Abstract
Vacuolar proton-translocating inorganic pyrophosphatase and H+-ATPase acidify the vacuoles and power the vacuolar secondary active transport systems in plants. Developmental changes in the transcription of the pyrophosphatase in growing hypocotyls of mung bean (Vigna radiata) were investigated. The cDNA clone for the mung bean enzyme contains an uninterrupted open reading frame of 2298 bp, coding for a polypeptide of 766 amino acids. Hypocotyls were divided into elongating and mature regions. RNA analysis revealed that the transcript level of the pyrophosphatase was high in the elongating region of the 3-d-old hypocotyl but was extremely low in the mature region of the 5-d-old hypocotyl. The level of transcript of the 68-kD subunit of H+-ATPase also decreased after cell maturation. In the elongating region, the proton-pumping activity of pyrophosphatase on the basis of membrane protein was 3 times higher than that of H+-ATPase. After cell maturation, the pyrophosphatase activity decreased to 30% of that in the elongating region. The decline in the pyrophosphatase activity was in parallel with a decrease in the enzyme protein content. These findings indicate that the level of the pyrophosphatase, a main vacuolar proton pump in growing cells, is negatively regulated after cell maturation at the transcriptional level.
The vacuole of higher plants is an enormous, acidic organelle that occupies a large part of the cell. Typical plant vacuoles are 50 to 100 μm in diameter. The acidic condition in the vacuole is maintained by the two distinct proton pumps, H+-ATPase and H+-PPase. The H+ gradient generated by the proton pumps powers the secondary active transporters of inorganic ions, sugars, and organic acids (Hedrich and Schroeder, 1989; Taiz, 1992).
The vacuolar-type H+-PPase attracts considerable attention from two viewpoints. The H+-PPase enzyme molecule is a great proton-pump model with which to study molecularly how the hydrolysis of phosphate ester is coupled with proton translocation across the membrane, because the enzyme consists of a single polypeptide of about 73 kD (Maeshima and Yoshida, 1989; Kim et al., 1994a), and its substrate is also a simple compound (PPi). Therefore, the biochemical properties of H+-PPase and the detailed mechanism of the enzyme reaction have been extensively analyzed (Baykov et al., 1993; Rea and Poole, 1993). Additionally, the cDNAs for H+-PPase have been cloned from several species of higher plants (Sarafian et al., 1992; Tanaka et al., 1993; Kim et al., 1994b; Lerchl et al., 1995; Sakakibara et al., 1996).
From the physiological aspect, an important question is why H+-PPase coexists with V-ATPase in the same vacuolar membrane. In other words, why does the H+-PPase exist in the vacuolar membranes of plants but not in yeast vacuoles or animal acidic compartments? The vacuolar membranes prepared from all of the plant species, including mosses, ferns, and algae, show H+-PPase activity in addition to V-ATPase activity (Takeshige et al., 1988; Ikeda et al., 1991; Maeshima et al., 1994). The existence of H+-PPase in plant cells may be related to the huge size of the plant vacuole. Quantitative analysis of the levels of enzymatic activity, enzyme protein, and the transcript of the H+-PPase under stress conditions may provide important information for understanding the physiological function of H+-PPase. Carystinos et al. (1995) reported that the relative transcript and enzyme activity of H+-PPase increased notably under anoxia and chilling in rice seedlings. They proposed that H+-PPase may replace V-ATPase under energy stress. An increase in the H+-PPase activity was also observed in mung bean (Vigna radiata) hypocotyls under low-temperature stress (Darley et al., 1995; Davies, 1997).
We examined the developmental change of H+-PPase in hypocotyls under normal conditions. The hypocotyl of dicotyledonous seedlings is widely used as a model for the study of cell elongation and its control mechanisms (Gen-dreau et al., 1997). Elongation of hypocotyl cells is accompanied by quick expansion of central vacuoles in their cells, and there may be a comparatively modest increase in the actual amount of cytoplasm during cell elongation. The expanding vacuoles in elongating cells must incorporate solutes and water. Therefore, the interior acidic condition of the expanding vacuoles must be maintained to support the secondary active transport systems. We are conducting a series of experiments to determine whether the vacuolar proton pumps are essential for quick expansion of vacuoles.
Previously we reported that, even in the young cells of the mung bean hypocotyl, the relative H+-PPase level on the basis of membrane protein content was the same as that of the mature cell (Maeshima, 1990). In this study we focused our attention on the developmental expression of the H+-PPase gene during expansion of vacuoles in elongating hypocotyls of mung bean. RNA-blot analysis revealed the down-regulation of H+-PPase gene expression in hypocotyl development, supporting the previous observations. Here we discuss the primary structure of H+-PPase and the physiological meanings of the regulation of H+-PPase level in growing plant cells.
MATERIALS AND METHODS
Seeds of mung bean (Vigna radiata cv Wilczek) were soaked in 1 mm CaSO4 and then germinated at 26°C in the dark. Hypocotyls from 3- and 5-d-old seedlings were cut into 1- and 1.5-cm segments, respectively. The segments were numbered from the hypocotyl apex, as shown in Figure 2A (3-d-old hypocotyl, segment nos. 31–36; 5-d-old hypocotyl, segment nos. 51–59). To obtain leaves, mung beans were grown in garden soil for 3 weeks under long-day conditions (16/8 h, 28/20°C, light/dark regime). The first, second, and third leaf pairs were harvested from the 3-week-old plants.
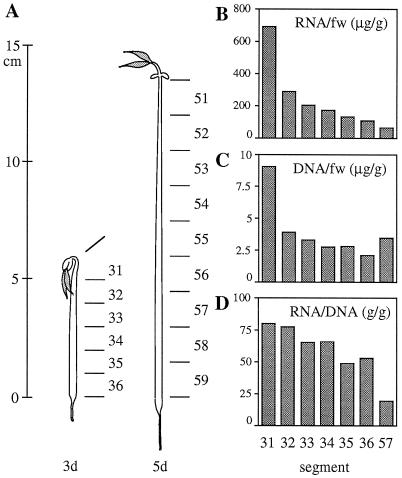
Figure 2.
Amounts of RNA and DNA in segments of etiolated mung bean hypocotyls. A, Mung bean seedlings germinated for 3 and 5 d. Three- and 5-d-old hypocotyls were cut into 1- and 1.5-cm segments, respectively. B and C, Amounts of total RNA and total DNA, calculated on the basis of fresh weight (fw) of segment. D, Relative content of RNA on the basis of DNA content.
cDNA Construction and Screening
Total RNA was isolated from the hook portion of 3-d-old hypocotyls for construction of a cDNA library. The hypocotyl segments were immediately frozen in liquid nitrogen, and then RNA was extracted by the phenol/SDS extraction method. RNA and DNA were precipitated by cold ethanol and resuspended in TE buffer (10 mm Tris-HCl, pH 8.0, and 1 mm EDTA). Then RNA was precipitated by 4 m LiCl (total RNA fraction). Poly(A+) RNA was purified with oligo(dT)-latex. A cDNA library was synthesized from 5 μg of the poly(A+) RNA, ligated into phage vector (Uni-ZAP XR, Stratagene), and then packaged with a packaging extract (Gigapack II Gold, Stratagene). Recombinant phages from the library were blotted onto nylon membranes and then screened with a 32P-labeled DNA probe according to standard hybridization protocol (Sambrook et al., 1989). Positive clones were excised in vivo into the pBluescript SK(−) plasmid (Stratagene).
DNA Preparation
DNA was isolated from hypocotyl segments by the same method as RNA extraction. After RNA was removed by LiCl precipitation, the remaining supernatant was used as the DNA fraction. Samples and standard DNA were diluted to 0.5 mL with TE buffer and 10 ng/mL 4′,6-diamidino-2-phenylindole dihydrochloride. Fluorescence of the DNA solution was measured with a fluorescence spectrophotometer set at 348 nm for excitation and 450 nm for emission, and then DNA content was calculated.
Preparation of DNA Probes
After treatment of mRNA with DNase, cDNAs were synthesized from the mRNAs with reverse transcriptase using oligo(dT)-16 primer. cDNA templates were amplified by PCR using pfu DNA polymerase and gene-specific primers (forward, 5′-ACTGGTTATGGTCTTGGTGGGT-3′; reverse, 5′-GGCAACATCTTGCACAGGGCTGT-3′). The primers correspond to the consensus nucleotide sequences of H+-PPase cDNA of barley (Tanaka et al., 1993) and Arabidopsis thaliana (Sarafian et al., 1992). The amplification protocol was 5 min at 95°C (once), 1 min at 95°C, 1 min at 55°C, 2 min at 72°C (40 cycles), and 7 min at 72°C (once). Amplified DNA fragments (630 bp) were purified and ligated into the HincII site of the pUC119 plasmid vector for transformation of Escherichia coli MV1184. The insert DNA was confirmed to be a cDNA fragment of H+-PPase by DNA sequencing.
A DNA probe for subunit A of V-ATPase was prepared by PCR using specific primers (forward, 5′-TCCTGATGCCATGGGAAAGAT-3′; reverse, 5′-CGCATCATCCAAACAGACTTGT-3′). Both primers correspond to the consensus nucleotide sequences of V-ATPase cDNA of carrot (Daucus carota, Zimniak et al., 1988) and cotton (Gossypium hirsutum, Wilkins et al., 1993). The cDNA fragment obtained showed 83 to 84% homology with the cDNAs for the V-ATPase subunit A from carrot and cotton. The DNA probes were radiolabeled with α-[32P]dCTP by the random-priming method. For northern analysis, a fragment of H+-PPase cDNA digested by restriction enzyme NcoI and a PCR product of DNA for the V-ATPase subunit A were used as DNA probes.
DNA Sequencing and Analysis
The DNA sequence was determined from single-strand plasmid DNAs by the dideoxy chain-termination method using T7 polymerase. Templates were prepared as single-strand DNA from the pBluescript SK(−) plasmid with helper phage M13K07. The DNA sequence was analyzed using a DNA sequencer (A.L.F., Pharmacia). Sequences were analyzed using the DNASIS program of Hitachi Software Engineering (Tokyo).
RNA Analysis
Total RNA was isolated from hypocotyl segments and leaves as described above. RNAs were electrophoresed in a 1.0% agarose gel, capillary transferred to a nylon membrane, and fixed to the membrane by UV irradiation. A nylon membrane was prehybridized in a hybridization buffer containing 0.02% denatured salmon sperm DNA at 65°C for 1 h and was then hybridized with 106 cpm/mL 32P-labeled denatured probe DNA at 65°C overnight. The blot membrane was washed twice in 2× SSC (0.3 m NaCl and 30 mm sodium citrate) containing 0.1% SDS at room temperature for 5 min and in 0.1× SSC and 0.1% SDS at 55°C for 1 h. The membrane was exposed to radiographic film or an imaging plate. The radioimage of the plate was analyzed (BAS2000, Fuji, Tokyo).
Vacuolar Membrane Preparation, Immunoblotting, and Enzyme Assay
Vacuolar membranes were prepared from hypocotyl segments as described previously (Maeshima and Yoshida, 1989). Antibodies against the putative substrate-binding site of mung bean H+-PPase (Takasu et al., 1997) and subunit A of the mung bean V-ATPase (Matsuura-Endo et al., 1992) were prepared as described previously. For immunoblotting, proteins were separated by SDS-PAGE (10% gel) and transferred to a PVDF membrane by standard procedures. Antibodies bound to the antigen were detected with horseradish peroxidase-coupled protein A and chemiluminescent reagents (Amersham). Levels of the antigens on immunoblots were quantified with an image analyzer (Bio-Rad). Activities of substrate hydrolysis and proton transport of H+-PPase and H+-ATPase were measured as described previously (Maeshima and Yoshida, 1989; Matsuura-Endo et al., 1990). Protein content was determined by the method of Bradford (1976).
RESULTS
Molecular Cloning of H+-PPase cDNA of Mung Bean
To analyze the expression of a gene for vacuolar H+-PPase during elongation of hypocotyl, we constructed a cDNA library for mRNAs in the hypocotyl. Of 120,000 recombinant phages, DNAs of 190 were hybridized with a radiolabeled H+-PPase cDNA probe, and the inserted DNAs of 20 positive clones were then sequenced. Ten of the clones encoded the same sequence of H+-PPase, and the remaining clones did not. The longest of the H+-PPase clones was denoted VVP2, and both of its strands were fully sequenced.
Figure 1 shows the nucleotide sequence and the deduced amino acid sequence of the clone VVP2. The cDNA consists of 2531 bp upstream of the polyadenylate tail, which includes a 13-bp 5′ leader sequence, followed by 2298 bp of an open reading frame encoding 766 amino acids, and, finally, a 220-bp 3′-noncoding region. The polyadenylate addition site lies 38 bp downstream from the AAGAAA polyadenylation signal. The presumed translation start site is the first ATG encountered downstream from the 5′ end. The deduced amino acid sequence of the N-terminal part showed agreement with the direct sequence of the purified H+-PPase, although the purified enzyme lacks the N-terminal Met residue (Maeshima and Yoshida, 1989). There is no cleaved signal peptide in the deduced H+-PPase sequence. From gel-blot analysis of the total RNA fraction of hypocotyls, the H+-PPase mRNA was estimated to be 2.7 kb (Fig. 3), and this size corresponds to that of the insert DNA. Hung et al. (1995) registered the cDNA sequence of mung bean H+-PPase without the triplet of GGT encoding the Gly-688 found in the present study. There is a possibility that the difference is derived from plant materials. The sequence of a part including Gly-688 in the present study was identical to that of the other H+-PPases from many plant species.
Figure 1.
Nucleotide sequence and deduced amino acid sequence of H+-PPase cDNA of mung bean (clone VVP2). The underlined amino acid residues were identical to those of the direct sequence obtained from H+-PPase purified from mung bean. Thirteen putative membrane-spanning domains predicted from the hydropathy profile are boxed. The termination codon is marked with an asterisk.
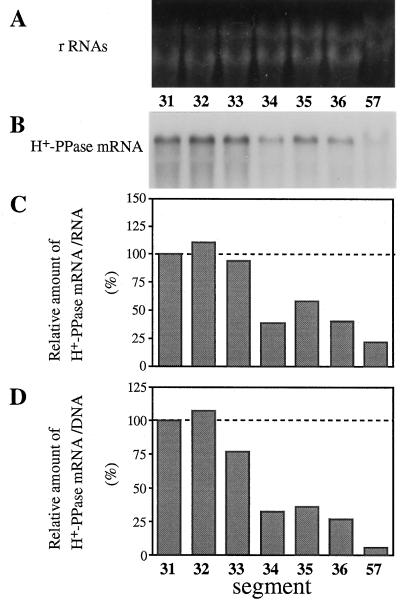
Figure 3.
Developmental change in the content of mRNA for H+-PPase in hypocotyls. Hypocotyls from more than 100 seedlings germinated for 3 or 5 d were cut into segments and RNA fractions were prepared. Aliquots (4.5 μg) of the total RNA were electrophoresed and blotted onto a nylon membrane. The membrane was subjected to northern analysis with a 32P-labeled cDNA fragment of H+-PPase, and the radioactivity was measured as described in Methods. A, Ethidium bromide staining of rRNAs (control). B, Northern blot with a DNA probe for H+-PPase. C and D, Relative amounts of H+-PPase mRNA on the basis of RNA or DNA content, respectively. Values shown are relative to that of segment no. 31.
Structural Characteristics of the H+-PPase Polypeptide
The calculated molecular mass of the derived polypeptide was 80,002 D, which was larger than the value of 73 kD for the purified H+-PPase on SDS-PAGE (Maeshima and Yoshida, 1989; Kim et al., 1994b). Rapid migration of the enzyme on the SDS gel may be due to the high hydrophobicity of the H+-PPase. Hydrophobic amino acids (481 residues) account for about 63% of the total 766 residues. In the sequence of mung bean H+-PPase, there are 13 hydrophobic domains of sufficient length and hydrophobicity to span a lipid bilayer (Fig. 1). The deduced sequence of mung bean H+-PPase has high identity to those of H+-PPases of other plant species: A. thaliana (87.5%; Sarafian et al., 1992), barley (88.6%; Tanaka et al., 1993), red beet (two isoforms, 89.2 and 89.3%; Kim et al., 1994b), tobacco (four isoforms, 88.6–91.5%; Lerchl et al., 1995), and rice (two isoforms; 88.1 and 86.9%; Sakakibara et al., 1996). The mung bean enzyme has a putative substrate-binding site (residues 253–263), as was also described for Arabidopsis H+-PPase by Rea and Poole (1993).
Decrease in mRNAs for H+-PPase and H+-ATPase Subunit A during Elongation of Hypocotyl
Mung bean hypocotyls can be separated into a hook part (the elongating region) and a lower part under the hook (the mature region). To compare the mRNA level of vacuolar proton pumps among hypocotyl segments, 1- or 1.5-cm segments were prepared from seedlings germinated for 3 and 5 d (Fig. 2A). The top segment, no. 31, consists of small, young cells; the lower segments, nos. 35 and 36, consist of large, mature cells (Maeshima, 1990). Rapid elongation took place at segment no. 31, in which the elongation rate was about 400%/d in 3-d-old seedlings. The lower part did not elongate so rapidly; the rate at segment no. 32 was about 30%. The amount of total RNA per gram fresh weight was highest in the elongating region (segment no. 31). Figure 2C shows the total amount of DNA in each segment on the basis of fresh weight. The level of DNA was highest in segment no. 31, which has a small cell volume. To compare the relative amounts of RNA in cells, the ratio of RNA to DNA was calculated. The RNA to DNA ratio decreased during hypocotyl elongation, as shown in Figure 2D.
The levels of H+-PPase mRNA during hypocotyl elongation were determined by northern analysis (Fig. 3). Figure 3, C and D, shows the H+-PPase mRNA levels on the basis of the amount of RNA and DNA, respectively. In both cases, the H+-PPase mRNA content was the highest in the elongating region (segment nos. 31 and 32) and lowest in the mature region (segment nos. 34 and 35). The level of H+-PPase mRNA per DNA content in segment no. 57 of 5-d-old hypocotyl was 5% of that in the elongating region.
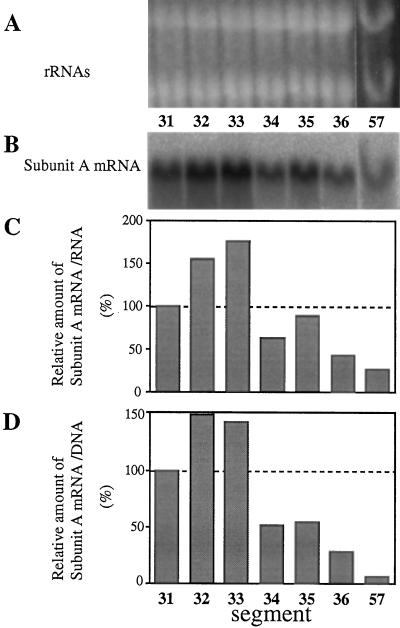
To determine the levels of mRNA for the V-ATPase subunit A, which is a nucleotide-binding subunit, its cDNA was prepared and used as a probe. In contrast to the H+-PPase mRNA, the level of subunit-A mRNA was highest in the middle part (segment nos. 32 and 33) of 3-d-old hypocotyls (Fig. 4). These findings suggest that the expression of genes for H+-PPase and the V-ATPase subunit A are regulated independently. Expression in the mature regions (segment nos. 34–36 and no. 57) was very low, as was the H+-PPase mRNA.
Figure 4.
Level of mRNA for the subunit A of V-ATPase in hypocotyl segments. Aliquots (20 μg) of the total RNA fractions from the hypocotyl segments were subjected to northern analysis with a 32P-labeled cDNA fragment of the V-ATPase subunit A. A, Ethidium bromide staining of rRNAs. B, Northern analysis. C and D, Relative amount of mRNA for the V-ATPase subunit A on the basis of RNA or DNA content, respectively. Values shown are relative to that of segment no. 31.
Change in Levels of mRNAs for H+-PPase and V-ATPase Subunit A in Leaves
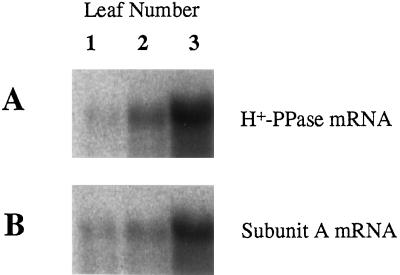
RNA was extracted from leaves of different ages to determine the levels of mRNA for the two vacuolar proton pumps. As shown in Figure 5, the levels of mRNAs for H+-PPase and V-ATPase subunit A were highest in the immature third leaf and decreased during leaf growth. The relative levels of the mRNAs for both proteins in the first, old leaf were less than 5% of that of the third, young leaf.
Figure 5.
Levels of mRNAs for H+-PPase and subunit A of V-ATPase in mung bean leaves. Total RNA fractions were prepared from eight pairs of the first, second, and third leaves of 3-week-old mung bean plants. RNAs (20 μg) were subjected to northern analysis with a radiolabeled DNA probe for H+-PPase (A) or the subunit A of V-ATPase (B).
Levels of H+-PPase and H+-ATPase in Vacuolar Membranes of Hypocotyl
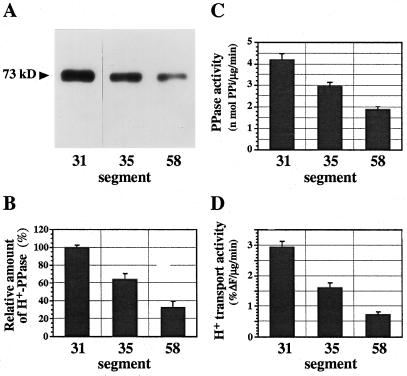
We determined the levels of H+-PPase and H+-ATPase in the vacuolar membranes of the hypocotyl segments. Immunoblot analysis with the antibody against mung bean H+-PPase revealed that the amount of H+-PPase molecules on the basis of membrane protein was highest in segment no. 31 and decreased during cell elongation (Fig. 6, A and B). As presumed from quantification of mRNA, the amount of H+-PPase protein in the mature part of the hypocotyl (segment no. 58) was less than one-third of that in the elongating region (segment no. 31). The activities of PPi hydrolysis and PPi-dependent H+ transport in the membrane vesicles on the basis of membrane protein decreased during cell elongation (Fig. 6, C and D). This also indicates that there was no inactivation or suppression of H+-PPase activity in the membranes during cell elongation. The data are the relative levels of the enzyme on the basis of the membrane protein and reflect the relative density of the enzyme in a unit area of vacuolar membrane. Since the amount of vacuolar membrane in each cell increases as cells grow, the total amount of H+-PPase in the cell increased during cell growth, although the relative content of the enzyme on the basis of membrane protein decreased markedly.
Figure 6.
Level of H+-PPase in vacuolar membranes of hypocotyl segments. Vacuolar membranes were prepared from more than 200 pieces of segment (segment nos. 31, 35, and 58). Aliquots (1 μg) of vacuolar membranes were subjected to SDS-PAGE and immunobloted with antibody to mung bean H+-PPase (73 kD). A, Immunoblot. B, Amounts of H+-PPase protein relative to segment no. 31. C and D, PPi-hydrolysis activity (C) and H+-transport activity (D) of H+-PPase in the vacuolar membranes. The data shown are means ± sd for two experiments, each with triplicate assays.
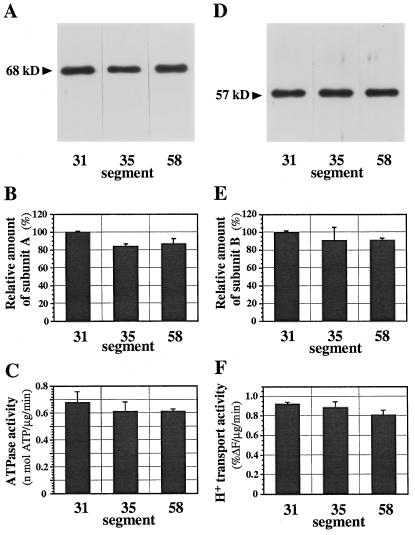
In contrast to H+-PPase, the relative content of V-ATPase subunit-A protein did not decrease in the mature part of hypocotyls (Figs. 7, A and B); neither did that of subunit B (Fig. 7, D and E). The activities of ATP hydrolysis and ATP-dependent H+ transport of the membranes were constant during growth (Fig. 7, C and F). Consequently, the V-ATPase activity in 5-d-old hypocotyls was slightly higher than the H+-PPase activity, although the H+-PPase activity in 3-d-old hypocotyls was several times greater than the ATPase activity. The present data suggest that the distribution density of V-ATPase on vacuolar membranes did not change during development of the vacuole.
Figure 7.
Levels of V-ATPase in vacuolar membranes of hypocotyl segments. Aliquots (1 μg) of vacuolar membranes of each segment were subjected to SDS-PAGE and immunoblotting with antibodies to subunits A (68 kD, A) and B (57 kD, D) of V-ATPase. B and E, Amounts of subunit A (B) and B (E) of V-ATPase relative to that in segment no. 31. C and F, ATP-hydrolysis activity (C) and H+-transport activity (F) of V-ATPase in the vacuolar membranes. The data shown are means ± sd for two experiments, each with triplicate assays.
DISCUSSION
We cloned the cDNA of the vacuolar membrane H+-PPase of mung bean and examined its expression in elongating hypocotyls. Many positive clones for H+-PPase obtained from a cDNA library of hypocotyl have the same sequence. Genomic Southern analysis suggested the presence of two copies of H+-PPase genes, and we sequenced about 1500 bp of the two genes. The nucleotide sequences of their protein-coding regions were identical to each other (data not shown). Furthermore, immunoblot analysis of the vacuolar membranes separated by two-dimensional PAGE showed only a single spot with the antibody to H+-PPase (Takasu et al., 1997). Therefore, we concluded that there may be only one species of mRNA for H+-PPase and no isoform of the enzyme in mung bean hypocotyl.
The cDNA VVP2 encodes a polypeptide of 80,002 with 766 amino acids. We compared it with several cDNA clones of H+-PPase among plant species. The deduced polypeptides consisted of 761 to 771 amino acids with a pI of approximately 5.0. Their molecular masses ranged from 79,841 to 80,800 D. A detailed amino acid comparison between the mung bean H+-PPase polypeptide and the published sequences from A. thaliana (Sarafian et al., 1992), Hordeum vulgare (Tanaka et al., 1993), Beta vulgaris (Kim et al., 1994b), Nicotiana tabacum (Lerchl et al., 1995), and Oryza sativa (Sakakibara et al., 1996) revealed that the polypeptides are well conserved, with 86 to 91% identity. The least conserved region is the amino-terminal part (the first 56 residues), with 29% homology among 11 H+-PPases. The carboxyl-terminal half from the 405th residue showed a high homology of 90%. The vacuolar H+-PPase is one of the most highly conserved polypeptides among higher plants.
Mung bean H+-PPase shows a higher similarity to H+-PPases of other plant species. The vacuolar H+-PPase is a common enzyme among green plants, including nonvascular and vascular plants, Chara corallina, and Acetabularia acetabulum (Ikeda et al., 1991; Maeshima et al., 1994). In addition to green plants, the H+-PPase was purified from a photosynthetic bacterium Rhodospirillum rubrum (Nyrén et al., 1991), and it has been shown to react with antibodies against mung bean H+-PPase (Nore et al., 1991). Molecular cloning of the H+-PPases of other organisms, such as mosses, ferns, C. corallina, A. acetabulum, and R. rubrum, may provide information to help gain an understanding of the molecular evolution of H+-PPase.
A comparison between vacuolar H+-PPase and soluble PPases suggested that the configuration (E/D)(X)7KXE is a putative catalytic site of the PPases (Rea et al., 1992; Kim et al., 1994b). Indeed, this motif, which is at position 253 of mung bean H+-PPase, is conserved among 11 H+-PPases from six plant species. The polyclonal antibodies specific to a peptide of DVGADLVGKVE, which corresponds to the motif, markedly inhibited the activities of PPi hydrolysis and the PPi-dependent H+ transport (Takasu et al., 1997). These findings support the hypothesis that the motif (E/D)(X)7KXE is involved in substrate binding and/or substrate hydrolysis at the cytosolic surface of H+-PPases.
The vacuolar H+-PPase is sensitive to the sulfhydryl reagent NEM. Free Mg2+ or substrate (Mg2+ plus PPi) protects the reversible inhibition by NEM (Zhen et al., 1994; Kim et al., 1995; Gordon-Weeks et al., 1996). Kim et al. (1995) demonstrated that substrate-protectable NEM inhibition is due to NEM binding to Cys-634, which is located on a cytoplasmic loop between membrane-spanning domains X and XI of Arabidopsis H+-PPase. Recently, Gordon-Weeks et al. (1996) investigated the Mg2+-protectable sensitivity of mung bean H+-PPase to NEM and proposed that Glu and Asp residues in the loop containing Cys-634 are involved in Mg2+ binding to the enzyme. The sequence of mung bean enzyme shows that Glu and Asp residues are conserved in the loop containing Cys-630 of mung bean enzyme. The hypothesis must be tested by site-directed mutagenesis and direct binding experiments.
The aim of this study was to examine whether the vacuolar H+-PPase gene is expressed constantly in the process of vacuole development. The hypocotyl of mung bean seedlings is a good system for the study of vacuole development and cell elongation. The amount of the vacuolar membranes, which reflects the surface area of vacuoles, increased at least four times during cell elongation in mung bean hypocotyls (Maeshima, 1996). High expression of the H+-PPase gene and its active biosynthesis are believed to be essential to quick expansion of the vacuole in growing cells (Maeshima et al., 1996). Measurements of enzymatic activities and the immunochemical quantification of the two proton pumps clearly showed that H+-PPase is the main proton pump of vacuoles in elongating hypocotyls. In 3-d-old hypocotyls, proton-pumping activity of H+-PPase per membrane protein content was 2 to 3 times greater than that of V-ATPase (Figs. 6 and 7). This is in agreement with a previous study (Maeshima, 1990). In 5-d-old hypocotyls, however, the proton-pumping activity of V-ATPase was slightly higher than that of H+-PPase. The change in ratio of the activity between the two proton pumps is due to the decrease in H+-PPase activity. A marked decrease in the amount of H+-PPase was also observed in radish roots (Maeshima et al., 1996).
In this study we determined the levels of transcripts of genes for H+-PPase and the V-ATPase subunit A in segments of hypocotyls. The decrease of the gene expression resulted in the low activity of H+-PPase. The genes for H+-PPase and the V-ATPase subunit A are highly transcribed in the elongating parts of the hypocotyl, and the levels of transcripts are lower in mature parts. In the same hypocotyls, the levels of mRNAs for H+-PPase and the V-ATPase subunit A in the mature part were, on the basis of DNA, less than 50% of that in the elongating part. A similar phenomenon was observed in the developing leaf. Immature leaves contained a high level of mRNAs for H+-PPase and the V-ATPase subunit A, but the levels decreased in mature leaves. Our findings are in agreement with those reported by Lerchl et al. (1995). In tobacco the expression of H+-PPase was active in young sink leaves, and the transcripts decreased in the mature source leaves. The decrease in transcription of the H+-PPase gene resulted in low levels of H+-PPase protein and activity. The H+-PPase may be gradually degraded after cell maturation.
Growth of hypocotyls is impossible without vacuole expansion, since the central vacuole occupies more than 80% of the volume of the mature cell. To maintain the high osmotic pressure of the expanding vacuole, the vacuole must actively incorporate solutes such as sugars and inorganic ions. Vacuolar H+-PPase and H+-ATPase provide the power for the secondary active transporters. Therefore, the vacuolar proton pumps are essential for vacuole enlargement. The physiological change in the H+-PPase level is efficient from the perspective of the cell's energetics. The substrate for H+-PPase, PPi, is produced as a by-product of several metabolic processes, such as polymerization of DNA and RNA and syntheses of aminoacyl-tRNA (protein synthesis), ADP-Glc (starch synthesis), UDP-Glc (cellulose synthesis), and fatty acyl-CoA (β-oxidation of fatty acid).
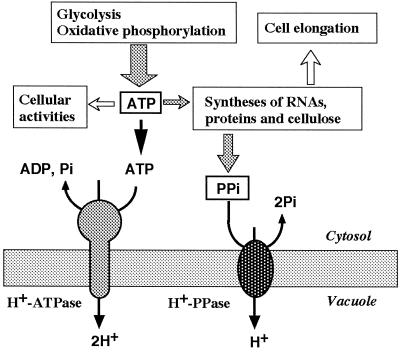
In growing cells, RNAs, proteins, and cellulose are actively synthesized for the construction of cells and as a result, a large amount of PPi is produced (Fig. 8). PPi accumulated in the cytosol at high concentrations inhibits the biosynthesis of macromolecules. In animal and bacteria cells, PPi is removed by soluble inorganic PPases. In plant cells, the vacuolar H+-PPase scavenges PPi in the cytosol and utilizes it as a source of energy for the active transport of protons into the vacuole. The vacuolar H+-PPase in growing cells helps to conserve ATP, which is a cell's energy currency. In mature cells metabolic activity decreases and PPi may not be available in such large amounts. Thus, the suppression of gene expression of H+-PPase after cell maturation is economical for the plant cell.
Figure 8.
PPi production and vacuolar membrane H+-ATPase and H+-PPase. PPi is supplied as a by-product of biosyntheses of macromolecules such as RNA, proteins, and cellulose in elongating cells.
The change in level of the V-ATPase subunit A did not parallel the change in the mRNA level. The relative content of the subunit A protein was the same after hypocotyl growth, although the relative mRNA content in the 5-d-old hypocotyl was about 30% of that in the elongating part of the 3-d-old hypocotyl (Fig. 4). The enzymatic activity of V-ATPase was also constant during hypocotyl growth (Fig. 7), suggesting that neither activation nor inactivation of V-ATPase occurred in hypocotyls. Probably, molecules of V-ATPase are more stable than those of H+-PPase. V-ATPase can operate constantly because ATP is maintained at a high concentration (a few millimolar). In conclusion, the H+-PPase is a major proton pump of vacuoles in growing hypocotyls, and its abundance is regulated at the transcriptional level in response to the supply of PPi. On the other hand, the V-ATPase activity remains relatively constant as a fundamental proton pump in the growing hypocotyl.
The level of H+-PPase is regulated under stress conditions. Carystinos et al. (1995) reported that the relative transcript and enzyme activity of H+-PPase increased notably under anoxia and chilling in rice seedlings. They proposed that H+-PPase may replace V-ATPase under energy stress. The H+-PPase activity was also increased in mung bean hypocotyls under low-temperature stress (Darley et al., 1995). It is obvious that gene expression of H+-PPase is regulated mainly in response to the demand of vacuolar acidification and is independent of V-ATPase. Transgenic plants that lack the H+-PPase gene(s) or the enzyme activity of H+-PPase should be analyzed to determine why plant vacuoles have H+-PPase in addition to V-ATPase. Also, the molecular mechanism involved in the regulation of the H+-PPase gene expression by the supply of ATP and PPi requires further study.
ACKNOWLEDGMENTS
We thank Dr. Yoshiyuki Tanaka of the National Institute of Agrobiological Resources and Dr. Kenzo Nakamura of Nagoya University for their kind advice and stimulating discussions.
Abbreviations:
- H+-PPase
H+-transporting inorganic pyrophosphatase
- NEM
N-ethylmaleimide
- V-ATPase
vacuolar H+-ATPase
Footnotes
Part of the research for this work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, and Culture of Japan to M.M. (nos. 09257221 and 09660114).
The DDBJ accession number for the DNA sequence reported in this article is DAB009077.
LITERATURE CITED
- Baykov AA, Bakuleva NP, Rea PA. Steady-state kinetics of substrate hydrolysis by vacuolar H+-pyrophosphatase: a simple three-state model. Eur J Biochem. 1993;217:755–762. doi: 10.1111/j.1432-1033.1993.tb18303.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Carystinos GD, MacDonald HR, Monroy AF, Dhindsa RS, Poole RJ. Vacuolar H+-translocating pyrophosphatase is induced by anoxia or chilling in seedlings of rice. Plant Physiol. 1995;108:641–649. doi: 10.1104/pp.108.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darley CP, Davies JM, Sanders D. Chill-induced changes in the activity and abundance of the vacuolar proton-pumping pyrophosphatase from mung bean hypocotyls. Plant Physiol. 1995;109:659–665. doi: 10.1104/pp.109.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM. Vacuolar energization: pumps, shunts and stress. J Exp Bot. 1997;48:633–641. [Google Scholar]
- Gendreau E, Traas J, Densons T, Grandjean O, Caboche M, Höfte H. Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 1997;114:295–305. doi: 10.1104/pp.114.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon-Weeks R, Steele SH, Leigh RA. The role of magnesium, pyrophosphate, and their complexes as substrates and activators of the vacuolar H+-pumping inorganic pyrophosphatase. Plant Physiol. 1996;111:195–202. doi: 10.1104/pp.111.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrich R, Schroeder JI. The physiology of ion channels and electrogenic pumps in higher plants. Annu Rev Plant Physiol. 1989;40:539–569. [Google Scholar]
- Hung S, Chiu S, Lin L, Pan R. Vacuolar H+-pyrophosphatase cDNA (accession no. U31467) from etiolated mung bean seedlings (PGR 95-082) Plant Physiol. 1995;109:1125. [Google Scholar]
- Ikeda M, Satoh S, Maeshima M, Mukohata Y, Moritani C. A vacuolar ATPase and pyrophosphatase in Acetabulariaacetabulum. Biochim Biophys Acta. 1991;1070:77–82. doi: 10.1016/0005-2736(91)90148-2. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Zhen R-G, Rea PA. Heterologous expression of plant vacuolar pyrophosphatase in yeast demonstrates sufficiency of substrate-binding subunit for transport. Proc Natl Acad Sci USA. 1994a;91:6128–6132. doi: 10.1073/pnas.91.13.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Zhen R-G, Rea PA. Site-directed mutagenesis of vacuolar H+-pyrophosphatase: necessity of Cys634for inhibition by maleimides but not catalysis. J Biol Chem. 1995;270:2630–2635. doi: 10.1074/jbc.270.6.2630. [DOI] [PubMed] [Google Scholar]
- Kim Y, Kim EJ, Rea PA. Plant Physiol. 1994b;106:375–382. doi: 10.1104/pp.106.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerchl J, König S, Zrenner R, Sonnewald U. Molecular cloning, characterization and expression analysis of isoforms encoding tonoplast-bound proton-translocating inorganic pyrophosphatase in tobacco. Plant Mol Biol. 1995;29:833–840. doi: 10.1007/BF00041172. [DOI] [PubMed] [Google Scholar]
- Maeshima M. Development of vacuolar membranes during elongation of cells in mung bean hypocotyls. Plant Cell Physiol. 1990;31:311–317. [Google Scholar]
- Maeshima M. H+-translocating inorganic pyrophosphatase of plant vacuoles: inhibition by Ca2+, stabilization by Mg2+and immunological comparison with other inorganic pyrophosphatases. Eur J Biochem. 1991;196:11–17. doi: 10.1111/j.1432-1033.1991.tb15779.x. [DOI] [PubMed] [Google Scholar]
- Maeshima M, Mimura T, Sato T. Distribution of vacuolar H+-pyrophosphatase and a membrane integral protein in a variety of green plants. Plant Cell Physiol. 1994;35:323–328. [Google Scholar]
- Maeshima M, Nakanishi Y, Matsuura-Endo C, Tanaka Y. Proton pumps of vacuolar membrane in growing plant cell. J Plant Res. 1996;109:119–125. [Google Scholar]
- Maeshima M, Yoshida S. Purification and properties of vacuolar membrane proton-translocating inorganic pyrophosphatase from mung bean. J Biol Chem. 1989;264:20068–20073. [PubMed] [Google Scholar]
- Matsuura-Endo C, Maeshima M, Yoshida S. Subunit composition of vacuolar membrane H+-ATPase from mung bean. Eur J Biochem. 1990;187:745–751. doi: 10.1111/j.1432-1033.1990.tb15362.x. [DOI] [PubMed] [Google Scholar]
- Matsuura-Endo C, Maeshima M, Yoshida S. Mechanism of the decline in vacuolar H+-ATPase activity in mung bean hypocotyls during chilling. Plant Physiol. 1992;100:718–722. doi: 10.1104/pp.100.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nore BF, Sakai-Nore Y, Maeshima M, Baltscheffsky M, Nyrén P. Immunological cross-reactivity between proton-pumping inorganic pyrophosphatases of widely phylogenic separated species. Biochem Biophys Res Commun. 1991;181:962–967. doi: 10.1016/0006-291x(91)92030-n. [DOI] [PubMed] [Google Scholar]
- Nyrén P, Nore BF, Strid A. Proton-pumping N,N′-dicyclohexylcarbodiimide-sensitive inorganic pyrophosphate synthase from Rhodospirillum rubrum: purification, characterization, and reconstitution. Biochemistry. 1991;30:2883–2887. doi: 10.1021/bi00225a022. [DOI] [PubMed] [Google Scholar]
- Rea PA, Kim Y, Sarafian V, Poole RJ, Davies JM, Sanders D. Vacuolar H+-translocating pyrophosphatases: a new category of ion translocase. Trends Biochem Sci. 1992;17:348–353. doi: 10.1016/0968-0004(92)90313-x. [DOI] [PubMed] [Google Scholar]
- Rea PA, Poole RJ. Vacuolar H+-translocating pyrophosphatase. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:157–180. [Google Scholar]
- Sakakibara Y, Kobayashi H, Kasamo K. Isolation and characterization of cDNAs encoding vacuolar H+-pyrophosphates isoforms from rice (Oryza sativaL.) Plant Mol Biol. 1996;31:1029–1038. doi: 10.1007/BF00040721. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sarafian V, Kim Y, Poole RJ, Rea PA. Proc Natl Acad Sci USA. 1992;89:1775–1779. doi: 10.1073/pnas.89.5.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz L. The plant vacuole. J Exp Biol. 1992;172:113–122. doi: 10.1242/jeb.172.1.113. [DOI] [PubMed] [Google Scholar]
- Takasu A, Nakanishi Y, Yamauchi T, Maeshima M. Analysis of the substrate binding site and carboxyl terminal region of vacuolar H+-pyrophosphatase of mung bean with peptide antibodies. J Biochem. 1997;122:883–889. doi: 10.1093/oxfordjournals.jbchem.a021837. [DOI] [PubMed] [Google Scholar]
- Takeshige K, Tazawa M, Hager A. Plant Physiol. 1988;86:1168–1173. doi: 10.1104/pp.86.4.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Chiba K, Maeda M, Maeshima M. Molecular cloning of cDNA for vacuolar membrane proton-translocating inorganic pyrophosphatase in Hordeum vulgare. Biochem Biophys Res Commun. 1993;190:1110–1114. doi: 10.1006/bbrc.1993.1164. [DOI] [PubMed] [Google Scholar]
- Wilkins TA. Vacuolar H+-ATPase 69-kilodalton catalytic subunit cDNA from developing cotton (Gossypium hirsutum) ovules. Plant Physiol. 1993;102:679–680. doi: 10.1104/pp.102.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen R-G, Kim EJ, Rea PA. Localization of cytosolically orientated maleimide-reactive domain of vacuolar H+-pyrophosphatase. J Biol Chem. 1994;269:23342–23350. [PubMed] [Google Scholar]
- Zimniak L, Dittrich P, Gogarten JP, Kibak H, Taiz L. The cDNA sequence of the 69-kDa subunit of the carrot vacuolar H+-ATPase. J Biol Chem. 1988;263:9102–9112. [PubMed] [Google Scholar]