Abstract
Background
Atopic dermatitis and psoriasis are common inflammatory diseases, canonically described as involving distinct T-helper polarization and granulocytic infiltration. Acute atopic dermatitis lesions are associated with TH2 and eosinophilic inflammation, while psoriasis lesions are associated with TH1/17 and neutrophilic inflammation. Despite intensive investigation, these pathways remain incompletely understood in vivo in human subjects.
Objective
Using atopic dermatitis and psoriasis lesional skin as exemplar TH2 and TH1/17 diseased tissue, we sought to clarify common and unique molecular and pathophysiologic features in inflamed skin with different types of inflammatory polarization.
Methods
We conducted gene expression microarray analyses to identify distinct and commonly dysregulated expression in atopic dermatitis (by Hanifin & Rajka criteria) and psoriasis lesions. We defined gene sets comprising genes encoding cytokines, chemokines, and growth factors that were uniquely or jointly dysregulated in atopic dermatitis and psoriasis, and calculated aggregate gene set expression scores for lesional skin of these dermatoses and healthy control skin.
Results
The atopic dermatitis gene set score correlated with systemic and local measures of allergic inflammation including serum IgE, blood eosinophil count, and tissue eosinophils. Unexpectedly, genes encoding neutrophil chemoattractants among the common gene set were highly expressed in atopic dermatitis lesional skin. H&E and immunohistochemical analyses showed the numbers of neutrophils in atopic dermatitis lesional skin were comparable to those in psoriasis lesional skin, and both were correlated with the extent of expression of neutrophil chemoattractant genes.
Conclusion
These data are evidence that neutrophilic inflammation is a feature of lesional atopic dermatitis pathology, comorbid with allergic inflammation.
Keywords: Atopic dermatitis, psoriasis, TH2, TH17, gene expression microarray, neutrophil, eosinophil
Introduction
Atopic Dermatitis (AD) and psoriasis are common and clinically distinct human diseases characterized by inflammatory skin lesions 1–3. These diseases have been associated with contrasting polarization of the adaptive immune system and distinct granulocytic infiltration in lesional tissue. AD is canonically described as featuring excessive T-helper type 2 (TH2) and eosinophilic infiltration in acute lesions and a mixed TH1 and TH2 pattern in chronic lesions 4, while psoriasis features excessive TH1/17 inflammation and neutrophilic infiltration 2, 5, 6. Investigative studies of therapies that specifically target canonical TH2 cytokines in AD 7 and TH1/17 cytokines in psoriasis 8–11 have provided direct evidence of the causal roles of these T-helper pathways in disease morbidity. However, variable therapeutic efficacy and the lack of sustained clinical response in the absence of continuous therapy underscore the need to better understand these types of inflammation. This concerns both the relationship between the activity of targeted pathways and specific pathologic measures and underlying bases of disease heterogeneity, which may directly influence clinical outcomes. Addressing this need may lead to the discovery of new therapeutic targets, and facilitate the development of selection strategies to appropriately pair patients with molecularly targeted therapies.
Molecular phenotyping of diseased tissues by high-dimensional gene expression technologies (e.g. microarrays) comprehensively surveys the transcriptome. However, the high dimensionality of gene expression microarrays requires appropriate considerations for the manifold nature of employing gene-by-gene hypothesis testing that may consequently reduce statistical power 12. Independent filtering, which aims to remove variables unlikely to be informative prior to testing, is a two-stage approach that filters variables by criteria independent of the test statistic (e.g. overall variance, gene annotation) and can increase detection rate without loss of type I error control 13. Cytokines, chemokines, and growth factors (CCGf) are potent molecular mediators involved in both homeostatic and aberrant processes of inflammation and tissue development. Given the well-characterized roles of CCGfs as mediators of inflammation and intercellular communication in inflammatory diseases 14, we have focused our analyses on CCGfs based on both biological relevance and attractive statistical properties. We have previously derived a quantitative gene expression signature of TH2 inflammation in asthmatic bronchial biopsies that correlated with physiologic measures of allergic inflammation, and evaluated this signature against a gene set comprising CCGfs to better understand phenotypic heterogeneity in asthma and the involvement of key mediators of inflammation, cellular migration, and tissue remodeling 15. We anticipated that CCGf expression analyses in inflamed tissues from AD and psoriasis would be particularly informative in understanding pathophysiologic features of these inflammatory skin diseases.
Herein, we investigated AD and psoriasis tissues by gene expression microarrays where we aimed to: 1) clarify T-helper-associated inflammatory expression patterns in the context of human skin disease, 2) gain insight into the degree to which T-helper type-associated inflammation and other pathways are involved in individual patients’ disease, and 3) relate pathway activity to specific pathologic measures. Leveraging the distinct inflammatory etiologies associated with AD and psoriasis, we performed comparative analyses to delineate common and unique expression patterns associated with each disease. We derived lesional AD, lesional psoriasis, and common lesional gene sets (GS) and observed dramatic TH2- and TH1/17- associated gene expression patterns in AD and psoriasis, respectively. We summarized these skin biopsy GSs into quantitative expression signature scores, and observed correlations with related physiologic measures of inflammation suggesting that transcriptional perturbations in lesional skin are involved in broader, related systemic inflammatory manifestations. Unexpectedly, we found that patterns of lesional gene expression common to both AD and psoriasis are enriched for genes encoding neutrophil chemoattractants, whose expression levels correlate with peripheral and immunohistochemical measures of neutrophilia. Collectively, these data suggest that neutrophilic inflammation is a pathologic feature of chronic AD co-existing with hallmarks of allergic inflammation.
Methods
Subjects
The subject population consisted of 12 and 14 individuals with atopic dermatitis and psoriasis, respectively, and 5 healthy controls. Hanifin & Rajka criteria 16 were used to identify adult subjects with atopic dermatitis. Subjects had active lesions and were not on systemic therapy. Patient demographics along with the disease severity and distribution of the affected skin are summarized in Table E1. All subjects had at least three of the four major features and at least three of the minor features defined in Hanifin & Rajka criteria. All except one subject (MB19) met the revised criteria by American Academy of Dermatology 17. However, this subject fulfills the criteria developed by Williams 18. Median total serum IgE levels of these subjects was 303 [527, IQR] kU/ml.
The biopsies were taken from established lesions in affected areas that exhibited characteristic eczematous lesions with signs of epithelial disruption, excoriations and crusts, but without clinical evidence of superinfection. Figure E1 depicts a representative clinical appearance of the AD lesions in this study.
Psoriasis participants consisted of adult subjects with chronic disease and active skin lesions with characteristic morphology and histologic presentation. Patient demographics are tabulated in Table E1. See Methods in this article’s Online Repository at www.jacionline.org for additional details.
RNA processing and microarray hybridization
Agilent two color Whole Human Genome (WHG) expression microarray analysis was conducted as described previously 15 on RNA isolated from biopsies preserved in RNALater (Invitrogen, Carlsbad, CA) (see Methods section in this article’s Online Repository).
Microarray data analysis and statistics
All statistical calculations were performed using the R Project software package, version 2.14.0 (http://www.R-project.org). R package hgug4112a.db, version 2.6.3 was utilized for microarray annotation. A moderated paired t-test was used to assess differential expression using the limma package of Bioconductor 19.
Correlation coefficients between gene expression and physiologic parameters were performed using Spearman’s method. Continuous versus categorical variable testing was implemented by Kruskal-Wallis ANOVA. Multiple testing for pairwise tests was addressed via the calculation of false discovery rate.
See the Methods section in this article’s Online Repository detailed statistics.
Histochemistry and Immunohistochemistry
Detection of inflammatory cells in paraffin-embedded skin biopsies was performed as described 20 (see Methods section in this article’s Online Repository). Detection of neutrophils was accomplished by optimized neutrophil lipocalin (lipocalin-2) and neutrophil elastase (elastase-2) staining supplemented with H&E, both in combination with observation of polymorphic nuclei. Initial neutrophilic assessment involved myeloperoxidase staining.
Results
AD, psoriasis, and common lesion gene sets are defined by comparative differential expression analyses of cytokine, chemokine, and growth factor (CCGf) genes
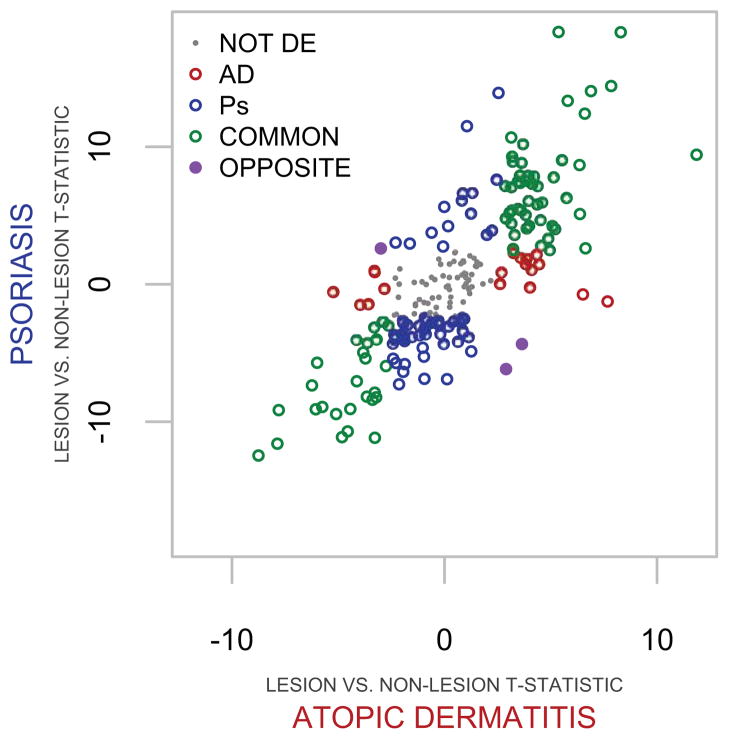
Leveraging the distinct inflammatory etiologies associated with AD and psoriasis, we performed comparative expression analyses to define disease-specific Gene Sets (GS) that were composed of distinguishing T-helper associated genes, and those which were dysregulated in common. We compared within-disease lesion versus non-lesion moderated paired t-tests (see Methods section in this article’s Online Repository) and defined AD, psoriasis, or common gene sets (AD-GS, Ps-GS, and C-GS) as being associated with lesional AD (red and purple genes), psoriasis (blue and purple genes), or commonly dysregulated in both diseases (green genes) (Figure 1 and Table E2). The t-statistic scatterplot of CCGf genes comparing diseased lesion versus non-lesion depicts how the intersection of magnitude and direction of dysregulation define the GSs (Figure 1). Differential expression analysis revealed a substantial number of genes that are significantly altered (false discovery rate < 0.05) commonly (C-GS) in the two diseases (75 of 217 CCGf, 36.6%), as compared to AD-GS (21 of 217 CCGf, 9.6%) and Ps-GS (59 of 217 CCGf, 27.2%).
Fig 1. Comparative expression analysis of cytokines, chemokines, and growth factors (CCGf) defines gene sets.
T-statistics of within disease lesional versus non-lesion comparisons are depicted by scatter plot. Atopic dermatitis gene set (AD-GS; red) and psoriasis (Ps)-GS (blue) comprise unique and opposingly differentially expressed genes (purple). Commonly and not differentially expressed (NOT DE) genes are represented by green and gray symbols, respectively.
Atopic dermatitis gene set (AD-GS) comprises TH2-associated genes and the signature expression intensity correlates with serum IgE, blood, and tissue eosinophil count
The atopic dermatitis gene set (AD-GS, Table E2 and Figure 2A) includes IL13, CCL13, CCL17, CCL26, and TNFSF4, which are established mediators of TH2 inflammation 5, 15, 21. IL13 has been under intense investigation as a therapeutic target in TH2-driven inflammatory diseases 22. We have observed CCL13 and CCL26 expression in bronchial biopsies to be associated with a “TH2 high” subphenotype of asthma 15. CCL17 is a chemoattractant for TH2 cells via CCR422 and is potently upregulated in TH2 central memory T cells stimulated by TSLP-DCs 23. TNFSF4 (also known as OX40L) has been demonstrated to be a key component in the initiation of thymic stromal lymphopoietin (TSLP)-dependent allergic inflammation 21, 24.
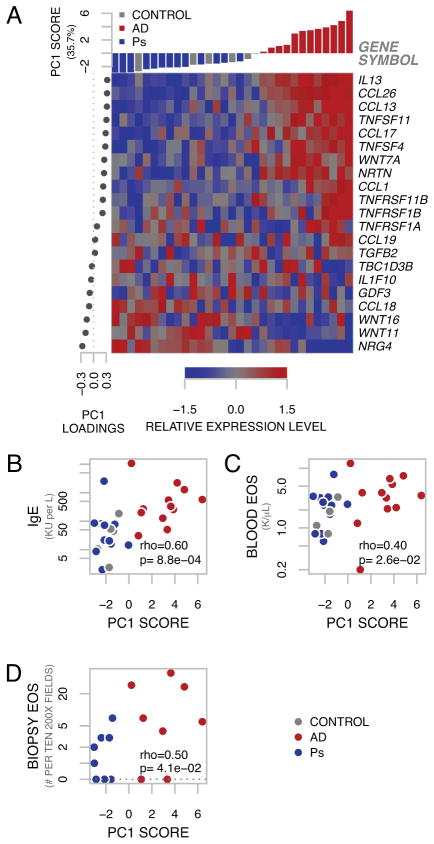
Fig 2. Atopic Dermatitis-Gene Set.
A, Genes comprising AD-GS are represented by expression heatmap, organized by PCA factors. Columns (samples) are arranged by PC1 score. Rows (genes) are organized by PC1 loadings. B, PC1 significantly correlates with serum IgE, C, blood eosinophils and D, biopsy eosinophils.
Given the strong representation of dysregulated TH2-associated genes in AD, we performed supervised principal component analysis (SPCA) on the 21 AD-GS genes. This allowed us to derive a quantitative signature 15, 25 to link pathway gene expression in lesional skin and skin from healthy controls to systemic clinical covariates related to allergy and inflammation. Expression intensity of these genes is represented by heatmap, organized by supervised PCA factors (Figure 2A). We found that PC1, representing 35.7% percent of the variance of these 21 genes, correlated (Spearman’s rank correlation) strongly with serum IgE (Figure 2B, rho=0.60, p-value=8.8×10−4). Statistically significant correlations with PC1 were also observed with blood eosinophil count (Figure 2C, rho=0.40, p-value=0.026) and with tissue eosinophils (Figure 2D, rho=0.50, p-value=0.041).
Psoriasis gene set (Ps-GS) comprises up-regulated TH17-associated and down-regulated TH2-associated CCGf genes
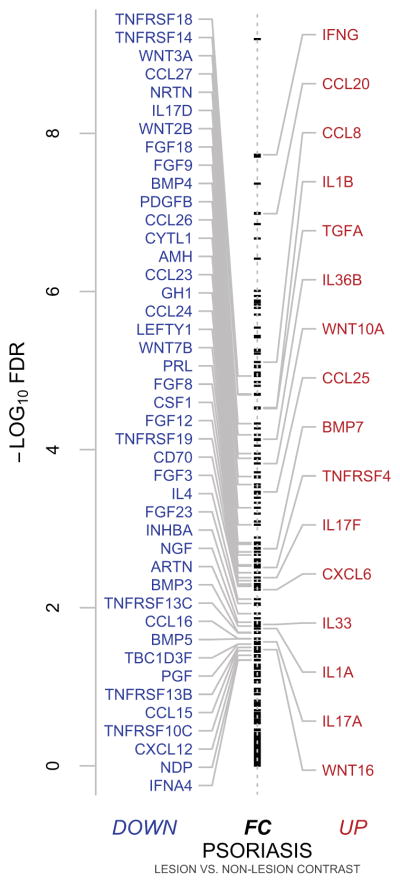
The psoriasis gene set (Ps-GS, Table E2 and Figure 3) comprises 59 CCGf genes. Upregulated Ps-GS genes included IL17A, IL17F, IL1β, and CCL20, all of which are associated with TH17 inflammation. IL17A and IL17F are expressed by TH17 cells whose normal functions are associated with defense against extracellular pathogens 26. IL1β and CCL20 have been reported to be significantly upregulated in lesional skin in psoriasis in vivo and in IL17A-stimulated keratinocytes in vitro, and synergistically upregulated by IL17A and TNF stimulation 27. These are expected and reassuring observations, as these genes whose expression are the result of TH17 inflammation have been previously implicated in pathophysiology of psoriasis 27.
Fig 3. Psoriasis-gene set expression.
Summary statistics (-log10 false discovery rates and fold-change direction) of moderated paired t-tests of lesional psoriasis versus non-lesional psoriasis is plotted by annotated stripchart for all 217 CCGf’s. Ps-GS genes, which are all differentially expressed with false discovery rates < 0.05 are annotated.
Among down-regulated Ps-GS genes were TH2 inflammation associated CCL16, CCL23, CCL26, CCL24, and IL4. CCL16 potently mediates eosinophil migration in vitro 28. Microarray analysis of peripheral blood mononuclear cells stimulated by IL13 identified CCL23 (MPIF-1) and CCL26 (eotaxin-3) as significantly upregulated genes 29. CCL24 (eotaxin-2), like CCL26, is a CCR3 agonist and a specific eosinophil chemoattractant 30. IL4 is produced by TH2 cells and drives B-cell differentiation to IgE producing plasma cells 5.
Common lesional gene set (C-GS) comprises genes encoding neutrophil chemoattractants and the signature expression intensity correlates with blood and tissue neutrophils
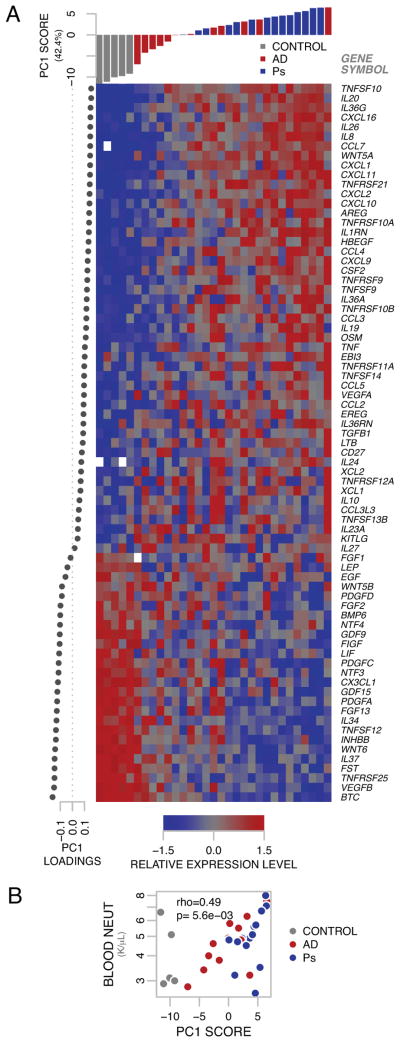
Among the common lesional gene set (C-GS) genes similarly dysregulated in AD and psoriasis as compared to non-lesional skin are CXCL1, CXCL2, IL8, and CSF2 (GM-CSF). These are all potent neutrophil chemoattractants 31, 32. We hypothesized that the dysregulation of neutrophil chemoattractant genes would have pathologic relevance and sought to relate the expression of the C-GS genes with local and systemic measures of neutrophilic inflammation. We performed supervised principal component analysis (SPCA) 15, 25 on the 75 C-GS genes to quantitatively relate pathway gene expression of lesional skin from subjects with AD or psoriasis and skin from healthy controls with measures of blood and lesional neutrophils. Expression intensity of these genes is represented by heatmap, organized by SPCA factors (Figure 4A). We found that PC1, representing 42.4% percent of the variance of these 75 genes, correlated (Spearman’s rank correlation) with blood neutrophils (Figure 4B, rho=0.49, p-value=5.6×10−3).
Fig 4. Common-gene set expression signature correlates with blood neutrophil count.
A Genes comprising C-GS are represented by expression heatmap, organized by PCA factors. Columns (samples) are arranged by PC1 score. Rows (genes) are organized by PC1 loadings. Missing expression values are represented in white. B PC1 significantly correlates with blood neutrophil count.
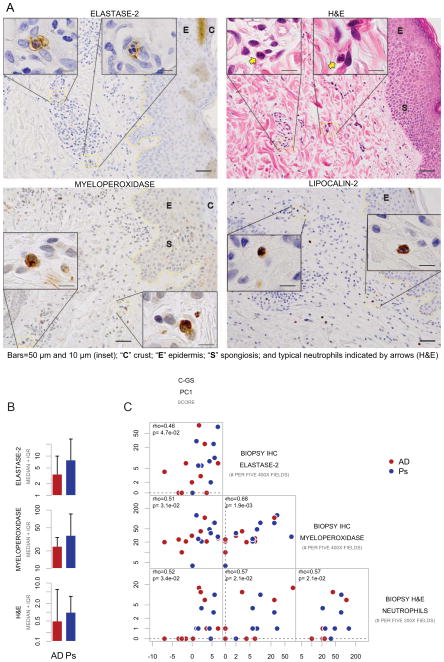
We detected neutrophil infiltration in AD and psoriasis lesional skin, but not in control or perilesional AD or psoriasis skin by histological and immunohistochemical analyses (Figure 5A, representative AD lesional skin). MPO-positive, elastase-2-positive, and neutrophil cell counts by H&E in the dermis are shown in Figure 5B.
Fig 5. Common-gene set expression signature correlates with neutrophil enumeration by histology.
A Detection of neutrophil elastase-2, lipocalin-2, and myeloperoxidase by immunohistochemistry (IHC), and H&E in representative atopic dermatitis lesional skin without excoriations from different individuals. B Cell counts for atopic dermatitis (AD) and psoriasis (Ps) lesions. C Scatterplot matrix of biopsy neutrophil indicators and C-GS PC1 score (all pairwise comparisons, p<0.05).
We initially observed a large number of MPO-positive cells (neutrophil marker 33) in both AD and psoriatic skin and the amounts of these cells were comparable between these two disease cases. These cells included intensely stained cells without distinct nuclear morphology typical of neutrophils, and may include immature neutrophils and/or monocytes.
We then used a more specific marker for neutrophils, elastase-2 (and in a limited subset with lipocalin-2), and confirmed that the amounts of positively stained cells were also comparable between AD and psoriasis. Other cell types are known to express elastase-2, although at lower levels relative to neutrophils 34, 35. However, we established that most cells exhibiting intense positivity for neutrophil elastase-2 and lipocalin-2 in both AD and psoriasis lesions were neutrophils on the basis of polymorphic nuclear morphology. These cells were largely present in the papillary dermis.
Importantly, by H&E staining we noted extravascular neutrophil counts with the characteristic morphology of neutrophils (i.e., multilobular nuclei, eosinophilic cytoplasm) were also comparable between lesional AD and psoriasis, although they were substantially lower than elastase-positive cells (Figure 5B).
Each of the three neutrophil indicators in lesional skin positively correlated (Spearman’s rank correlation) with PC1 of the C-GS (positive elastase-2 staining and cell morphologies consistent with neutrophils, rho=0.46, p-value=4.7×10−2; positive MPO staining, rho=0.51, p-value=3.1×10−2, ; and characteristic morphology by H&E analysis, rho=0.52, p-value=3.4×10−2, Figure 5C). Finally, we found that these three indicators of neutrophils were positively intercorrelated (all pairwise correlations, p<0.05, Spearman’s).
At the microscopic level, most biopsies contained surface crust, consistent with a typical clinical feature of AD. Figure E1 depicts the maximum crust identified microscopically. The numbers of elastase-2-positive cells correlated with the presence of surface crust in the stratum corneum. None of the AD cases exhibited prominent crust or microscopic evidence of bacterial colonization, or clinical or microscopic evidence of secondary infection (impetiginization)(Table E1), and neutrophilic infiltration was clearly evident in the dermis deep to the crusts in all cases.
Discussion
Genome wide association studies have identified susceptibility loci implicating TH2 cytokines IL4 and IL13 for both AD and psoriasis but with opposing effects for the same alleles in each disease 2, 36, 37, suggesting that these diseases represent opposing extremes of TH2 dysregulation. An investigational study of 20 psoriasis subjects treated with recombinant human IL4 demonstrated clinical efficacy, further suggesting that psoriasis may involve a relative deficiency in TH2 activity 38. CCL26 (eotaxin-3) was one of three CCGf genes to be differentially expressed (false discovery rate < 0.05) in opposite directions in AD and psoriasis (upregulated in lesional AD and down-regulated in lesional psoriasis, Figure E2 and Table E2). CCL26 expression is induced by IL13 or IL4 in vitro and due to its function as a CCR3 agonist CCL26 is a potent eosinophil chemoattractant 29, 39. Its expression has been observed to be upregulated in vivo in other TH2-associated human diseases, including bronchial biopsies of asthmatics and esophageal biopsies from eosinophilic esophagitis 15, 40. CCL26 expression can be induced via activation of STAT6, which is the predominant signal transducer downstream of IL4 and IL13 receptor complexes 15, 29, 39, 40. Therefore, if CCL26 expression is taken as an indicator of IL4 and/or IL13 activity, we found that the expression patterns of CCL26 in AD and psoriasis were consistent with relatively increased IL4/IL13 activity in AD and relatively decreased IL4/IL13 activity in psoriasis.
Supporting the relevance of CCGfs in this study, principal component analysis (PCA) (Figure E3) revealed a prominent relationship between the study factors of lesion status and diagnosis with respect to CCGf expression. Principal component (PC) 1 accounted for 24% of all gene expression variance. As illustrated by arrows connecting individual subjects’ non-lesional and lesional skin samples, PC1 clearly separates sample lesion status, regardless of diagnosis. A smaller proportion of gene expression variance was explained by disease diagnosis. PC2 (accounting for 10.1% of all gene expression variance) effectively separates lesional AD and psoriasis samples. These data suggest that CCGf expression is highly enriched with informative gene expression variance for the assessment of lesional AD and psoriasis.
The findings that AD-GS expression in AD and psoriasis lesional skin is correlated with serum IgE and biopsy eosinophil count is analogous to our previous description of a correlation between a quantitative TH2 gene signature in asthmatic bronchial biopsies and local and systemic pathophysiologic features of allergic inflammation: serum IgE, bronchial alveolar lavage (BAL) eosinophils, and peripheral blood eosinophils 15. The findings that the magnitude of TH2-associated gene expression in asthmatic airways as well as lesional skin in AD and psoriasis is mirrored by relevant local and systemic physiologic measures of allergic inflammation suggest a direct relationship between gene expression patterns and disease phenotype, and that the pattern of expression is useful in understanding the heterogeneity of pathway involvement. These findings are reconcilable with the expected function of genes implicated in these analyses. TH2-associated cytokines IL4 and IL13 drive immunoglobulin class switching in B cells and the production of IgE 5. Clinically, atopy is an important criterion in the diagnosis of AD 41 and IgE is thought to play a key role in the atopic response 42. TH2 cells also produce IL5, a dominant eosinophilopoietic factor whose activity directly affects peripheral blood eosinophil levels 43. These observations suggest that serum IgE and/or blood eosinophils may be useful as biomarkers for assessing the extent of TH2 inflammation in diseased tissues, which may be directly relevant when considering patient selection strategies for therapies targeting TH2 inflammation in allergic disorders. Accordingly, we have found in the context of bronchial allergen challenge that asthma patients with elevated serum IgE and/or blood eosinophil levels experienced enhanced clinical benefit from therapeutic IL13 blockade compared to patients with lower IgE and/or blood eosinophil levels 44.
As expected, the Ps-GS was enriched with upregulated TH17 associated genes, e.g. IL17A and F. Interestingly, we observed TH2 associated genes among down-regulated Ps-GS genes. The observation of down-regulated IL4 was notable with respect to the previously mentioned study in 20 psoriasis subjects effectively treated with recombinant IL4 38. Since allergic disease is characterized by excessive TH2 inflammation 5, these findings further support previous genetic 37, 44 and therapeutic studies 38 that psoriasis may be characterized by a deficiency of TH2 pathway involvement.
Our comparative transcriptomic analyses of AD and psoriasis yielded the unexpected finding that commonly differentially expressed genes (C-GS) are strongly enriched for those coding for neutrophil chemoattractants and the extent of expression of the C-GS correlates with physiologic measures of neutrophilic inflammation. Highlighting an advantage of our study design (patient-matched lesional and non-lesional skin), these findings can be reconciled with previous smaller studies, such as a genome-wide expression analysis of skin lesions that have described IL8 and CXCL2 to be among other genes differentially expressed among six AD versus seven psoriasis patients 6. As expected for the C-GS signature score, which are derived from strongly differentially expressed lesional versus non-lesional skin comparisons common to both dermatoses, we found these values to be significantly elevated in AD versus control (FDR=4.8×10−4, Figure E4) and in psoriasis versus control (FDR=4.8×10−4, Figure E4). However, lesional psoriasis C-GS scores were also significantly elevated versus lesional AD (FDR=6.4×10−3). Taken together, this suggests that a similar mechanism of neutrophilic inflammation operates in both diseases, though factors relating to the recruitment of neutrophils may be involved to a more variable extent in AD than in Ps.
In support of our transcriptomic observations is the remarkable and important finding that numbers of neutrophils present in the dermis of AD lesions are comparable to those found in psoriasis lesions. While the presence of neutrophils in lesional skin is characteristic of psoriasis, this is not typically considered as a feature associated with AD 2, 45–47. The unexpected finding of the presence of neutrophils in AD lesions at levels comparable to those found in psoriasis lesions was prompted by our observation of commonly differentially expressed neutrophil chemoattractant genes in both AD and psoriasis.
We noted that substantial numbers of MPO-positive cells possessed morphologic characteristics of neutrophils. In addition, we definitively identified neutrophils by positive lipocalin-2 and elastase-2 staining and the presence of characteristic neutrophil morphologies in those positively stained cells. Thus, we believe that the numbers of neutrophils may be higher than is evident by H&E staining alone. To our knowledge, immunodetection of neutrophils in human AD lesions has not been previously described. However, neutrophilia in the dermis has been described in antigen-specific dermatitis driven by a Th2-polarized response in a mouse model of AD 48, although mouse models by no means duplicate the human disease.
We found a correlation between the number of neutrophils and the presence of crusts in the AD lesions. The presence of crusts as well as excoriations is characteristic, and diagnostic features of AD. They reflect epithelial disruption and, in the case of AD, are likely associated with scratching that results from itching, which is a prominent and essential clinical feature of AD. Thus, a contributing factor to neutrophilia may be tissue damage and the resultant release of neutrophil-chemotactic factors 49, as has been observed in skin wounding when neutrophils are the initial dermal infiltrating cell type 50. In fact, mRNAs for neutrophil chemoattractants, including CXCL1, CXCL2, IL8, and CSF2 (GM-CSF), were detected by our microarray analyses in AD lesions. These chemokines were also detected by our microarray in psoriasis lesions, as expected. However, the psoriasis lesions are typically devoid of encrustation, and thus the neutrophil infiltrations are likely not due to tissue damage but represent a part of the immune-mediated inflammatory response specific for this skin disease 51, 52. It is noteworthy that AD lesions are characterized by S. aureus colonization, which may be facilitated by reduced levels of β-defensin 2 as a consequence of attenuated IL-17 in the Th2 milleu 53. Although the lesions we studied did not have clinical evidence of frank infection and bacteria were not detected by H&E staining of the biopised tissues, we cannot rule out bacterial colonization as a contributing factor to tissue neutrophilia in AD and indeed, this feature of AD may provide another mechanism by which neutrophil infiltration in these lesions could occur. Specifically, scratching might introduce colonizing bacteria into the affected skin, leading to a neutrophilic response
While neutrophils were identified in the majority of cases, none exhibited zones of dermal neutrophilia, i.e., dense neutrophilic infiltrates or even areas where neutrophils predominated over lymphocytes. Thus, there were no dermal neutrophil “hot spots”. In all cases the infiltrates were confined to the papillary and upper reticular dermis, as is typical of AD and Ps, and were entirely sampled in five nonoverlapping fields. The fields that were not selected were consistently areas of the reticular dermis that contained minimal or absent inflammatory cells, as expected in AD and Ps.
We believe that our comparative analyses in AD and psoriasis provide a precise transcriptomic description in the context of primary human tissue, which may provide insights into understanding the bases of disease heterogeneity. However, there are limitations to the design and execution of this study that may potentially be subjects of further investigation. Though focusing our analyses on CCGf genes offered greater statistical power for detecting differential expression, it precludes the discovery of genes outside of our search space that may have uncharacterized relevance. The subjects in our cohorts discontinued systemic and topical anti-inflammatory therapy for at least two weeks prior to biopsy. Though this measure potentially reduces the confounding effects of treatment, it may also accentuate the magnitude of inflammatory patterns that are ameliorated by available topical therapies. Refractory AD is often associated with concomitant herpesvirus and/or staphylococcal infections 2; thus the extent of neutrophilic infiltration in AD should be evaluated with respect to infection in future studies. Future studies in AD and psoriasis patients who are refractory to topical anti-inflammatory therapies may build on the observations presented here to further pinpoint potential therapeutic targets most relevant to the residual unmet medical need in these dermatoses. Intriguingly, a subset of asthma patients refractory to inhaled corticosteroid therapy also exhibits mixed eosinophilic and neutrophilic airway inflammation 54, which may reflect a common pathogenic mechanism to steroid-refractory AD with mixed eosinophilic and neutrophilic skin inflammation. Finally, the cross-sectional nature of this study does not elucidate the mechanism of neutrophil infiltration in AD, and while a neutrophil response may not be a primary event, whether these cells contribute to the augmentation and perpetuation of the primary inflammatory response is worthy of further investigation.
Supplementary Material
Key Messages.
Lesional atopic dermatitis is enriched for transcripts encoding TH2 associated factors, which correlate with tissue and systemic eosinophilia and IgE.
Lesional psoriasis exhibits upregulated transcripts encoding TH17 associated factors and downregulated TH2-associated factors.
Expression of neutrophil chemoattractants is enriched in both atopic dermatitis and psoriasis, and the magnitude of gene expression correlates with the degree of lesional neutrophilia.
Acknowledgments
We thank Dr. Sepideh Bagheri for her excellent assistance in evaluation of the subjects and obtaining skin biopsies. We also thank Lan Yu and Diane Yamamoto for expert technical assistance.
Funding: Research was supported by NIH-NIAID grant RO1 AI20958 and NIAMS grant R01AR056343 to FTL.
Abbreviations used
- AD
Atopic Dermatitis
- AUC
Area Under the Curve
- BAL
Bronchial Alveolar Lavage
- CCGf
Cytokines, Chemokines, and Growth factors
- DE
Differential Expression or Differentially Expressed
- FDR
False Discovery Rate
- GS
Gene Set
- IHC
Immunohistochemistry
- IQR
Inter-Quartile Range
- MPO
myeloperoxidase
- PCA
Principal Component Analysis
- Ps
Psoriasis
- TH1/2/17
T-helper type 1/2/17
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bieber T. Atopic dermatitis. N Engl J Med. 2008;358:1483–94. doi: 10.1056/NEJMra074081. [DOI] [PubMed] [Google Scholar]
- 2.Guttman-Yassky E, Nograles KE, Krueger JG. Contrasting pathogenesis of atopic dermatitis and psoriasis--part I: clinical and pathologic concepts. The Journal of allergy and clinical immunology. 2011;127:1110–8. doi: 10.1016/j.jaci.2011.01.053. [DOI] [PubMed] [Google Scholar]
- 3.Schon MP, Boehncke WH. Psoriasis. N Engl J Med. 2005;352:1899–912. doi: 10.1056/NEJMra041320. [DOI] [PubMed] [Google Scholar]
- 4.Leung DY, Boguniewicz M, Howell MD, Nomura I, Hamid QA. New insights into atopic dermatitis. J Clin Invest. 2004;113:651–7. doi: 10.1172/JCI21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galli SJ, Tsai M, Piliponsky AM. The development of allergic inflammation. Nature. 2008;454:445–54. doi: 10.1038/nature07204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nomura I, Gao B, Boguniewicz M, Darst MA, Travers JB, Leung DY. Distinct patterns of gene expression in the skin lesions of atopic dermatitis and psoriasis: a gene microarray analysis. J Allergy Clin Immunol. 2003;112:1195–202. doi: 10.1016/j.jaci.2003.08.049. [DOI] [PubMed] [Google Scholar]
- 7.Antoniu SA. Pitrakinra, a dual IL-4/IL-13 antagonist for the potential treatment of asthma and eczema. Curr Opin Investig Drugs. 2010;11:1286–94. [PubMed] [Google Scholar]
- 8.Hueber W, Patel DD, Dryja T, Wright AM, Koroleva I, Bruin G, et al. Effects of AIN457, a fully human antibody to interleukin-17A, on psoriasis, rheumatoid arthritis, and uveitis. Science translational medicine. 2010;2:52ra72. doi: 10.1126/scitranslmed.3001107. [DOI] [PubMed] [Google Scholar]
- 9.Leonardi C, Matheson R, Zachariae C, Cameron G, Li L, Edson-Heredia E, et al. Anti-interleukin-17 monoclonal antibody ixekizumab in chronic plaque psoriasis. N Engl J Med. 2012;366:1190–9. doi: 10.1056/NEJMoa1109997. [DOI] [PubMed] [Google Scholar]
- 10.Leonardi CL, Kimball AB, Papp KA, Yeilding N, Guzzo C, Wang Y, et al. Efficacy and safety of ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with psoriasis: 76-week results from a randomised, double-blind, placebo-controlled trial (PHOENIX 1) Lancet. 2008;371:1665–74. doi: 10.1016/S0140-6736(08)60725-4. [DOI] [PubMed] [Google Scholar]
- 11.Papp KA, Leonardi C, Menter A, Ortonne JP, Krueger JG, Kricorian G, et al. Brodalumab, an anti-interleukin-17-receptor antibody for psoriasis. N Engl J Med. 2012;366:1181–9. doi: 10.1056/NEJMoa1109017. [DOI] [PubMed] [Google Scholar]
- 12.Noble WS. How does multiple testing correction work? Nat Biotechnol. 2009;27:1135–7. doi: 10.1038/nbt1209-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9546–51. doi: 10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopf M, Bachmann MF, Marsland BJ. Averting inflammation by targeting the cytokine environment. Nature Reviews Drug Discovery. 2010;9:703–18. doi: 10.1038/nrd2805. [DOI] [PubMed] [Google Scholar]
- 15.Choy DF, Modrek B, Abbas AR, Kummerfeld S, Clark HF, Wu LC, et al. Gene expression patterns of Th2 inflammation and intercellular communication in asthmatic airways. Journal of Immunology. 2011;186:1861–9. doi: 10.4049/jimmunol.1002568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanifin JM, Rajka G. Diagnostic features of atopic dermatitis. Acta Derm Venereol Suppl (Stockh) 1980;10:44–7. [Google Scholar]
- 17.Eichenfield LF, Hanifin JM, Luger TA, Stevens SR, Pride HB. Consensus conference on pediatric atopic dermatitis. J Am Acad Dermatol. 2003;49:1088–95. doi: 10.1016/s0190-9622(03)02539-8. [DOI] [PubMed] [Google Scholar]
- 18.Williams HC. Clinical practice. Atopic dermatitis. N Engl J Med. 2005;352:2314–24. doi: 10.1056/NEJMcp042803. [DOI] [PubMed] [Google Scholar]
- 19.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 20.Saegusa J, Hsu DK, Chen HY, Yu L, Fermin A, Fung MA, et al. Galectin-3 is critical for the development of the allergic inflammatory response in a mouse model of atopic dermatitis. Am J Pathol. 2009;174:922–31. doi: 10.2353/ajpath.2009.080500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu YJ. Thymic stromal lymphopoietin and OX40 ligand pathway in the initiation of dendritic cell-mediated allergic inflammation. J Allergy Clin Immunol. 2007;120:238–44. doi: 10.1016/j.jaci.2007.06.004. quiz 45–6. [DOI] [PubMed] [Google Scholar]
- 22.Holgate ST, Polosa R. Treatment strategies for allergy and asthma. Nat Rev Immunol. 2008;8:218–30. doi: 10.1038/nri2262. [DOI] [PubMed] [Google Scholar]
- 23.Wang YH, Ito T, Homey B, Watanabe N, Martin R, Barnes CJ, et al. Maintenance and polarization of human TH2 central memory T cells by thymic stromal lymphopoietin-activated dendritic cells. Immunity. 2006;24:827–38. doi: 10.1016/j.immuni.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 24.Seshasayee D, Lee WP, Zhou M, Shu J, Suto E, Zhang J, et al. In vivo blockade of OX40 ligand inhibits thymic stromal lymphopoietin driven atopic inflammation. J Clin Invest. 2007;117:3868–78. doi: 10.1172/JCI33559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2:E108. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miossec P, Korn T, Kuchroo VK. Interleukin-17 and type 17 helper T cells. N Engl J Med. 2009;361:888–98. doi: 10.1056/NEJMra0707449. [DOI] [PubMed] [Google Scholar]
- 27.Chiricozzi A, Guttman-Yassky E, Suarez-Farinas M, Nograles KE, Tian S, Cardinale I, et al. Integrative responses to IL-17 and TNF-alpha in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. The Journal of investigative dermatology. 2011;131:677–87. doi: 10.1038/jid.2010.340. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama T, Kato Y, Hieshima K, Nagakubo D, Kunori Y, Fujisawa T, et al. Liver-expressed chemokine/CC chemokine ligand 16 attracts eosinophils by interacting with histamine H4 receptor. J Immunol. 2004;173:2078–83. doi: 10.4049/jimmunol.173.3.2078. [DOI] [PubMed] [Google Scholar]
- 29.Syed F, Huang CC, Li K, Liu V, Shang T, Amegadzie BY, et al. Identification of interleukin-13 related biomarkers using peripheral blood mononuclear cells. Biomarkers. 2007;12:414–23. doi: 10.1080/13547500701192652. [DOI] [PubMed] [Google Scholar]
- 30.Rothenberg ME. Eotaxin. An essential mediator of eosinophil trafficking into mucosal tissues. Am J Respir Cell Mol Biol. 1999;21:291–5. doi: 10.1165/ajrcmb.21.3.f160. [DOI] [PubMed] [Google Scholar]
- 31.Colobran R, Pujol-Borrell R, Armengol MP, Juan M. The chemokine network. I. How the genomic organization of chemokines contains clues for deciphering their functional complexity. Clin Exp Immunol. 2007;148:208–17. doi: 10.1111/j.1365-2249.2007.03344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Commins SP, Borish L, Steinke JW. Immunologic messenger molecules: cytokines, interferons, and chemokines. J Allergy Clin Immunol. 2010;125:S53–72. doi: 10.1016/j.jaci.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Nichols BA, Bainton DF. Differentiation of human monocytes in bone marrow and blood. Sequential formation of two granule populations. Lab Invest. 1973;29:27–40. [PubMed] [Google Scholar]
- 34.Pulford KA, Erber WN, Crick JA, Olsson I, Micklem KJ, Gatter KC, et al. Use of monoclonal antibody against human neutrophil elastase in normal and leukaemic myeloid cells. J Clin Pathol. 1988;41:853–60. doi: 10.1136/jcp.41.8.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seveus L, Amin K, Peterson CG, Roomans GM, Venge P. Human neutrophil lipocalin (HNL) is a specific granule constituent of the neutrophil granulocyte. Studies in bronchial and lung parenchymal tissue and peripheral blood cells. Histochem Cell Biol. 1997;107:423–32. doi: 10.1007/s004180050129. [DOI] [PubMed] [Google Scholar]
- 36.Chien YH, Hwu WL, Chiang BL. The genetics of atopic dermatitis. Clin Rev Allergy Immunol. 2007;33:178–90. doi: 10.1007/s12016-007-0041-8. [DOI] [PubMed] [Google Scholar]
- 37.Elder JT. Genome-wide association scan yields new insights into the immunopathogenesis of psoriasis. Genes Immun. 2009;10:201–9. doi: 10.1038/gene.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghoreschi K, Thomas P, Breit S, Dugas M, Mailhammer R, van Eden W, et al. Interleukin-4 therapy of psoriasis induces Th2 responses and improves human autoimmune disease. Nat Med. 2003;9:40–6. doi: 10.1038/nm804. [DOI] [PubMed] [Google Scholar]
- 39.Komiya A, Nagase H, Yamada H, Sekiya T, Yamaguchi M, Sano Y, et al. Concerted expression of eotaxin-1, eotaxin-2, and eotaxin-3 in human bronchial epithelial cells. Cell Immunol. 2003;225:91–100. doi: 10.1016/j.cellimm.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–47. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu FT, Goodarzi H, Chen HY. Clinical reviews in allergy & immunology. 2011. IgE, Mast Cells, and Eosinophils in Atopic Dermatitis. [DOI] [PubMed] [Google Scholar]
- 42.Gould HJ, Sutton BJ. IgE in allergy and asthma today. Nat Rev Immunol. 2008;8:205–17. doi: 10.1038/nri2273. [DOI] [PubMed] [Google Scholar]
- 43.Oldhoff JM, Darsow U, Werfel T, Katzer K, Wulf A, Laifaoui J, et al. Anti-IL-5 recombinant humanized monoclonal antibody (mepolizumab) for the treatment of atopic dermatitis. Allergy. 2005;60:693–6. doi: 10.1111/j.1398-9995.2005.00791.x. [DOI] [PubMed] [Google Scholar]
- 44.Scheerens H, Arron JR, Su Z, Zheng Y, Putnam W, Erickson RW, et al. Predictive and Pharmacodynamic Biomarkers of Interleukin-13 Blockade: Effect of Lebrikizumab on Late Phase Asthmatic Response to Allergen Challenge. The Journal of allergy and clinical immunology. 2011;127:AB164. (Abstr.) [Google Scholar]
- 45.Abramovits W. Atopic dermatitis. J Am Acad Dermatol. 2005;53:S86–93. doi: 10.1016/j.jaad.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 46.Novak N, Bieber T, Leung DY. Immune mechanisms leading to atopic dermatitis. J Allergy Clin Immunol. 2003;112:S128–39. doi: 10.1016/j.jaci.2003.09.032. [DOI] [PubMed] [Google Scholar]
- 47.Simpson EL, Hanifin JM. Atopic dermatitis. J Am Acad Dermatol. 2005;53:115–28. doi: 10.1016/j.jaad.2005.01.130. [DOI] [PubMed] [Google Scholar]
- 48.Spergel JM, Mizoguchi E, Brewer JP, Martin TR, Bhan AK, Geha RS. Epicutaneous sensitization with protein antigen induces localized allergic dermatitis and hyperresponsiveness to methacholine after single exposure to aerosolized antigen in mice. J Clin Invest. 1998;101:1614–22. doi: 10.1172/JCI1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rose FR, Hirschhorn R, Weissmann G, Cronstein BN. Adenosine promotes neutrophil chemotaxis. J Exp Med. 1988;167:1186–94. doi: 10.1084/jem.167.3.1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baum CL, Arpey CJ. Normal cutaneous wound healing: clinical correlation with cellular and molecular events. Dermatol Surg. 2005;31:674–86. doi: 10.1111/j.1524-4725.2005.31612. discussion 86. [DOI] [PubMed] [Google Scholar]
- 51.Gillitzer R, Ritter U, Spandau U, Goebeler M, Brocker EB. Differential expression of GRO-alpha and IL-8 mRNA in psoriasis: a model for neutrophil migration and accumulation in vivo. J Invest Dermatol. 1996;107:778–82. doi: 10.1111/1523-1747.ep12371803. [DOI] [PubMed] [Google Scholar]
- 52.Pietrzak AT, Zalewska A, Chodorowska G, Krasowska D, Michalak-Stoma A, Nockowski P, et al. Cytokines and anticytokines in psoriasis. Clin Chim Acta. 2008;394:7–21. doi: 10.1016/j.cca.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 53.Eyerich K, Pennino D, Scarponi C, Foerster S, Nasorri F, Behrendt H, et al. IL-17 in atopic eczema: linking allergen-specific adaptive and microbial-triggered innate immune response. J Allergy Clin Immunol. 2009;123:59–66. e4. doi: 10.1016/j.jaci.2008.10.031. [DOI] [PubMed] [Google Scholar]
- 54.Fahy JV. Eosinophilic and neutrophilic inflammation in asthma: insights from clinical studies. Proc Am Thorac Soc. 2009;6:256–9. doi: 10.1513/pats.200808-087RM. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.