Abstract
The mammalian host has developed a long-standing symbiotic relationship with a considerable number of microbial species. These include the microbiota on environmental surfaces, such as the respiratory and gastrointestinal tracks1, and also endogenous retroviruses (ERVs), comprising a substantial fraction of the mammalian genome2,3. The long-term consequences for the host of interaction with these microbial species can range from mutualism to parasitism and are not always completely understood. The potential impact of one microbial symbiont on another is even less clear. We have studied the control of ERVs in the commonly-used C57BL/6 (B6) mouse strain, which lacks endogenous murine leukaemia viruses (MLVs) able to replicate in murine cells. We demonstrate the spontaneous emergence of fully infectious ecotropic4 MLV (eMLV) in B6 mice with a range of distinct immune deficiencies affecting antibody production. These recombinant retroviruses establish infection of immunodeficient mouse colonies, and ultimately result in retrovirus-induced lymphomas. Notably, ERV activation in immune-deficient mice is prevented in husbandry conditions associated with reduced or absent intestinal microbiota. Our results shed light onto a previously unappreciated role for immunity in the control of ERVs and provide a potential mechanistic link between immune activation by microbial triggers and a range of pathologies associated with ERVs, including cancer.
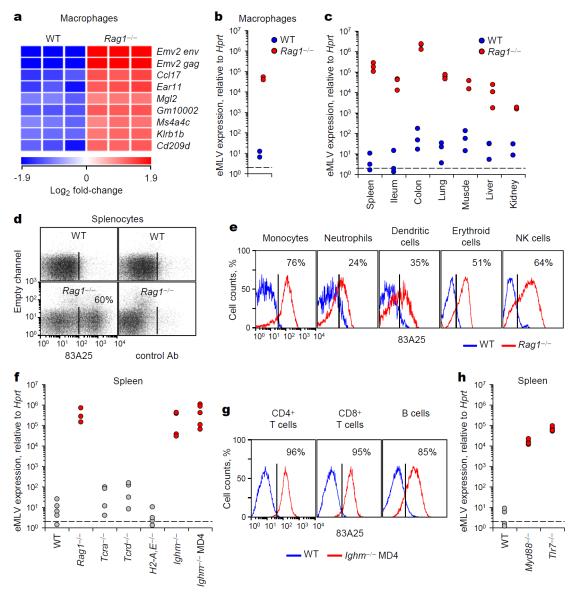
Retroviruses can establish germline infection and become part of the host genome2,3. Most, if not all ERVs have become inactive due to mutations or transcriptionally silenced through the action of diverse mechanisms2,3. However, RNA and protein expression of replication-defective ERVs is frequently elevated in infection, autoimmunity and cancer2,3. Whether or not the immune system defends against potential threats posed by ERVs is currently unclear. To address the role of adaptive immunity in this process, we assessed ERV expression in B6 mice. We initially compared the transcriptional profiles of purified macrophages from B6 wild-type (WT) and T and B lymphocyte-deficient Rag1−/− mice. The transcripts with the highest increase in expression levels in macrophages from Rag1−/− mice (Fig. 1a) correspond to the env and gag genes, respectively (Supplementary Table 1), of an endogenous eMLV locus, Emv2, a replication-defective single-copy ERV present in B6 mice5. Differential expression of eMLV was confirmed by qRT-PCR for spliced env mRNA in macrophages (Fig. 1b), and in multiple tissues (Fig. 1c).
Figure 1. eMLV activation in antibody-deficient mice.
a, Significantly upregulated (>4-fold) genes in CD11b+MHC-IIhiB220−Gr1− macrophages from Rag1−/− mice compared with macrophages from WT mice. Triplicate microarrays from cells isolated from 40 mice are shown. b, eMLV spliced env mRNA expression in the same cells as in a. Each symbol represents macrophages from 20 mice (p=0.024; paired Student’s t-test). c, eMLV spliced env mRNA expression in indicated organs from WT or Rag1−/− mice (spleen: p=0.020; small intestine: p=0.032; large intestine: p=0.004; lung: p= 0.001; muscle: p=0.016; and kidney: p=0.009; unpaired Student’s t-test). d, e, MLV SU expression in splenocytes (d) or in indicated cell types (e) from WT or Rag1−/− mice. f, eMLV spliced env mRNA expression in the spleens of the indicated strains (p<0.001 between WT and either Ighm−/− or Ighm−/− MD4 mice; one-way ANOVA). g, MLV SU expression in splenic lymphocytes from WT or Ighm−/− MD4 mice. h, eMLV spliced env mRNA expression in the spleens of the indicated strains (p<0.001 between WT and either Myd88−/− or Tlr7−/− mice; one-way ANOVA). In c, f and h, each symbol is an individual mouse. In d, e and g, plots are representative of 4 mice per group. In f and h, values above 103 were considered high and are indicated with red-filled symbols.
Other ERV families were also differentially expressed in macrophages, albeit less strongly (1.7 – 2.1-fold) (Supplementary Table 1). Not distinguishing between members of multi-copy families, expression of polytropic MLVs (pMLVs), xenotropic MLVs (xMLVs) and of the MusD family of retrotransposons was also elevated in the lungs of Rag1−/− mice (Supplementary Fig. 1). In line with increased MLV mRNA expression, 60-80% of total splenocytes and all hematopoietic lineages analysed from Rag1−/− but not from WT mice, expressed MLV surface glycoprotein (SU) (Fig. 1d, e).
Tcra−/− or Tcrd−/− mice, lacking TCRαβ and TCRγδ T cells, respectively, showed low eMLV expression (Fig. 1f). eMLV expression was similarly low in H2-A,E−/− mice, deficient in MHC II, MHC II-restricted T cells and T cell-dependent antibodies (Fig. 1f). In contrast, mice lacking B cells (Ighm−/− mice) or only the ability to produce polyclonal antibodies (Ighm−/− MD4 mice) expressed substantially higher eMLV levels than WT mice (Fig. 1f), demonstrating that high eMLV expression characterised mice lacking antigen-specific antibodies. In addition to splenocytes from Rag1−/− mice (Fig. 1e), T and B cells from Ighm−/− MD4, but not from WT control mice, expressed MLV SU (Fig. 1g), indicating that eMLV can be highly expressed also in lymphocytes.
Toll-like receptors (TLRs) have been implicated in the control of B cell responses, and both Myd88−/− and Tlr7−/− mice have significantly reduced serum levels of natural antibodies and a defective antibody response to immunisation or infection, including with retroviruses6-9. Notably, expression of eMLV was dramatically elevated in Myd88−/− and Tlr7−/− mice in comparison with WT mice (Fig. 1h). Similarly elevated eMLV expression was observed in Tlr7−/−, but not Tlr9−/− mice housed in a different facility (Supplementary Fig. 2).
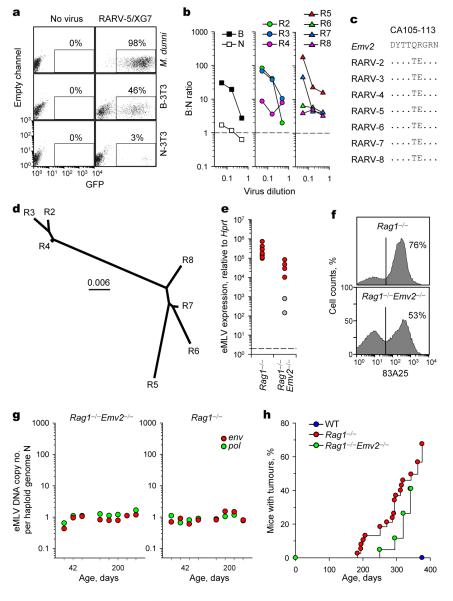
To investigate the mechanistic link between antibody deficiencies and elevated MLV expression, we examined the origin of eMLV transcription. The B6 genome does not contain replication-competent eMLV proviruses and, although the Emv2 locus can produce mRNA, it is unable to produce infectious virus due to an inactivating G to C mutation at position 3576 of the pol region5,10. In addition, Emv2 gag encodes an N-tropic capsid, which would be restricted by the Fv1b restriction factor in B6 mice10. However, it was theoretically possible that recombination between replication-defective Emv2 and non-ecotropic MLVs resulted in an MLV with full infectivity11 that could spread in Rag1−/− mice. Remarkably, plasmas of young and old Rag1−/− mice, but not of WT control mice, contained retroviruses that were capable of replicating in murine cells in vitro (Fig. 2a), which we refer to as Rag1−/− mouse-associated retroviruses (RARVs). Sequencing of the pol region demonstrated repair of the Emv2-inactivating mutation in all RARV isolates (Supplementary Fig. 3). Functional in vitro assays (Fig. 2b) and sequencing of the gag region (Fig. 2c) showed that RARVs also exhibited B-tropism. Genome sequence comparisons between RARVs revealed that young Rag1−/− mice harboured highly similar viruses, which diverged substantially in old Rag1−/− mice (Fig. 2d). All RARVs were recombinants between Emv2 and endogenous non-ecotropic MLVs (Supplementary Fig. 4). The pol defect of Emv2 was likely restored in RARVs by recombination with Xmv43 (Supplementary Fig. 4), an ERV that contains a functional pol region but is unable to infect mouse cells due to polymorphisms in the murine cellular receptor2. Recombination events involving Xmv43 have also been found responsible for the emergence of leukaemogenic MLVs in AKR mice12. However, the switch in capsid tropism resulted from recombination with other endogenous xMLVs (Supplementary Fig. 4). Notably, the divergence of RARVs isolated from old Rag1−/− mice was due to further recombination replacing the ecotropic env with polytropic env either from Pmv1/5 or Pmv16 (Supplementary Fig. 4). Together, these findings indicated the emergence of infectious eMLVs that could have infected Rag1−/− mice and gave further rise to infectious pMLVs. Supporting this notion, the majority of gag/pol eMLV mRNA detected in Rag1−/− mice seemed to be transcribed from integrated RARVs, rather than the germline copy of Emv2 (Supplementary Fig. 5).
Figure 2. Retroviremia and leukaemias/lymphomas in antibody-deficient mice.
a, Detection of infectious MLV (RARV-5/XG7) from the plasma of a representative Rag1−/− mouse by restoring infectivity of the green fluorescent protein (GFP)-expressing XG7 retroviral vector in the indicated cell type. Numbers within the plots denote the percentage of retrovirally-transduced (GFP+) cells. b, Fv1 tropism of RARVs isolated from 6- (R2-R4) or 25- (R5-R8) week-old healthy Rag1−/− mice, shown as the ratio of infectivity in B-3T3 to N-3T3 cells (B:N ratio). B- and N-tropic strains of Friend MLV (F-MLV) are shown for comparison. c, Amino acid residues of capsid positions 105-113 deduced from the nucleotide sequence of Emv2 and the same RARVs as in b. Dots indicate identities. d, Phylogenetic tree of the same RARVs as in b. The scale indicates the probability of base substitution per site. e, eMLV spliced env mRNA expression in the spleens of Rag1−/− mice or vertically-infected Rag1−/−Emv2−/− mice. Each dot is an individual mouse (p<0.001; unpaired Student’s t-test). Values above 103 were considered high and are indicated with red-filled symbols. f, MLV SU expression in splenocytes from Rag1−/− or vertically-infected Rag1−/−Emv2−/− mice (representative of 9 mice per group). g, eMLV DNA copy numbers per haploid genome, determined by qPCR for the pol or ecotropic env gene, in DNA from the spleens of healthy Rag1−/− (right) or vertically-infected Rag1−/− Emv2−/− mice (left). Symbols represent individual mice, grouped according to their age. The sensitivity limit of this PCR method was determined as a median of 0.0003 copies per haploid genome, using Emv2−/− mice. eMLV DNA copy numbers for Rag1−/− mice include Emv2 (1/N). h, Tumour (leukaemias/lymphomas) incidence in cohorts of WT (n=37), Rag1−/− (n=38) or vertically-infected Rag1−/−Emv2−/− mice (n=23) at the NIMR SPF facility (p<0.000001 between WT and Rag1−/− mice; p=0.00025 between WT and Rag1−/−Emv2−/− mice; Log-Rank Survival analysis).
Emv2 and non-ecotropic MLV recombination events resulting in infectious eMLV generation might occur de novo in individual Rag1−/− mice. Alternatively, these RARVs might have been vertically transmitted through successive generations. To directly test the latter possibility and assess the capacity of RARVs to spread in immunodeficient mice, we established a colony of Rag1−/−Emv2−/− mice by firstly crossing an Emv2−/− male mouse13 with a Rag1−/− female mouse, and then intercrossing selected progeny to homozygosity. These mice lack the germline copy of Emv2, meaning any infectious eMLV present would have been vertically transmitted. Both eMLV spliced env mRNA (Fig. 2e) and MLV SU expression (Fig. 2f) was readily detected in the spleens of Rag1−/−Emv2−/− mice in this colony. Furthermore, analysis of eMLV env and pol DNA copies indicated extensive replication of vertically transmitted RARVs in Rag1−/−Emv2−/− mice (Fig. 2g). In contrast, sexual or in utero infection was not observed in separate crosses of either male or female virus-positive Rag1−/−Emv2−/− mice with virus-free Emv2−/− mice (Supplementary Fig. 6).
To further examine the potential of RARVs to replicate in Rag1−/− mice, we assessed the frequency of tumours characteristic of retroviral infection2,3 in cohorts of Rag1−/− mice. Notably, starting from 180 days and affecting 67% of the animals by 380 days, Rag1−/− mice, but not WT control mice, showed signs of morbidity (Fig. 2h). Upon examination, large tumours, often associated with anaemia, were observed in all morbid Rag1−/− mice (Fig. 2h and Supplementary Fig. 7). The pathogenic potential of infection with RARVs was established in aged cohorts of vertically-infected Rag1−/− Emv2−/− mice, which developed tumours at a comparable incidence rate (Fig. 2h).
Thymic or splenic tumours in Rag1−/− mice consisted mainly of a single MLV SU-expressing cell type, which differed between animals, and had the histological appearance of lymphoblastic lymphosarcomas (Supplementary Fig. 8). Discrete chromosomal aberrations were found in the majority of tumours analysed (Supplementary Fig. 8), suggestive of clonal origin. Consistent with MLV production by tumour cells, we observed an abundance of MLV-type particles in the extracellular space of tumour samples, but not in a spleen sample from a healthy Rag1−/− mouse (Supplementary Fig. 9). Furthermore, a significant increase in both eMLV env and pol DNA copy numbers was detected in all tumour samples, with one exception where only pol DNA copies were increased (Supplementary Fig. 9), indicating that RARVs had extensively infected the cells that gave rise to lymphomas. Together these results support a model where multiple recombination events restore Emv2 infectivity, leading to spontaneous retroviremia and vertical transmission to progeny, and eventually drive an oncogenic process similar to that extensively described in mouse strains carrying fully infectious ERVs2,3.
Our results associated lack of antibodies with establishment of infectious eMLVs in mouse colonies. Next, we investigated the potential mode of antibody action. Antiretroviral antibodies have a long-established role in limiting the spread of infectious endogenous retroviruses14, both within and between animals. However, it was also possible that antibodies were preventing a step prior to the emergence of infectious eMLV recombinants. Rescue of Emv2 infectivity by recombination with a non-ecotropic MLV necessitates co-expression of both proviruses in the same cell at sufficient levels for co-packaging into the same virion. Low expression of these proviruses in WT mice could be a rate-limiting factor in the emergence of infectious eMLVs. However, expression of certain endogenous MLVs in mouse cells is known to be inducible, notably, by microbial products2,3,15. For example, bacterial lipopolysaccharide (LPS) stimulation activates non-ecotropic MLVs, and Xmv43 in particular15-19. To examine the responsiveness of ERVs and other retroelements (REs) to microbial stimulation, we took advantage of probes or probesets in standard microarray platforms that report ERV or RE expression. Analysis of a publicly-available dataset20 uncovered specific induction of non-ecotropic MLV transcripts by LPS and poly(I:C), and suppression by Pam3CSK4 (Fig. 3a, b). Early Transposon (ETn) transcripts were also induced by poly(I:C) (Fig. 3a). These findings were confirmed by LPS stimulation, which induced high MLV SU expression in WT and Emv2−/− mouse splenocytes, also in a MyD88-independent manner (Fig. 3c and Supplementary Fig. 10). These in vitro conditions did not significantly induce eMLV expression (Fig. 3a). However, ex vivo analysis revealed higher expression of eMLV (Fig. 1c), as well as of other ERVs/REs (Supplementary Fig. 1), specifically in the colon, suggestive of microbial involvement.
Figure 3. Murine ERV activation by microbial products.
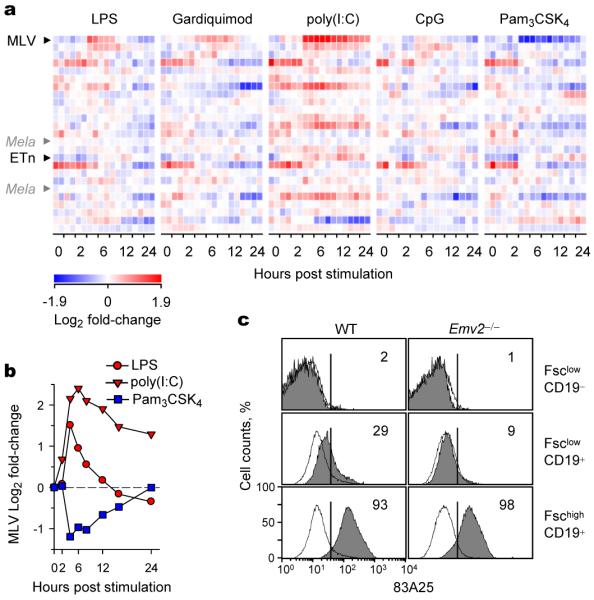
a, ERV/RE-reporting probeset (Supplementary Table 3) signals in a publicly-available Affymetrix HT Mouse Genome 430A microarray dataset20 (E-GEOD-17721) of WT B6 bone marrow-derived dendritic cells following stimulation with microbial products. Black arrows indicate the probesets that are significantly regulated (p<0.05) more than 2-fold by at least one stimulus. Mela (Emv2)-specific probesets are also indicated by grey arrows for comparison. b, Mean log2 fold-change in MLV-reporting probeset in the same dataset. c, MLV SU expression in WT or Emv2−/− splenocytes before (open histograms) and after stimulation with 10 μg/ml LPS for 48 hrs (grey-shaded histograms), according to Forward scatter (Fsc) and CD19 expression. Numbers with the plots denote the percentage of cells within each gate and represent 2 donors each analysed in duplicate.
Antibodies have established roles in controlling intestinal bacteria and neutralising their products, such as LPS, in the gut lumen or the systemic circulation, and antibody-deficient mice are known to display increased microbial trasnslocation1,9,21-23. It was, therefore, possible that antibody deficiency allowed microbial products to induce expression of MLV proviruses in Rag1−/− mice, including the parents of recombinant RARVs (Supplementary Fig 1). In support of this notion, and in agreement with the established role of natural IgM in systemic clearance of bacterial LPS and protection from endotoxemia9,21,22, production of non-hypermutated IgM alone was sufficient for eMLV control (Supplementary Fig. 11). If antibodies required for preventing MLV expression were, indeed, against multiple microbial products, then antibody deficiency should not result in elevated MLV expression in the absence of microbial triggers (Supplementary Fig 12).
To begin to examine the contribution of microbial triggers, we measured eMLV expression in specific pathogen-free (SPF) Rag1−/− mice from colonies that differed in intestinal microbiota. Importantly, the use of embryo transfer for the rederivation of these independent colonies removes adventitious organisms, including vertically-transmitted eMLVs. Therefore, any RARVs found in these rederived Rag1−/− mouse colonies must be generated de novo in the life-history of each colony. In stark contrast to Rag1−/− mice that were maintained on neutral pH water at NIMR, colonies of Rag1−/− mice that were maintained on acidified water expressed minimal eMLV levels (Fig. 4a). Water acidification reduced overall bacterial diversity in the colons of Rag1−/− mice (Supplementary Fig. 13 and Supplementary Table 2) and is a common precautionary measure used in many animal facilities that reduces bacterial colonisation within the intestinal tract and translocation into the circulation24. Lack of eMLV expression was noted in Rag1−/− mice obtained from The Jackson Laboratory (JAX), also maintained on acidified water (Fig. 4a). Furthermore, minimal levels of eMLV were detected in Rag1−/− mice on neutral pH water at the Rodent Center CHCI, ETH Zurich (RCHCI) (Fig. 4a), which harboured distinct bacterial genera in comparison with Rag1−/− mice at NIMR (Supplementary Fig. 13 and Supplementary Table 2). Lastly, negligible levels of eMLV were expressed in Rag1−/− mice in germ-free facilities at the University of Michigan (UMICH) and offered neutral pH water (Fig. 4a). The latter two conditions also distinguished between effects of acidified water on intestinal flora and effects on other physiological processes. Thus, high eMLV expression in independently-rederived colonies of Rag1−/− mice correlated with the presence of the normal SPF microbiota.
Figure 4. eMLV activation in antibody-deficiency depends on husbandry conditions.
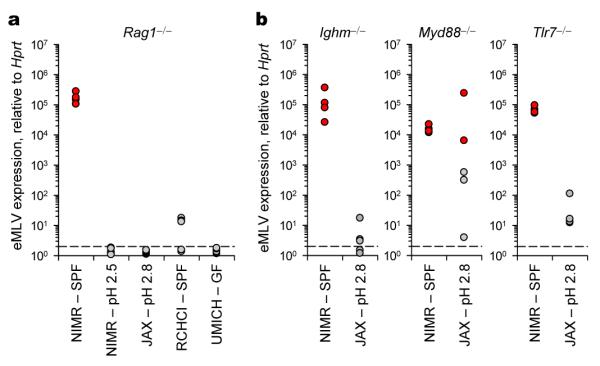
a, eMLV spliced env mRNA expression in the spleens of Rag1−/− mice on neutral pH (SPF) or acidified water (pH 2.5) at NIMR, on acidified water (pH 2.8) at JAX, on neutral pH at RCHCI or in germ-free (GF) facilities at UMICH (p<0.016 between Rag1−/− mice at NIMR SPF and all other groups; one-way ANOVA). b, eMLV spliced env mRNA expression in the spleens of Ighm−/−, Myd88−/− or Tlr7−/− mice on neutral pH water (SPF) at NIMR or on acidified water (pH 2.8) at JAX (p=0.005 and p=0.029 for Ighm−/− and Tlr7−/− mice, respectively; unpaired Student’s t-test). Each dot is an individual mouse and values above 103 were considered high and are indicated with red-filled symbols.
Husbandry conditions contributed to high eMLV expression also in independently-rederived strains with distinct immunodeficiencies. Colonies of Ighm−/− and Tlr7−/− mice maintained on acidified water at JAX expressed minimal levels of eMLV (Fig. 4b). Variable eMLV expression levels were detected in Myd88−/− mice at JAX (Fig. 4b), likely because, once generated, vertical transmission of infectious eMLVs was unaffected by water acidification. Although defining the precise role of the microbiota on eMLV induction will require further investigation, collectively our results demonstrate that the high eMLV expression phenotype, in genotypes causing immunodeficiency, requires environmental interaction.
High eMLV expression in independently-rederived immunodeficient colonies reveals de novo eMLV induction in each colony. It does not, however, indicate when or how often in a colony’s life-history infectious eMLV may emerge. Adult RARV-free Rag1−/− mice, previously on acidified water, maintained low eMLV expression following switch to neutral pH water, indicating low probability of RARV emergence and spread in an individual mouse. To examine if this probability is low in general or could be higher during early mouse development, we monitored successive generations of Ighm−/− mouse colonies from two independent rederivations, one of which was recent, into the NIMR SPF facility. This analysis suggested that eMLV induction occurred during the first few filial generations (F) (Supplementary Fig. 14). Moreover, eMLV-expressing Myd88−/− mice at JAX (Fig. 4b), were at F5 of homozygous breeding. Thus, although low in individual mice, there is high cumulative probability of infectious eMLV emergence, involving a sequence of recombination events similar to those seen in Rag1−/− mice (Supplementary Fig. 12), and subsequent establishment in an antibody-deficient colony over a few generations.
The B6 strain historically dominated many research fields partly due to their resistance to retrovirally-induced tumours. Our results demonstrate that this important attribute of B6 mice is conditional on their immune competence, with significant implications both for the design and interpretation of mouse studies. Furthermore, our results also show that ERV activation is determined by husbandry conditions, thus accentuating potential differences in ERV expression between animal facilities.
Although well-established in the mouse2,3, an oncogenic potential of ERVs in humans has not been observed25,26. However, non-LTR RE families have been documented to have caused human cancers by insertional mutagenesis at the somatic level25,26. Interestingly, we found that TLR stimulation of human cells induced expression of distinct ERVs and REs, including the mammalian apparent long terminal repeat (LTR) retrotransposon (MaLR) family (Supplementary Fig. 15), previously implicated in the pathogenesis of human lymphomas27. Transcription of human ERVs and REs can also be induced by physiological activation of both adaptive and innate immune cells28. Moreover, increased risk of lymphomas in humans is linked to infection or inflammation29 and also to antibody deficiencies30. Thus, interplay between microbial symbionts, leading to ERV/RE activation, may provide a mechanistic link between cancer and stimulation of the immune system by microbiota or pathogenic infections.
Methods
Mice
Inbred C57BL/6J (B6) and B6-backcrossed Rag1-deficient B6.129S7-Rag1tm1Mom (Rag1−/−) mice31, T cell receptor (TCR) α-deficient B6.129P2-Tcratm1Mjo (Tcra−/−) mice32, TCR δ-deficient B6.129S7-Tcrdtm1Mom (Tcrd−/−) mice33, MHC class II-deficient mice B6.129S2-H2dlAb1-Ea (H2-A,E−/−) mice34, B cell-deficient, Ig heavy constant chain μ-targeted B6.129S2-Ighmtm1Cgn (Ighm−/−) mice35, hen-egg lysozyme (HEL)-specific B cell receptor-transgenic (Ighm−/− MD4) mice36, MyD88-deficient B6.129P2-Myd88tm1Aki (Myd88−/−) mice37, toll-interleukin 1 receptor (TIR) domain-containing adaptor protein (TIRAP)-deficient B6.129P2-Tiraptm1Aki (Tirap−/−) mice38, and toll-like receptor adaptor molecule 1 (TICAM-1)-deficient B6.129P2-Ticam1tm1Aki (Ticam1−/−) mice39 have been described. B6-congenic mice lacking Emv2 (Emv2−/−) were generated by 12 nuclear generations onto the B6 genetic background of the respective site on chromosome 8 from the A/J strain, which lacks the Emv2 integration, and have been previously described40. Emv2−/− mice were subsequently crossed with B6-backcrossed Rag1−/− mice to create Rag1−/−Emv2−/− mice. These strains were maintained in individually ventilated cages (IVCs) in specific pathogen-free (SPF) facilities at NIMR, and kept on UV-irradiated and filtered neutral pH water. In addition, separate colonies of Rag1−/− mice, Ighm−/− mice, Myd88−/− mice, Tirap−/− mice and Tcam1−/− mice were maintained in the same facilities and constantly kept on a diet of acidified water (pH 2.5).
B6 and B6-backcrossed B6.129S7-Rag1tm1Mom/J (Rag1−/−) mice, B6.129S2-Ighmtm1Cgn/J (Ighm−/−) mice, B6.129S1-Tlr7tm1Flv/J (Tlr7−/−) mice41, and B6.129P2-Myd88tm1.1Defr/J (Myd88−/−) mice42 were also maintained in SPF facilities at The Jackson Laboratory, MA, USA (JAX), and were kept on acidified water (pH 2.8-3.2).
B6 and B6-backcrossed B6.129S7-Rag1tm1Mom (Rag1−/−) were also maintained in germ-free (GF) facilities at the Unit for Laboratory Animal Medicine, University of Michigan, MI, USA (UMICH) and kept on autoclaved distilled water.
B6 and B6-backcrossed B6.129S1-Tlr7tm1Flv (Tlr7−/−) mice, and B6.129P2-Tlr9tm1Aki (Tlr9−/−) mice43 were also maintained in SPF facilities at the Centre d’Immunologie de Marseille-Luminy, Marseille, France (CIML) and kept on autoclaved water.
B6-backcrossed TLR7-deficient B6.129P2-Tlr7tm1Aki (Tlr7−/−) mice44 were also maintained in SPF facilities at the London Research Institute (LRI), Cancer Research UK and kept on autoclaved water. These mice were received into the NIMR quarantine facility and bred before testing.
B6 and B6-backcrossed B6.129S7-Rag1tm1Mom (Rag1−/−) mice, B cell-deficient, Ig heavy chain joining region-targeted B6.129S7-Igh-Jtm1Dhu (Igh-j−/−) mice45, IgA-deficient, Ig heavy constant α chain-targeted B6.129S7-Ighatm1Grh (Igha−/−) mice46, and polymeric Ig receptor-deficient B6.129P2-Pigrtm1Rast (Pigr−/−) mice47 were maintained in IVC cages in SPF facilities at Rodent Center HCI, ETH Zürich, Zürich, Switzerland (RCHCI) and kept on autoclaved water.
C57BL/6NCrl-backcrossed B6.129S2-Ighmtm1Cgn (Ighm−/−) and activation-induced cytidine deaminase-deficient B6.CBA-Aicdatm1Hon (Aicda−/−) mice48, were also maintained in SPF facilities at the Center for Cancer Research, National Cancer Institute, Frederick, MD, USA (NCI) and kept on autoclaved water.
All colonies of Rag1−/− mice tested in this study (NIMR, JAX, UMICH and RCHCI) were established by rederivation of a common JAX stock into each of these facilities. This process ensures the removal of adventitious agents, including infectious eMLVs, and leads to colonisation of the newly-rederived mice with the specific microbiota of each facility (or the lack of colonisation in the case of the germ-free facility). These independent colonies were maintained by homozygous breeding for at least 12 and up to 45 filial generations (F12-F45) prior to testing.
Colonies of Ighm−/− mice at NIMR and JAX were also established by rederivation. The Ighm−/− mouse colony at JAX was at least the F15 of homozygous breeding prior to testing. Two colonies of Ighm−/− mice, established at NIMR by two independent rederivations (as part of the unit reorganisation), were tested at different points following rederivation. The first was maintained for 16 months (F4) on a diet of neutral pH water (before switching to acidified water for F12), and the second, more recent colony was maintained on a diet of neutral pH water for only 7 months (F1-F2) prior to testing.
Colonies of Myd88−/− mice at NIMR and JAX were also established by rederivation and maintained for approximately F16 and F5 of homozygous breeding, respectively, prior to testing. Colonies of Tlr7−/− mice at LRI/NIMR, JAX and CIML were also established by rederivation and maintained for approximately F11, F5 and F9 of homozygous breeding, respectively, prior to testing.
Unless otherwise indicated (e.g. aged mice) all mice were used between 4 and 8 weeks after birth. All animal experiments were approved by ethical committees of respective Institutes, and conducted according to local guidelines and regulations.
Expression analyses
For transcriptional analysis, peritoneal exudate cells were isolated from B6 or Rag1−/− mice and were subsequently purified by cell sorting, performed on MoFlo cell sorters (Dako, Fort Collins, CO, USA), as CD11b+MHC-IIhiB220−Gr1− macrophages. RNA was isolated from the cells using TRI reagent (Sigma-Aldrich, St. Louis, US), according to manufacturer’s instructions. Purified RNA samples were checked for quality using the Agilent bioanalyzer (Agilent, Santa Clara, CA, USA). Synthesis of cDNA, probe labelling and hybridisation were performed using Affymetrix MouseGene 1.0 ST oligonucleotide arrays (Affymetrix, Santa Clara, CA, USA). Primary microarray data from triplicate arrays were analysed with GeneSpring GX (Agilent Technologies, Inc., Santa Clara, CA, USA) and deposited at http://www.ebi.ac.uk/arrayexpress (E-MEXP-3623).
ERV/RE transcription was quantified in various tissues by real-time quantitative reverse transcription-based PCR (qRT-PCR). Briefly, organs were removed from mice and placed in TRI reagent or RNAlater reagent (Invitrogen, Carlsbad, CA, USA) at each animal facility. For mice bred outside NIMR, tissues were then shipped to NIMR for further processing. Total RNA was isolated with TRI reagent, precipitated with isopropanol and washed in 75% ethanol before being dissolved in water. DNase digestion and clean-up was performed with the RNeasy Mini Kit (Qiagen, Hilden, Germany) and cDNA produced with the High Capacity Reverse Transcription kit (Applied Biosystems, Carlsbad, US) with an added RNase-inhibitor (Promega Biosciences, Madison, US). A final clean-up was performed with the QIAquick PCR purification kit (Qiagen). RNase-free water (Qiagen) and later nuclease-free water (Qiagen) was used throughout the protocol. Purified cDNA was then used as template for the amplification of target gene transcripts with SYBR Green PCR Master Mix (Applied Biosystems), run on the ABI Prism SDS 7000 and 7900HT (Applied Biosystems) cyclers, with the following primers, produced by Eurofins MWG Operon (Ebersberg, Germany):
eMLV spliced env cDNA – 116 bp product. Primers previously described40.
Forward 5′-CCAGGGACCACCGACCCACCGT-3′
Reverse 5′-TAGTCGGTCCCGGTAGGCCTCG-3′
Mouse mammary tumour virus (MMTV) spliced env cDNA – 116 bp product
Forward 5′-AGAGCGGAACGGACTCACCA-3′
Reverse 5′-TCAGTGAAAGGTCGGATGAA-3′
Hprt cDNA – 92 bp product
Forward 5′-TTGTATACCTAATCATTATGCCGAG-3′
Reverse 5′-CATCTCGAGCAAGTCTTTCA-3′
Primers specific for polytropic MLV (pMLV), modified polytropic MLV (mpMLV), xenotropic MLV (xMLV), Mus musculus type D retrovirus (MusD), murine retroviruses that use tRNAGln (GLN), murine endogenous retrovirus-like (MuERV-L) and intracisternal A-type particle elements (IAP) have been previously described49-51. Data are plotted as expression of the target transcript, relative to expression of Hprt in the same sample, derived using the algorithm:
A theoretical detection limit of 2 arbitrary units is also displayed as dashed horizontal lines. For eMLV expression in particular, values above 103 were considered as high and are indicated with red-filled symbols.
eMLV mRNA transcripts originating from the Emv2 locus or from potentially integrated RARV proviruses were distinguished by RT-PCR on cDNA reverse-transcribed using a reverse primer specific to the ecotropic env gene (5′-TTCTGGACCACCACACGAC-3′) and amplified using unique transcript-specific forward primers in the pol region together with a common reverse primer in the same region. The 3′ nucleotides of the transcript-specific forward primers correspond to position 3576 of the pol gene, which is a C in Emv2, but a required G in all sequenced RARVs. These primers (Eurofins MWG Operon) were:
Emv2 specific mRNA forward primer
Forward 5′-CCTGGGTTTGCGGAAATGGCAC-3′
RARV specific mRNA forward primer
Forward 5′-CCTGGGTTTGCGGAAATGGCGG-3′
Common reverse primer
Reverse 5′-TTTGGCGTAGCCCTGCTTCTCG-3′
Both PCRs produce a 192 bp product.
Microarray-based analyses of retroelement expression
Expression levels of ERVs and retroelements (RE) were determined in publicly-available microarray data sets, by assigning ERV- or RE-reporting probes to the correct ERV or RE family they are reporting. Individual probe sequences were obtained for mouse and human microarray platforms from the vendors’ websites. Nucleotide sequence data for mouse and human REs (including elements present in ancestral species) were downloaded from RepBase Update52 (http://www.girinst.org/repbase/) (v25/11/2011) and additional human REs from dbRIP database53 (http://www.dbrip.org) (v2h). Low-copy mouse ERV sequences were obtained from the literature54,55, database searches and mining of the C57BL/6J RefSeq assembly (v37) sequence. Local BLAST libraries were produced with the NCBI C++ Toolkit (http://blast.ncbi.nlm.nih.gov/) and a Python 3.2 (http://www.python.org/) script was used to run and query BLASTn (‘-task blastn-short’ optimised) for the obtained probe sequences. Three separate stringencies were used in the screening: 90% (>90% length, >90% nucleotide homology), 95% (>95% length, >95% nucleotide homology) and 100% (identical matching required). For microarray platforms where single transcripts are represented by multiple probes, an additional screen was imposed to ensure >75% of probes gave appropriate BLASTn hits. Under this threshold, probes were assumed to be divided across a RE-gene boundary and were excluded from further analysis. Probes were compiled from the 90% stringency analysis for human and mouse REs and from the 100% stringency analysis for low-copy C57BL/6J ERVs. These were used to produce a Python tool, REsearch, which will be described in detail elsewhere, and the code of which is available upon request, to mine RE expression from text-based microarray analysis output.
Emv2/eMLV copy number analysis
DNA copy numbers of Emv2/eMLV were determined by real-time quantitative PCR (qPCR) on DNA samples isolated from the indicated organs or tumours using the following primers (Eurofins MWG Operon):
Emv2/eMLV env DNA – 169 bp product. Primers modified from published sequences49.
Forward 5′-AGGCTGTTCCAGAGATTGTG-3′
Reverse 5′-TTCTGGACCACCACACGAC-3′
Emv2/eMLV pol DNA – 76 bp product. Primers previously described56.
Forward 5′-CACTTTGAGGGATCAGGAGCC-3′
Reverse 5′-CTTCTAGGTTTAGGGTCAACACCTGT-3′
Signals were normalised for the amount of DNA used in the reactions based on amplification of the single-copy Ifnar1 gene with the following primers:
Ifnar1 DNA – 150 bp product
Forward 5′-AAGATGTGCTGTTCCCTTCCTCTGCTCTGA-3′
Reverse 5′-ATTATTAAAAGAAAAGACGAGGCGAAGTGG-3′
Copy numbers were calculated with a ΔΔCT method, using splenic DNA from B6 mice as control, which was assigned a value of 1 copy per haploid genome, N.
Sequencing and sequence analyses
Bacterial genera diversity was determined in faecal samples from the large intestines of mice, by high-throughput sequencing of amplicons of bacterial DNA encoding 16S ribosomal RNA, using a the Roche FLX Genome Sequencer. Samples were either collected at NIMR or at RCHCI and were subsequently transported frozen to NIMR, where DNA was isolated, using the QIAamp DNA Stool Mini Kit (Qiagen). DNA amplification, sequencing and metagenomic analysis was performed by DNAVision (Gosselies, Belgium).
The region of the pol gene carrying the inactivating mutation in Emv2 was sequenced in isolated RARVs as described below. A 708bp fragment spanning this region was amplified from genomic DNA isolated from Mus dunni cells infected with the respective RARV using the following primers (Eurofins MWG Operon):
Emv2/eMLV pol DNA – 708 bp product.
Forward 5′-ATCGGGCCTCGGCCAAGAAAG-3′
Reverse 5′-CCGGGAGAGGGAGTAAGGTGGC-3′
RARV genomes were amplified from the same DNA template in two halves using the following primers (Eurofins MWG Operon):
RARV first half – 4074 bp product.
Forward 5′-GCGCCAGTCCTCCGATAGACT-3′
Reverse 5′-CCGGGAGAGGGAGTAAGGTGGC-3′
RARV second half – 4909 bp product.
Forward 5′-ATCGGGCCTCGGCCAAGAAAG-3′
Reverse 5′-TGCAACAGCAAAAGGCTTTATTGG-3′
PCR products were purified with the QIAquick PCR purification kit (Qiagen) and subjected to sequencing at Source BioScience (Cambridge, UK).
Sequence analyses, comparisons and alignments were performed with Vector NTI v11.5 (Invitrogen). RARV contigs were aligned against B6 MLVs as previously defined54using MAFFT within UGENE software57,58. Distance plots were calculated with RDP (Recombination Detection Program) v4.16, using a 100 bp window and a 10 bp shift58. RARV phylogenetic analyses were performed with PHYLIP (Phylogeny inference package) v3.2 within UGENE software.
FACS
Cell suspensions from spleens, lymph nodes or peritoneal exudates were stained with directly-conjugated antibodies to surface markers, obtained from eBiosciences (San Diego, CA, USA), CALTAG/Invitrogen (Carlsbad, CA, USA), BD Biosciences (San Jose, CA, USA) or BioLegend (San Diego, CA, USA). MLV SU was detected using the 83A25 monoclonal antibody59 (rat IgG2a, anti-MLV SU) as the primary reagent, followed by staining with a biotinylated anti-rat IgG2a antibody (clone RG7/1.30, BD Biosciences) as the secondary reagent, and a streptavidin-phycoerythrin (PE) conjugate (BioLegend) or a streptavidin-PE Texas Red conjugate (CALTAG/Invitrogen) as the tertiary reagent. Four- and 8-color cytometry was performed on FACSCalibur (BD Biosciences) and CyAn (Dako, Fort Collins, CO) flow cytometers, respectively, and analysed with FlowJo v8.7 (Tree Star Inc., Ashland, OR, USA) or Summit v4.3 (Dako) analysis software, respectively.
Retroviral isolation and assays
For the isolation of infectious MLVs plasma samples obtained from Rag1−/− mice were incubated with Mus dunni cells transduced with the XG7 replication-defective retroviral vector, expressing green fluorescent protein (GFP) from a human cytomegalovirus (hCMV) promoter and a neomycin-resistance gene under the control of the LTR60. The presence of infectious MLVs was examined after 10-14 days of culture by testing for the presence of pseudotyped XG7 vector in culture supernatant. For this, culture supernatant of XG7-transduced Mus dunni cells that had been incubated with plasma samples was subsequently added onto untransduced Mus dunni cells, which were then assessed for GFP expression by flow cytometry 3 days later. The Fv1 tropism of infectious MLVs was determined by adding serial dilutions of culture supernatant of XG7-transduced Mus dunni cells that had been incubated with plasma samples onto B-3T3 or N-3T3 cells. Infection was quantified by GFP expression in these cells 3 days later. B-tropic and N-tropic stocks of F-MLV, obtained as culture supernatant of Mus dunni cells chronically infected with these viruses, were used as controls. Results were expressed as B:N ratios, which were the percentage of GFP+ cells in B-3T3 cultures, divided by the percentage of GFP+ cells in N-3T3 cultures.
Histology
For histological analysis, organs were collected in formalin immediately after culling of donor mice. Histological sections were prepared and stained with haematoxylin and eosin and assessed by Dr Mark Stidworthy at IZVG Pathology, Leeds, UK.
Transmission electron microscopy
Samples were immersion fixed in 2% glutaraldehyde/2% paraformaldehyde and post-fixed in 1% osmium tetroxide using 0.1 M sodium cacodylate buffer pH 7.2. Aqueous uranyl acetate was followed by dehydration through a graded ethanol series and propylene oxide. Samples were then embedded in Epon and 50nm sections mounted on pioloform coated grids and stained with ethanolic uranyl acetate followed by Reynold’s lead citrate. They were viewed with a JEOL 100EX transmission electron microscope (JEOL, Tokyo, Japan) equipped with an ORIUS 1000 CCD camera (Gatan, Pleasanton, CA, USA).
Statistical analyses
Statistical comparisons were made using SigmaPlot 12.0 (Systat Software Inc., Germany). Parametric comparisons of normally-distributed values that satisfied the variance criteria were made by unpaired Student’s t-tests. Pairwise comparisons of data sets that did not pass the variance test were compared with non-parametric two-tailed Mann-Whitney Rank Sum. Bacterial diversity was compared with Analysis of variance (ANOVA) tests with Bonferroni correction for multiple comparisons. Tumour incidence rates were compared by Log-Rank Survival analysis of Kaplan-Meier curves. ANOVA and statistical comparisons of microarray data were made using GeneSpring GX, with Benjamini–Hochberg false discovery rate (FDR) correction for multiple comparisons.
Supplementary Material
Acknowledgments
We wish to thank Prof. Wolf-Dietrich Hardt for mouse samples and helpful discussion, Ms Lara Sellés Vidal for technical assistance, and colleagues for critical reading of the manuscript. We also wish to thank the staff of the Unit for Laboratory Animal Medicine, University of Michigan for the provision of germ-free mice. We are grateful for assistance from the Division of Biological Services, the Flow Cytometry, Electron Microscopy and Microarray Facilities at NIMR. This work was supported by the UK Medical Research Council (U117581330 and U117512710).
References
- 1.Honda K, Littman DR. The Microbiome in Infectious Disease and Inflammation. Annu. Rev. Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stocking C, Kozak CA. Murine endogenous retroviruses. Cell. Mol. Life Sci. 2008;65:3383–3398. doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat. Rev. Microbiol. 2012;10:395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- 4.Stoye JP, Coffin JM. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J. Virol. 1987;61:2659–2669. doi: 10.1128/jvi.61.9.2659-2669.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King SR, Berson BJ, Risser R. Mechanism of interaction between endogenous ecotropic murine leukemia viruses in (BALB/c X C57BL/6) hybrid cells. Virology. 1988;162:1–11. doi: 10.1016/0042-6822(88)90388-1. [DOI] [PubMed] [Google Scholar]
- 6.Demaria O, et al. TLR8 deficiency leads to autoimmunity in mice. J. Clin. Invest. 2010;120:3651–3662. doi: 10.1172/JCI42081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeFranco AL, Rookhuizen DC, Hou B. Contribution of Toll-like receptor signaling to germinal center antibody responses. Immunol. Rev. 2012;247:64–72. doi: 10.1111/j.1600-065X.2012.01115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Browne EP. Regulation of B cell responses by Toll-like receptors. Immunology. 2012 doi: 10.1111/j.1365-2567.2012.03587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirkland D, et al. B cell-intrinsic MyD88 signaling prevents the lethal dissemination of commensal bacteria during colonic damage. Immunity. 2012;36:228–238. doi: 10.1016/j.immuni.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Huang X, Zhu Z, Gorelik E. Sequence and Insertion Sites of Murine Melanoma-Associated Retrovirus. J. Virol. 1999;73:9178–9186. doi: 10.1128/jvi.73.11.9178-9186.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pothlichet J, Mangeney M, Heidmann T. Mobility and integration sites of a murine C57BL/6 melanoma endogenous retrovirus involved in tumor progression in vivo. International Journal of Cancer. 2006;119:1869–1877. doi: 10.1002/ijc.22066. [DOI] [PubMed] [Google Scholar]
- 12.Stoye JP, Moroni C, Coffin JM. Virological events leading to spontaneous AKR thymomas. J. Virol. 1991;65:1273–1285. doi: 10.1128/jvi.65.3.1273-1285.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young GR, et al. Negative selection by an endogenous retrovirus promotes a higher-avidity CD4+ T cell response to retroviral infection. PLoS Pathog. 2012;8:e1002709. doi: 10.1371/journal.ppat.1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melamedoff M, Lilly F, Duran-Reynals ML. Suppression of endogenous murine leukemia virus by maternal resistance factor. J. Exp. Med. 1983;158:506–514. doi: 10.1084/jem.158.2.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoye JP, Moroni C. Endogenous retrovirus expression in stimulated murine lymphocytes. Identification of a new locus controlling mitogen induction of a defective virus. J. Exp. Med. 1983;157:1660–1674. doi: 10.1084/jem.157.5.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak CA, Rowe WP. Genetic mapping of xenotropic murine leukemia virus-inducing loci in five mouse strains. J. Exp. Med. 1980;152:219–228. doi: 10.1084/jem.152.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCubrey J, Risser R. Genetic interactions in induction of endogenous murine leukemia virus from low leukemic mice. Cell. 1982;28:881–888. doi: 10.1016/0092-8674(82)90067-8. [DOI] [PubMed] [Google Scholar]
- 18.Moroni C, Schumann G. Lipopolysaccharide induces C-type virus in short term cultures of BALB/c spleen cells. Nature. 1975;254:60–61. doi: 10.1038/254060a0. [DOI] [PubMed] [Google Scholar]
- 19.Greenberger JS, Phillips SM, Stephenson JR, Aaronson SA. Induction of mouse type-C RNA virus by lipopolysaccharide. J. Immunol. 1975;115:317–320. [PubMed] [Google Scholar]
- 20.Amit I, et al. Unbiased Reconstruction of a Mammalian Transcriptional Network Mediating Pathogen Responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim A, et al. Antibody and B-cell responses may control circulating lipopolysaccharide in patients with HIV infection. AIDS. 2011;25:1379–1383. doi: 10.1097/QAD.0b013e328348a789. [DOI] [PubMed] [Google Scholar]
- 22.Reid RR, et al. Endotoxin shock in antibody-deficient mice: unraveling the role of natural antibody and complement in the clearance of lipopolysaccharide. J. Immunol. 1997;159:970–975. [PubMed] [Google Scholar]
- 23.Shulzhenko N, et al. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat. Med. 2011;17:1585–1593. doi: 10.1038/nm.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu L, et al. Chronic acid water feeding protects mice against lethal gut-derived sepsis due to Pseudomonas aeruginosa. Curr. Issues Intest. Microbiol. 2006;7:19–28. [PubMed] [Google Scholar]
- 25.Belancio VP, Roy-Engel AM, Deininger PL. All y’all need to know ‘bout retroelements in cancer. Semin. Cancer Biol. 2010;20:200–210. doi: 10.1016/j.semcancer.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romanish MT, Cohen CJ, Mager DL. Potential mechanisms of endogenous retroviral-mediated genomic instability in human cancer. Semin. Cancer Biol. 2010;20:246–253. doi: 10.1016/j.semcancer.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Lamprecht B, et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat. Med. 2010;16:571–579. doi: 10.1038/nm.2129. [DOI] [PubMed] [Google Scholar]
- 28.Bannert N, Kurth R. Retroelements and the human genome: New perspectives on an old relation. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trinchieri G. Cancer and Inflammation: An Old Intuition with Rapidly Evolving New Concepts. Annu. Rev. Immunol. 2012;30:677–706. doi: 10.1146/annurev-immunol-020711-075008. [DOI] [PubMed] [Google Scholar]
- 30.Park MA, et al. Common variable immunodeficiency: a new look at an old disease. Lancet. 2008;372:489–502. doi: 10.1016/S0140-6736(08)61199-X. [DOI] [PubMed] [Google Scholar]
- 31.Mombaerts P, et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 32.Philpott KL, et al. Lymphoid development in mice congenitally lacking T cell receptor alpha beta-expressing cells. Science. 1992;256:1448–1452. doi: 10.1126/science.1604321. [DOI] [PubMed] [Google Scholar]
- 33.Itohara S, et al. T cell receptor delta gene mutant mice: independent generation of alpha beta T cells and programmed rearrangements of gamma delta TCR genes. Cell. 1993;72:337–348. doi: 10.1016/0092-8674(93)90112-4. [DOI] [PubMed] [Google Scholar]
- 34.Cosgrove D, et al. Mice lacking MHC class II molecules. Cell. 1991;66:1051–1066. doi: 10.1016/0092-8674(91)90448-8. [DOI] [PubMed] [Google Scholar]
- 35.Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 36.Goodnow CC, et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 1988;334:676–682. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 37.Adachi O, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto M, et al. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature. 2002;420:324–329. doi: 10.1038/nature01182. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 40.Young GR, et al. Negative selection by an endogenous retrovirus promotes a higher-avidity CD4+ T cell response to retroviral infection. PLoS Pathog. 2012;8:e1002709. doi: 10.1371/journal.ppat.1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lund JM, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou B, Reizis B, DeFranco AL. Toll-like receptors activate innate and adaptive immunity by using dendritic cell-intrinsic and -extrinsic mechanisms. Immunity. 2008;29:272–282. doi: 10.1016/j.immuni.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 44.Hemmi H, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat. Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 45.Chen J, et al. Immunoglobulin gene rearrangement in B cell deficient mice generated by targeted deletion of the JH locus. Int. Immunol. 1993;5:647–656. doi: 10.1093/intimm/5.6.647. [DOI] [PubMed] [Google Scholar]
- 46.Harriman GR, et al. Targeted deletion of the IgA constant region in mice leads to IgA deficiency with alterations in expression of other Ig isotypes. J. Immunol. 1999;162:2521–2529. [PubMed] [Google Scholar]
- 47.Uren TK, et al. Role of the polymeric Ig receptor in mucosal B cell homeostasis. J. Immunol. 2003;170:2531–2539. doi: 10.4049/jimmunol.170.5.2531. [DOI] [PubMed] [Google Scholar]
- 48.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 49.Yoshinobu K, et al. Selective Up-Regulation of Intact, but Not Defective env RNAs of Endogenous Modified Polytropic Retrovirus by the Sgp3 Locus of Lupus-Prone Mice. J. Immunol. 2009;182:8094–8103. doi: 10.4049/jimmunol.0900263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karimi M, et al. DNA Methylation and SETDB1/H3K9me3 Regulate Predominantly Distinct Sets of Genes, Retroelements, and Chimeric Transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macfarlan TS, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, et al. dbRIP: a highly integrated database of retrotransposon insertion polymorphisms in humans. Hum. Mutat. 2006;27:323–329. doi: 10.1002/humu.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jern P, Stoye JP, Coffin JM. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 2007;3:2014–2022. doi: 10.1371/journal.pgen.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bromham L, Clark F, McKee JJ. Discovery of a novel murine type C retrovirus by data mining. J. Virol. 2001;75:3053–3057. doi: 10.1128/JVI.75.6.3053-3057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lotscher M, et al. Induced Prion Protein Controls Immune-Activated Retroviruses in the Mouse Spleen. PLoS One. 2007;2:e1158. doi: 10.1371/journal.pone.0001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin DP, et al. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Evans LH, et al. A neutralizable epitope common to the envelope glycoproteins of ecotropic, polytropic, xenotropic, and amphotropic murine leukemia viruses. J. Virol. 1990;64:6176–6183. doi: 10.1128/jvi.64.12.6176-6183.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bock M, Bishop KN, Towers G, Stoye JP. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J. Virol. 2000;74:7422–7430. doi: 10.1128/jvi.74.16.7422-7430.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.