HC, Uzan G, Jasmin C, et al. (2008) Does primary myelofibrosis involve a defective stem cell niche? From concept to evidence. Blood 112: 3026–3035. [DOI] [PubMed] [Google Scholar]
- 36. Spoo AC, Lubbert M, Wierda WG, Burger JA (2007) CXCR4 is a prognostic marker in acute myelogenous leukemia. Blood 109: 786–791. [DOI] [PubMed] [Google Scholar]
- 37. Uy GL, Rettig MP, Motabi IH, McFarland K, Trinkaus KM, et al. (2012) A phase 1/2 study of chemosensitization with the CXCR4 antagonist plerixafor in relapsed or refractory acute myeloid leukemia. Blood 119: 3917–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The alignment of nucleotide and protein sequences of wild-type and mutant NPM1 of human and mice. CDS, coding sequence.
(TIF)
The cytoplasmic localization of mutated nucleophosmin protein from human and mice. Upper panel: GFP fused at the N terminal end of human wild-type NPM1 (left) localizes mainly in the nucleoli as punctate pattern. In contrast, GFP fused with human mutant NPM1 protein mainly resides in cytoplasm, although some appears in the nucleoli. Lower panel: GFP fused at the N terminal end of wild-type mouse Npm1 protein (left) and expected mutant Npm1 protein (right) exhibit similar patterns as the human condition.
(TIF)
Updated defined gene sets for GSEA.
(XLS)
Parameters and settings of GSEA in this Study.
(DOCX)
Summary of mouse and human gene expression data in gene set enrichment analysis.
(DOCX)
Complete summary of GSEA.
(XLS)
Leading-edge genes from GSEA.
(XLSX)