SUMMARY
Pyruvate kinase M2 (PKM2) is upregulated in multiple cancer types and contributes to the Warburg effect by unclarified mechanisms. Here we demonstrate that EGFR-activated ERK2 binds directly to PKM2 I429/L431 via the ERK2 docking groove and phosphorylates PKM2 Ser37 but not PKM1. Phosphorylated PKM2 Ser37 recruits PIN1 for cis-trans isomerization of PKM2, which leads to PKM2 binding to importin α5 and nuclear translocation. Nuclear PKM2, acting as a coactivator of β-catenin, induces c-Myc expression, resulting in the upregulation of GLUT1, LDHA, and, in a positive feedback loop, PTB-dependent PKM2 expression. Replacement of wild type PKM2 with a nuclear translocation-deficient mutant (S37A) blocks the EGFR-promoted Warburg effect and brain tumor development. In addition, levels of PKM2 S37 phosphorylation correlate with EGFR and ERK1/2 activity in human glioblastoma specimens. Our findings highlight the importance of nuclear functions of PKM2 in the Warburg effect and tumorigenesis.
Many cancer cells exhibit elevated glucose uptake and lactate production, regardless of oxygen availability. This phenomenon, known as aerobic glycolysis or the Warburg effect, facilitates tumor cell growth.1–3 Glucose-metabolizing enzymes, including pyruvate kinase, are often upregulated in cancer cells.4, 5 PKM2 arises from alternate splicing of PKM (formerly PKM2) pre-mRNA leading to the inclusion of exon 10 (PKM2) and the exclusion of exon 9 (PKM1) mediated by polypyrimidine-tract binding protein (PTB) and hnRNP A1/2 splicing factors.6, 7 Replacing PKM2 with the more active PKM1 isozyme in human lung cancer cells fails to support the Warburg effect and inhibits tumor formation.8, 9 However, the mechanisms underlying the distinct role of PKM2 and PKM1 in the Warburg effect remain unclear.
Work from our laboratory and others has demonstrated that PKM2 can function as a transcriptional coactivator.10,11, 12 Under hypoxic conditions, prolyl-hydroxylated PKM2 interacts with HIF-1α to induce glycolytic gene expression, which promotes glucose metabolism in cancer cells.11 Nevertheless, we are far from a complete understanding of the mechanism that underlies PKM2-mediated glycolysis in normoxia. We recently reported that the binding of nuclear PKM2 to phosphorylated β-catenin Y333 is required for epidermal growth factor receptor (EGFR)-induced β-catenin transactivation and cell cycle progression.10, 13 Importantly, PKM2 functions as a protein kinase, phosphorylating histone H3-T11 and leading to H3-K9 acetylation and transcription of genes, including CCND1 (coding for cyclin D1) and MYC.14
In this report, we demonstrate that extracellular signal-regulated kinase (ERK) phosphorylation-dependent nuclear translocation of PKM2 is required for the autoregulation of PKM2 expression and PKM2-dependent expression of glycolytic genes, which are essential for the EGFR-promoted Warburg effect and tumorigenesis.
RESULTS
ERK is required for PKM2 nucleus translocation
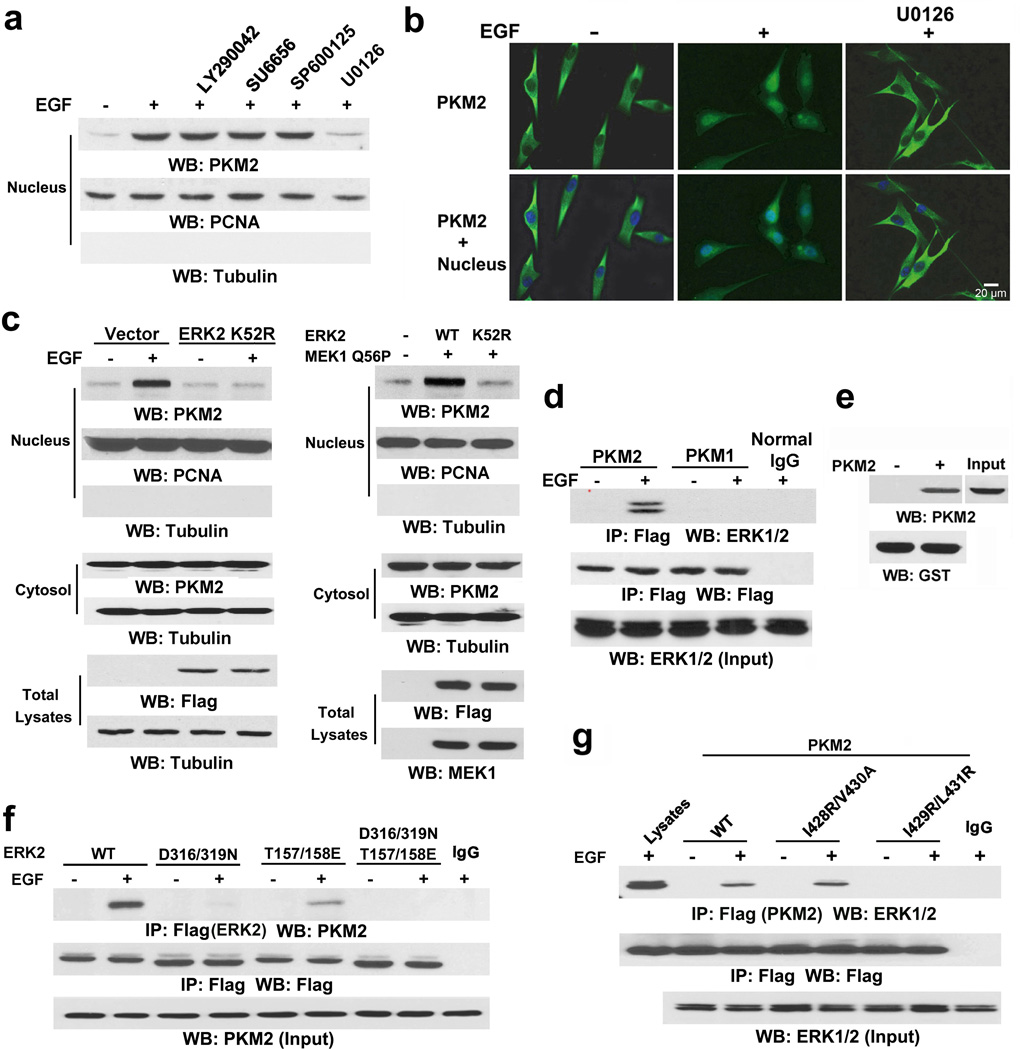
To understand the mechanism of PKM2 accumulation in the nucleus, we performed immunofluorescence analysis and showed that PKM2, a primarily cytosolic protein, translocated into the nucleus upon EGF stimulation in U251 human glioblastoma multiforme (GBM) cells (Supplementary Fig. 1a, left panel). This result is in line with our findings that EGFR activation by EGF or expression of the constitutively activated EGFRvIII mutant promotes nuclear translocation of PKM2.10 In contrast, Flag-PKM1 did not show any subcellular redistribution upon EGF stimulation (Supplementary Fig. 1a, right panel). Treatment with EGFR inhibitor AG1478, which blocked EGF-induced phosphorylation of EGFR and ERK1/2, abrogated EGF-induced nuclear accumulation of PKM2 (Supplementary Fig. 1b). Compared to the amount of cytosolic PKM2, nuclear PKM2 is a small portion (Supplementary Fig. 1C), which may account for no reduction of cytoplasmic PKM2 expression upon EGF treatment. Pretreatment of U87/EGFR (Fig. 1a) and U251 cells (Supplementary Fig. 1d) with the phosphoinositide 3-kinase inhibitor LY290042, Src inhibitor SU6656, JNK inhibitor SP600125, and MEK/ERK inhibitor U0126 blocked EGF-induced phosphorylation of AKT, c-Src, c-Jun, and ERK1/2, respectively (Supplementary Fig. 1e). Immunoblotting analyses showed that only inhibition of MEK/ERK abrogated EGF-induced nuclear translocation of PKM2 (Fig. 1a and Supplementary Fig. 1d). These results were further supported by immunofluorescence analyses (Fig. 1b). In addition, expression of the Flag-ERK2 K52R kinase-dead mutant blocked EGF-induced nuclear accumulation of PKM2 (Fig. 1c, left panel). Furthermore, coexpression of the constitutively active MEK1 Q56P mutant with Flag-tagged wild-type (WT) ERK2 or ERK2 K52R in U251 cells (Fig. 1c, right panel) showed that expression of WT ERK2, but not ERK2 K52R, induced nuclear translocation of PKM2. These results indicate that ERK activation is required for EGF-induced nuclear translocation of PKM2.
Figure 1. ERK is required for PKM2 nucleus translocation.
Immunoblotting and immunoprecipitation analyses were performed with the indicated antibodies.
a, Nuclear fractions were prepared from U87/EGFR cells pretreated with LY290042 (30 µM), SU6656 (4 µM), SP600125 (25 µM), and U0126 (20 µM) for 30 min before EGF (100 ng/ml) for 6 h. Nuclear PCNA and cytoplasmic tubulin were used as controls.
b, U87/EGFR cells were pretreated with or without U0126 (20 µM) for 30 min before EGF (100 ng/ml) for 6 h. Immunofluorescence analyses were performed with the indicated antibodies. Nuclei were stained with Hoechst 33342 (blue).
c, U251 cells were stably transfected with a vector with or without expressing Flag-ERK2 K52R (left panel) or transiently transfected with vectors expressing MEK1 Q56P and indicated Flag-tagged ERK2 proteins (right panel). The cells were treated with or without EGF (100 ng/ml) for 6 h, and the total cell lysates and nuclear fractions were prepared.
d, Flag-PKM2 or Flag-PKM1 was immunoprecipitated with an anti-Flag antibody from U87/EGFR cells treated with or without EGF (100 ng/ml) for 30 min.
e, Bacterially purified GST-ERK2 immobilized on glutathione agarose beads was mixed with or without purified His-PKM2. GST pull-down analyses were performed. 10% of total His-PKM2 was used as an input.
f, U87/EGFR cells transfected with vectors expressing the indicated Flag-tagged ERK proteins were treated with or without EGF (100 ng/ml) for 30 min.
g, U87/EGFR cells expressing the indicated Flag-PKM2 proteins were treated with or without EGF (100 ng/ml) for 30 min.
To further determine the relationship between ERK1/2 and PKM2, we performed a co-immunoprecipitation assay and revealed that EGF treatment resulted in ERK1/2 binding to FLAG-PKM2 but not FLAG-PKM1 (Fig. 1d). Moreover, an in vitro GST pull-down assay with mixed purified GST-ERK2 and His-PKM2 showed that these two proteins interacted directly (Fig. 1e). MAP kinases bind to their substrates through a docking groove comprised of an acidic common docking (CD) domain and glutamic acid-aspartic acid (ED) pockets.15 Immunoblotting of the immunoprecipitated Flag-ERK2 proteins with an anti-PKM2 antibody showed that mutation of either the ERK2 CD domain (D316/319N) or the ED pocket (T157/158E) reduced binding to endogenous PKM2, as compared with the WT ERK2 control. Combined mutations at both the CD domain and ED pocket (T/E-D/N) abrogated the binding of ERK2 to PKM2 entirely (Fig. 1f), indicating that ERK2 binds to PKM2 through its docking groove.
ERK substrates often have a docking (D) domain, which is characterized by a cluster of basic residues followed by an LXL motif (L represents Leu, but can also be Ile or Val; X represents any amino acid).15 Analysis of the PKM2 amino acid sequence with the Scansite program identified the putative ERK-binding sequence 422-KCCSGAIIVLTKSGR-436 in the aa 380–434 region, which contains LXL motifs at I428/V430 and I429/L431. This series of amino acids is encoded by the PKM2-specific exon 10 and is thus unique to PKM2. Immunoblotting of the immunoprecipitated Flag-PKM2 proteins with an anti-ERK1/2 antibody showed that a PKM2 I429/L431A mutant, but not a PKM2 I428R/V430A mutant, drastically reduced its binding to ERK1/2 (Fig. 1g). These results indicate that the ERK2 docking groove binds to a D domain in PKM2 at I429/L431.
ERK2 phosphorylates PKM2 S37
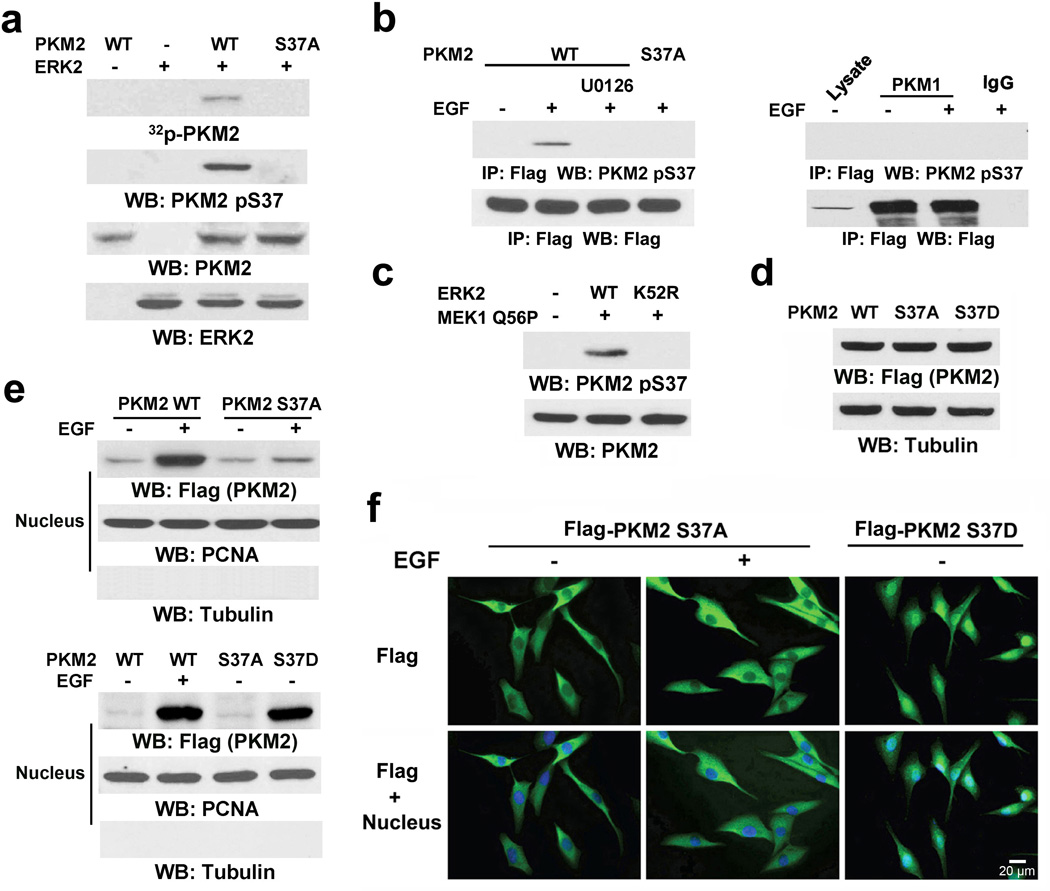
An in vitro kinase assay by mixing purified PKM2 with active ERK2 showed that ERK2 phosphorylated PKM2 (Fig. 2a). Sequence analysis of PKM2 revealed that it contains an ERK consensus phosphorylation motif (Ser-Pro)15 at the S37/P38 residues. The S37A mutation completely abrogated the ERK2-dependent phosphorylation of PKM2 in vitro; this finding was further validated by a specific phospho-PKM2 S37 antibody (Fig. 2a).
Figure 2. ERK2 phosphorylates PKM2 S37.
Immunoblotting and immunoprecipitation analyses were performed with the indicated antibodies.
a, In vitro kinase assays were performed with purified active ERK2, WT PKM2, and PKM2 S37A mutant.
b, Flag-PKM1 or Flag-PKM2 was immunoprecipitated with anti-Flag antibody from U87/EGFR cells pretreated with or without U0126 (20 µM) for 30 min before EGF (100 ng/ml) for 30 min.
c, MEK1 Q56P and the indicated ERK2 proteins were transiently expressed in U87/EGFR cells.
d, e, f, Total cell lysates (d) or nuclear fractions (e) were prepared from U87/EGFR cells expressing Flag-tagged WT PKM2, PKM2 S37A, or PKM2 S37D, which had been treated with or without EGF (100 ng/ml) for 6 h. (f) Immunofluorescence analyses were performed with an anti-Flag antibody. Nuclei were stained with Hoechst 33342 (blue).
Because PKM1 and PKM2 share the identical N-terminal amino acid sequence (including S37), we then tested whether ERK2-mediated phosphorylation of S37 is restricted to PKM2. Immunoblotting of the immunoprecipitated Flag-tagged PKM1 or PKM2 with the anti-phospho-PKM2 S37 antibody showed that EGF treatment resulted in phosphorylation of WT PKM2 (Fig. 2b, left panel) but not PKM1 (Fig. 2b, right panel) or the PKM2 S37A mutant. Consistently, pretreatment with U0126 blocked EGF-induced S37 phosphorylation and nuclear translocation of PKM2 (Fig. 2b, left panel; Supplementary Fig. 2a). In addition, expression of constitutively active MEK1 Q56P with WT ERK2, but not with the ERK2 K52R mutant, induced PKM2 S37 phosphorylation (Fig. 2c). These results indicate that ERK2 specifically phosphorylates PKM2 but not PKM1. This observation is consistent with the presence of the D domain in PKM2 (but not in PKM1) that allows unique ERK1/2 binding (Figs. 1d, 1g).
Expression of Flag-tagged WT PKM2, the PKM2 S37A mutant, or a phosphorylation-mimic PKM2 S37D mutant in U87/EGFR cells (Fig. 2d) showed that the Flag-PKM2 S37A mutant was resistant to EGF-induced nuclear translocation, as determined by immunoblotting analyses (Fig. 2e, top panel) or immunofluorescence analysis (Fig. 2f, left and middle panels). Similar results were observed in U87/EGFR cells with depleted endogenous PKM2 and reconstituted expression of RNAi-resistant rPKM2 S37A (Supplementary Fig. 2b). In contrast, the phosphorylation-mimic PKM2 S37D mutant had a higher level of nuclear accumulation in the absence of EGF treatment than did WT PKM2 or the S37A mutant (Figs. 2e, bottom panel and 2f right panel). These results indicate that PKM2 phosphorylation at S37 is required for nuclear translocation of PKM2.
PKM2 S37 phosphorylation recruits PIN1
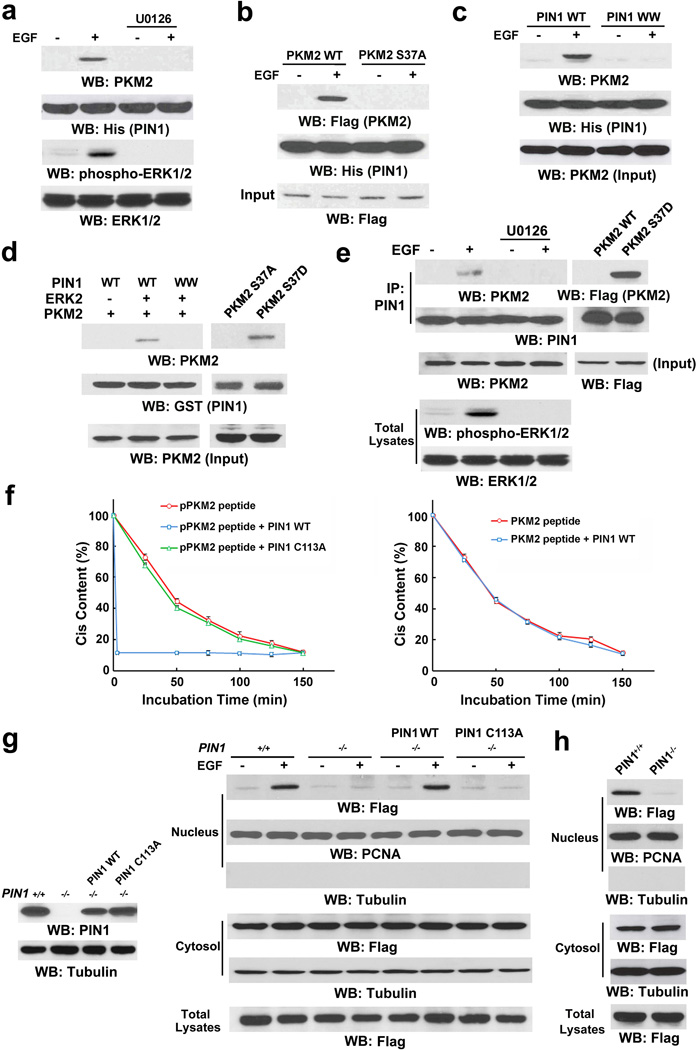
The peptidyl-proline isomerase protein interacting with never in mitosis A (NIMA)-1 (PIN1) recognizes phosphorylated pS/TP-peptide sequences and catalyzes their cis-trans isomerization.16, 17 EGF treatment induced a strong binding of endogenous PKM2 to His-PIN1 immobilized on nickel agarose beads (Fig. 3a), whereas this binding was abrogated by pretreatment of cells with U0126 (Fig. 3a) or S37A mutation of PKM2 (Fig. 3b). In addition, a His-PIN1 WW domain mutant, which prevents the binding of PIN1 to a pS/TP substrate,17 failed to bind to endogenous PKM2 in U87/EGFR cells upon EGF treatment (Fig. 3c). These results indicate that the PIN1 WW domain binds to the ERK1/2-phosphorylated S37 of PKM2.
Figure 3. PKM2 S37 phosphorylation recruits PIN1.
Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies.
a, His-PIN1 immobilized on nickel agarose beads was incubated with lysate of U87/EGFR cells treated with or without U0126 (20 µM) for 30 min before EGF (100 ng/ml) stimulation for 30 min.
b, U87/EGFR cells transiently expressing WT Flag-PKM2 or Flag-PKM2 S37A were treated with or without EGF (100 ng/ml) for 30 min. His-PIN1 immobilized on nickel agarose beads was incubated with the cell lysates.
c, His-PIN1 or His-PIN1 WW domain mutant (with substitutions at W11A, W34A, R14A, and R17A) immobilized on nickel agarose beads was incubated with lysate of U87/EGFR cells treated with or without EGF (100 ng/ml) for 30 min.
d, Purified GST-PIN1 on glutathione agarose beads was mixed with purified His-PKM2 with or without purified active ERK2 (left panel) or incubated with purified His-PKM2 S37A or His-PKM2 S37D mutant (right panel).
e, PIN1 was immunoprecipitated from U87/EGFR cells pretreated with or without U0126 (20 µM) for 30 min before EGF (100 ng/ml) stimulation for 30 min (left panel) or from U87/EGFR cells expressing Flag-PKM2 or Flag-PKM2 S37D (right panel).
f, Cis-trans isomerization assays were performed by mixing synthesized phosphorylated or nonphosphorylated oligopeptide of PKM2 containing the S37P motif with purified WT GST-PIN1 or GST-PIN1 C113A mutant. Data represent the means ± SD of three independent experiments.
g, PIN1-/- cells were reconstituted to express the indicated PIN1 proteins (left panel). The total cell lysates and nuclear fractions were prepared from the indicated cells treated with or without EGF (100 ng/ml) for 6 h (right panel).
h, Total cell lysates and nuclear fractions of PIN1-/- cells or PIN1-/- cells expressing Flag-PKM2 S37D were prepared.
To determine whether ERK1/2-dependent PKM2 phosphorylation is sufficient for PIN1 binding to PKM2, we mixed His-PKM2 with WT GST-PIN1 or a GST-PIN1 WW domain mutant in the presence or absence of active ERK2. As shown in Fig. 3d (left panel), phosphorylated PKM2, but not its nonphosphorylated counterpart, bound to WT PIN1, but not the PIN1 WW domain mutant. In addition, the purified phosphorylation-mimic His-PKM2 S37D mutant, but not His-PKM2 S37A, was able to interact with GST-PIN1 in the absence of ERK2 (Fig. 3d, right panel). These in vitro results were validated by a co-immunoprecipitation assay showing that EGF stimulation greatly increased the binding of endogenous PIN1 to endogenous PKM2, which was blocked by U0126 pretreatment (Fig. 3e, left panel). In contrast to WT Flag-PKM2, the Flag-PKM2 S37D mutant was co-immunoprecipitated with PIN1 in the absence of EGF treatment (Fig. 3e, right panel). These results indicate that PKM2 S37 phosphorylation is sufficient for PIN1 binding to PKM2.
To further examine whether the phosphorylated S37/P38 motif of PKM2 is a PIN1 substrate, we synthesized oligopeptides of PKM2 containing phosphorylated or nonphosphorylated S37/P38. As demonstrated in Fig. 3f, GST-WT PIN1 isomerized the phosphorylated S37/P38 peptide much more efficiently than did a catalytically inactive GST-PIN1 C113A mutant (left panel), whereas WT PIN1 failed to isomerize the nonphosphorylated counterpart (right panel). These results strongly suggest that PIN1 specifically isomerizes the phosphorylated S37/P38 within PKM2.
To determine the role of PIN1 in nuclear translocation of PKM2, we used EGF to treat PIN1+/+, PIN1-/-, or PIN1-/- mouse embryonic fibroblasts (MEFs) with reconstituted expression of WT PIN1 or a PIN1 C113A mutant (Fig. 3g, left panel). PIN1 deficiency completely blocked EGF-induced nuclear translocation of human Flag-PKM2 whereas this block was rescued by re-expression of WT PIN1 but not by expression of the PIN1 C113A mutant (Fig. 3g, right panel). These results indicate that the catalytic activity of PIN1 is required for EGF-induced nuclear translocation of PKM2. Furthermore, the nuclear translocation of the phosphorylation-mimic Flag-PKM2 S37D mutant (Fig. 2e) was inhibited by PIN1 deficiency (Fig. 3h). This observation indicate that phosphorylation of PKM2 by itself is not sufficient for PKM2 nuclear translocation, further supporting a role for PIN1 in EGF-induced nuclear translocation of PKM2.
PIN1 regulates binding of PKM2 to importin α5
PIN1 contains an NLS18. We next tested whether PKM2 translocates together with PIN1 into the nucleus in a PIN1 NLS-dependent manner. A PIN1 NLS (L60/61A) mutant, which was expressed in PIN1-/- cells, failed to accumulate in the nucleus (Supplementary Fig. 2c). However, expression of this PIN1 mutant, whose catalytic activity was intact (Supplementary Fig. 2d), still permitted EGF-induced nuclear translocation of Flag-PKM2 (Supplementary Fig. 2e). These results indicate that the nuclear translocation of PKM2 is not mediated by the NLS of PIN1 and suggest that these two proteins separately translocate into the nucleus. Furthermore, depletion of HIF1α, which is a PKM2-associated protein,11 did not affect EGF-induced nuclear translocation of PKM2 and β-catenin Y333 phosphorylation (which is required for β-catenin transactivation)10 (Supplementary Fig. 2f), suggesting separate nuclear translocation processes for these two proteins and β-catenin regulation independent of HIF1α.
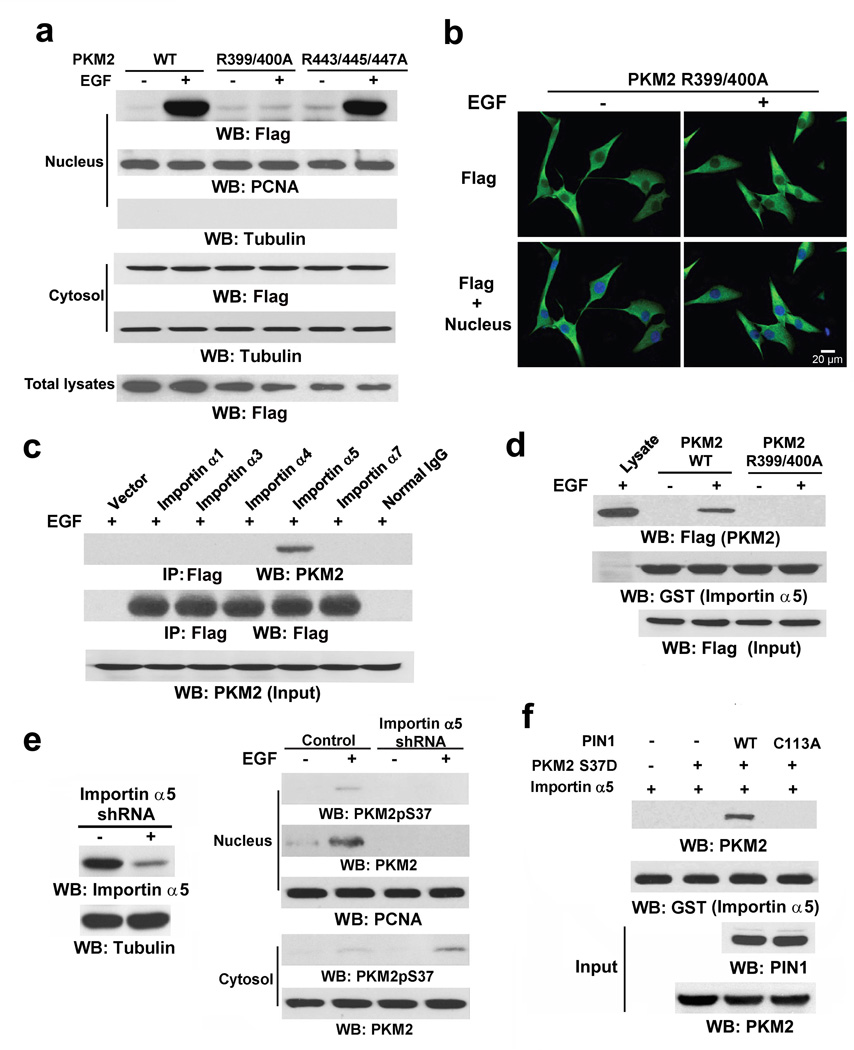
To test whether PKM2 contains a sterically inaccessible NLS that is exposed for importin binding after PIN1-mediated cis-trans isomerization, we mutated the R399/400 and R443/445/447 residues in the putative NLS sequences of the C-domain (aa 393–531) encoded by PKM2-specific exon 1019 into alanine. Cell fractionation (Fig. 4a) and immunofluorescence (Fig. 4b) analyses showed that Flag-PKM2 R399/400A, unlike the R443/445/447A mutant or WT PKM2, was unable to translocate into the nucleus upon EGF treatment. These results indicate that the NLS containing R399/400 in PKM2, but not in PKM1, is essential for EGF-induced nuclear translocation of PKM2.
Figure 4. PIN1 regulates binding of PKM2 to importin α5.
Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies.
a, U87/EGFR cells expressing WT Flag-PKM2 or the indicated Flag-PKM2 mutants were treated with or without EGF (100 ng/ml) for 6 h, and the total cell lysates and nuclear fractions were prepared.
b, Immunofluorescence analyses of U87/EGFR cells expressing Flag-PKM2 R399/400A were performed with an anti-Flag antibody. Nuclei were stained with Hoechst 33342.
c, The indicated Flag-importin α proteins and EGFR were coexpressed in 293T cells, which were treated with EGF (100 ng/ml) for 30 min. Flag-importin α proteins were immunoprecipitated with an anti-Flag antibody.
d, U87/EGFR cells expressing WT Flag-PKM2 or Flag-PKM2 R399/400A were treated with or without EGF (100 ng/ml) for 30 min. The cell lysate was mixed with purified GST-importin α5 for a GST pull-down assay.
e, U87/EGFR cells with or without depletion of importin α5 were treated with or without EGF (100 ng/ml) for 6 h. The cytosolic and nuclear fractions were prepared.
f, Purified His-PKM2 S37D on nickel agarose beads was mixed with purified GST-importin α5 in the presence or absence of purified WT His-PIN1 or His-PIN1 C113A. A GST pull-down assay was performed.
Importin α functions as an adaptor and links NLS-containing proteins to importin β, which then docks the ternary complex at the nuclear-pore complex (NPC) to facilitate the translocation of these proteins across the nuclear envelope. Six importin α family members (α1, α3, α4, α5, α6 and α7) have been identified in humans.20 Fig. 4c shows that EGF treatment resulted in endogenous PKM2 binding to importin α5, but not to α1, α3, α4, and α5 family members. In addition, purified GST-importin α5 was able to pull down WT Flag-PKM2, but not Flag-PKM2 R399/400A, from EGF-treated U87/EGFR cells (Fig. 4d). Furthermore, depletion of importin α5 with KPNA1 (coding for importin α5) shRNA (Fig. 4e) largely blocked EGF-induced nuclear translocation of PKM2 and resulted in accumulated phosphorylated PKM2 S37 in the cytosol (Fig. 4e).
To determine whether PIN1 plays a role in the binding of PKM2 to importin α5, we mixed purified phosphorylation-mimic His-PKM2 S37D and GST-importin α5 in the presence or absence of WT His-PIN1 or the His-PIN1 C113A mutant. As shown in Fig. 4f, PKM2 S37D alone did not bind to importin α5. However, inclusion of WT PIN1, but not of the inactive PIN1 C113A mutant, enabled PKM2 S37D to interact with importin α5. These results strongly suggest that phosphorylation of PKM2 at S37, which by itself is not sufficient for PKM2 nuclear translocation (Fig. 3h), leads to cis-trans isomerization of PKM2 by PIN1 to expose intermolecularly- or intramolecularly-masked R399/400 of the PKM2 NLS for binding to importin α5. In combination with our finding that the recruitment of PIN1 to PKM2 is mediated by both the binding of the ERK2 docking groove to PKM2-specific exon 10-encoded I429/L431 region and the phosphorylation of PKM2 at S37, these results highlight the significance of precise and sequential post-translational modifications of PKM2 and its interactions with ERK1/2, PIN1, and importin α5 in its nuclear translocation.
Nuclear PKM2 regulates glycolytic gene expression
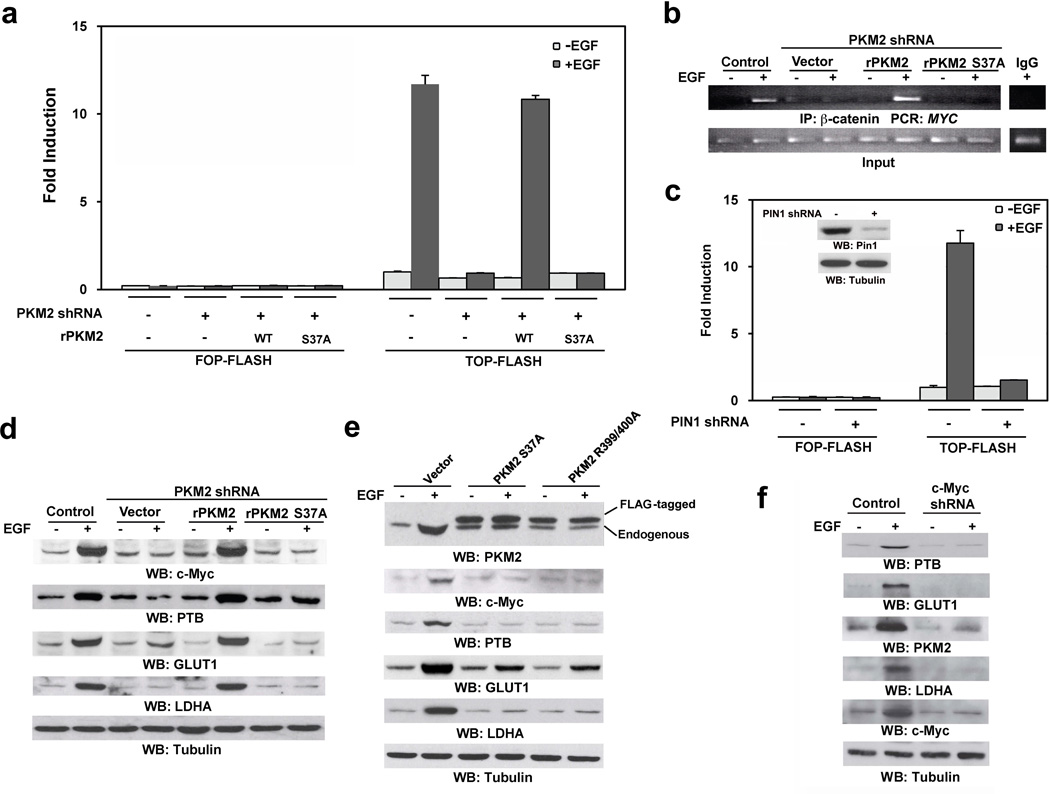
Previously, we demonstrated that nuclear PKM2 interacts with phosphorylated β-catenin Y333 for β-catenin transactivation.10 The TOP-FLASH TCF/LEF-1 luciferase reporter analyses showed that PKM2 depletion significantly inhibited EGF-induced β-catenin transactivation, which was largely rescued by reconstituted expression of RNAi-resistant WT rPKM2 but not those of rPKM2 S37A (Fig. 5a), WT Flag-PKM1, or Flag-PKM1 S37A (Supplementary Fig. 3a). In addition, chromatin immunoprecipitation (ChIP) with an anti-β-catenin antibody showed that EGFR activation resulted in the binding of β-catenin to the MYC promoter, which was inhibited by PKM2 depletion (Fig. 5b, Supplementary Fig. 3b). Notably, this inhibition was abrogated by expression of WT rPKM2 but not that of rPKM2 S37A. In addition, PIN1 depletion by PIN1 shRNA also blocked EGF-induced β-catenin transactivation (Fig. 5c). These results indicate that PIN1-dependent nuclear translocation of PKM2 plays a critical role in EGF-induced β-catenin transactivation.
Figure 5. Nuclear PKM2 regulates glycolytic gene expression.
Immunoblotting analyses were performed with the indicated antibodies.
a, c, U87/EGFR cells with or without PKM2 depletion and reconstituted expression of the indicated PKM2 proteins (a) or U87/EGFR cells with or without PIN1 depletion (c) were transfected with either TOP-FLASH or FOP-FLASH (control vector), which was followed by EGF (100 ng/ml) treatment for 12 h. The relative levels of luciferase activity were normalized to the levels of untreated cells and to the levels of luciferase activity of the Renilla control plasmid. Data represent the means ± SD of three independent experiments.
b, U87/EGFR cells with or without PKM2 depletion and reconstituted expression of the indicated PKM2 proteins were treated with or without EGF (100 ng/ml) for 12 h. ChIP assay was performed with an anti-β-catenin antibody for immunoprecipitation followed by PCR with MYC promoter–specific primers.
d, U87/EGFR cells with or without PKM2 depletion and reconstituted expression of WT rPKM2 or rPKM2 S37A mutant were treated with or without EGF (100 ng/ml) for 24 h.
e, U87/EGFR cells expressing Flag-PKM2 S37A or Flag-PKM2 R399/400A were treated with or without EGF (100 ng/ml) for 24 h.
f, U87/EGFR cells with or without c-Myc depletion were treated with or without EGF (100 ng/ml) for 24 h.
c-Myc expression is known to be upregulated by PKM2-dependent β-catenin transactivation.21,14 c-Myc transcriptionally induces expression of GLUT1 and lactate dehydrogenase A (LDHA)22, 23 and upregulates PTB expression, thereby regulating PKM pre-mRNA splicing for generation of PKM2 mRNA.6, 7 We next examined whether nuclear translocation of PKM2 plays a role in c-Myc-dependent expression of downstream genes. As shown in Fig. 5d, PKM2 depletion blocked EGF-enhanced expression of c-Myc, PTB, GLUT1, and LDHA, which was rescued by reconstituted expression of WT rPKM2 but not those of rPKM2 S37A, WT Flag-PKM1, or Flag-PKM1 S37A (Supplementary Fig. 3c). Of note, EGF treatment resulted in PKM2 upregulation, which is in agreement with the enhanced expression of PTB by EGF (Fig. 5e). Expression of Flag-PKM2 S37A or R399/400A blocked EGF-induced upregulation of c-Myc, PTB, PKM2, GLUT1, and LDHA. In line with these findings, c-Myc depletion blocked EGF-induced upregulation of PTB, PKM2, GLUT1, and LDHA (Fig. 5f). These results indicate that nuclear translocation of PKM2 plays a pivotal role in EGF-induced β-catenin transactivation, which results in a subsequent upregulation of PTB, PKM2, LDHA, and GLUT1 expression in a c-Myc-dependent manner.
Nuclear PKM2 is required for the Warburg effect
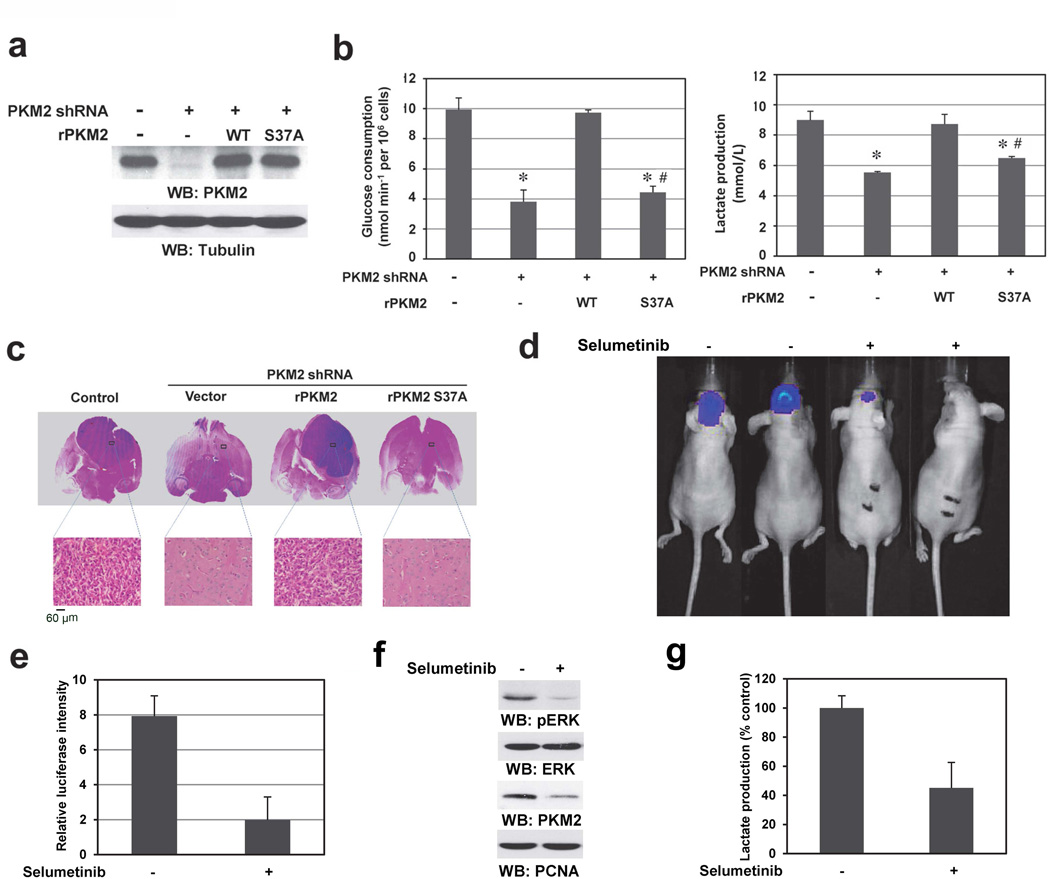
GLUT1 and LDHA are required for glucose uptake and the conversion of pyruvate to lactate, respectively.8 To investigate the role of nuclear translocation of PKM2 in EGFR-regulated tumor cell glycolysis, we depleted PKM2 with PKM2 shRNA in EGFRvIII-expressing U87 cells and reconstituted these cells with WT rPKM2, rPKM2 S37A (Fig. 6a), and WT Flag-PKM1 (Supplementary Fig. 4a). PKM2 depletion significantly reduced glucose consumption and lactate production (Fig. 6b), which were rescued by reconstituted expression of WT PKM2, but not that of PKM2 S37A (Fig. 6b) or WT Flag-PKM1 (Supplementary Fig. 4b). Because the S37A mutation did not affect PKM2 kinase activity (Supplementary Fig. 4c), these results indicate that nuclear translocation of PKM2, which results in upregulation of PKM2 itself, GLUT1, and LDHA, is essential for the EGFR-induced Warburg effect. These findings were further supported by the results showing that depletion of GLUT1, LDHA, and PTB largely reduced glucose consumption and lactate production (Supplementary Figs. 4d-4f).
Figure 6. Nuclear PKM2 is required for the Warburg Effect.
Immunoblotting analyses were performed with the indicated antibodies.
a, b, U87/EGFRvIII cells with or without PKM2 depletion and reconstituted expression of WT rPKM2 or rPKM2 S37A mutant (a) were cultured in no-serum DMEM. The media were collected for analysis of glucose consumption (b, left panel) or lactate production (b, right panel). Data represent the means ± SD of three independent experiments. *p < 0.01: statistically significant value in relation with U87/EGFRvIII cells without PKM2 depletion. #p <0.01: statistically significant value in relation with U87/EGFRvIII cells with PKM2 depletion and reconstituted expression of WT rPKM2.
c, U87/EGFRvIII cells (5 × 105) with or without RNAi-depleted PKM2 and reconstituted expression of WT rPKM2 or rPKM2 S37A were intracranially injected into athymic nude mice. After two weeks, mice were sacrificed for examining the tumor growth. H&E-stained coronal brain sections are representative tumor xenografts.
d, U87/EGFRvIII (5 × 105) cells expressing luciferase were intracranially injected into athymic nude mice. After 3 days, selumetinib (50 mg/ml in 5 µl of DMSO) or DMSO was intracranially and intratumorally injected every 3 days. Bioluminescence imaging of mice was taken at day 9.
e, Tumor volumes were measured using length (a) and width (b) and calculated using the equation: V = ab2/2. Data represent the means ± SD of three independent experiments.
f, Immunoblotting analyses of the tumor tissues received with or without selumetinib treatment were performed with the indicated antibodies.
g, Lactate production of the tumor tissues received with or without selumetinib treatment was analyzed. Data represent the means ± SD of three separate samples.
Acting like EGF, platelet-derived growth factor (PDGF) induced nuclear translocation of PKM2, which was blocked by pretreatment with U0126 (Supplementary Fig. 5a). In addition, U0126 treatment blocked PDGF-induced glucose uptake and lactate production (Supplementary Fig. 5b), suggesting that activation of both EGFR and PDGFR promote the Warburg effect mediated by ERK1/2-dependent nuclear translocation of PKM2.
To determine the role of nuclear translocation of PKM2 in brain tumorigenesis, we intracranially injected U87/EGFRvIII, U87/EGFRvIII-PKM2 shRNA, and U87/EGFRvIII-PKM2 shRNA cells with reconstituted expression of WT rPKM2, rPKM2 S37A, WT Flag-PKM1, and Flag-PKM1 S37A into athymic nude mice. Depletion of PKM2 abrogated the growth of brain tumors, which was rescued by reconstituted expression of WT rPKM2, but not that of rPKM2 S37A (Fig. 6c, Supplementary Fig. 6a) or WT Flag-PKM1 (Supplementary Fig. 6b). Similar results on PKM2-dependent Warburg effect and tumorigenesis were also obtained using human primary GSC11 GBM cells (Supplementary Fig. 7a-c). In addition, depletion of GLUT1, LDHA, and PTB inhibited EGFRvIII-induced brain tumor growth (Supplementary Fig. 7d). Furthermore, intratumoral injection of the MEK inhibitor selumetinib inhibited tumor growth (Figs. 6d, e) and reduced ERK1/2 phosphorylation, PKM2 expression (Fig. 6f), and lactate production in tumor tissue (Fig. 6g). These results highlight the significance of ERK-dependent nuclear translocation of PKM2 and PKM2-dependent gene transcription in the Warburg effect and brain tumor development.
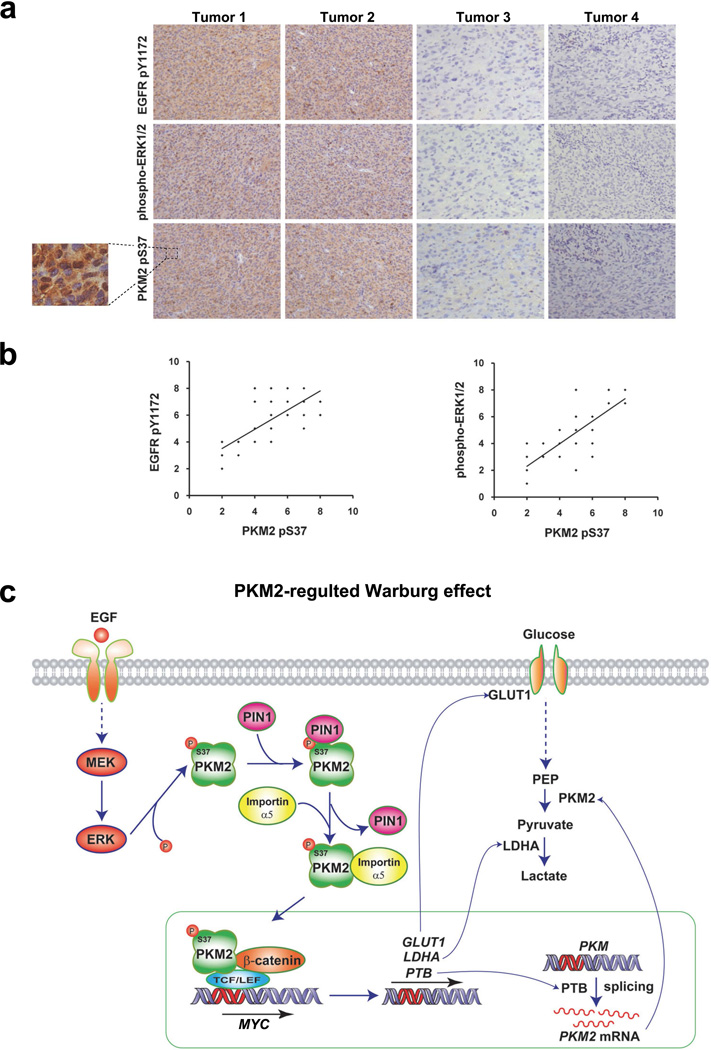
To further determine whether our findings have clinical relevance, we examined the activity of EGFR and ERK1/2 with PKM2 S37 phosphorylation in serial sections of 48 human primary GBM specimens by immunohistochemical (IHC) analyses by using antibodies with validated specificities (Supplementary Fig. 7e). As shown in Fig. 7a, the levels of EGFR and ERK1/2 activities correlated with the levels of PKM2 S37 phosphorylation. Quantification of the staining showed that this correlation was statistically significant among different specimens (Fig. 7b: r = 0.77, p < 0.001, top panel; r = 0.78, p < 0.001, bottom panel). These results strongly suggest that EGFR- and ERK1/2-induced PKM2 S37 phosphorylation occurs in human GBM.
Figure 7. Levels of PKM2 S37 phosphorylation correlate with EGFR and ERK1/2 activity in human GBM specimens.
a, b, IHC staining with anti–phospho-EGFR Y1172, anti–phospho-ERK1/2, and anti–phospho-PKM2 S37 antibodies was performed on 48 GBM specimens. Representative photos of four tumors are shown (a). Semi-quantitative scoring (using a scale from 0 to 8) was performed (Pearson product moment correlation test, r = 0.77, p < 0.001, top panel; r = 0.78, p < 0.001, bottom panel). Note that some of the dots on the graphs represent more than one specimen (i.e., some scores overlapped) (b).
c, A mechanism of PKM2-regulated Warburg effect. EGFR activation in human cancer cells induces PKM2 nuclear translocation, which is mediated by post-translational modification of PKM2 by ERK-dependent phosphorylation and PIN1-regulated cis-trans isomerization and subsequent importin α5 binding. Nuclear PKM2 is required for EGF-induced β-catenin transactivation, which in turn induces c-Myc expression and thereby upregulatesing GLUT1, LDHA, and PTB-dependent PKM2 expression. Nuclear PKM2-dependent expression of these glycolytic enzymes promotes the Warburg effect.
DISCUSSION
The Warburg effect, which was first reported in 1924,24 is a near-universal feature in tumors. High aerobic glycolysis by malignant tumors is used clinically to diagnose and monitor treatment responses of cancers by imaging uptake of 2-18F-deoxyglucose (FDG) with positron emission tomography (PET).1 Despite its widespread appreciation and clinical application, the mechanisms underlying the Warburg effect remain largely unclear.1, 25 Here, we demonstrate that nuclear translocation of PKM2 is a cause of the Warburg effect. Our study reveals an important mechanism underlying the Warburg effect: EGFR activation promotes aerobic glycolysis by means of a PKM2-dependent positive feedback loop on its own expression as well as the expression of GLUT1 and LDHA. EGFR activation results in nuclear translocation of PKM2, which is mediated by the ERK1/2-dependent phosphorylation of PKM2 S37 and consequently PIN1-catalized cis-trans isomerization of PKM2 for binding to importin α5. Nuclear PKM2 regulates β-catenin transactivation-dependent MYC transcription and, subsequently, the expression of GLUT1, LDHA, and PTB-mediated PKM2 expression. The elevated expression of these rate-limiting glycolytic genes plays a critical role in EGF-induced Warburg effect, featured by elevated glucose uptake and higher lactate production in the presence of oxygen, which leads to enhanced brain tumor development (Fig. 7c). Moreover, PKM2 S37 phosphorylation correlates with EGFR and ERK1/2 activity in human GBM, implying that PKM2 nuclear translocation play a significant role in the progression of these tumors in humans.
The mechanisms that accounted for the finding that PKM1 could not compensate for the loss of PKM2 in the Warburg effect and tumorigenesis were not well understood.8 We demonstrate here that PKM1, which lacks both a D domain and the NLS encoded by exon 10, did not bind activated ERK2 and was not phosphorylated at S37. PKM1 remained in cytosol after EGF treatment. Although PKM1 and PKM2 are both capable of converting phosphoenolpyruvate (PEP) to pyruvate, only PKM2 is able to translocate to the nucleus to conduct its unique nuclear functions. These functions include, but may not be limited to, transactivation of β-catenin phosphorylated at Y333; histone H3 phosphorylation and subsequent transcription of its downstream genes, such as MYC and CCND1; and promotion of cell cycle progression10,14 and the Warburg effect. Given that c-Myc and cyclin D1 play instrumental roles in cell proliferation, survival, and metabolism, our findings underscore the significant role of the metabolic and nonmetabolic functions of PKM2 in a very broad area of cellular activities. These findings also highlight the distinct functions of PKM2 relative to those of PKM1 in tumor development. Compared with the relatively well-studied cytosolic functions of PKM2, the illustrated nuclear functions of PKM2 in tumor development and its regulatory mechanisms for expression of glycolic enzymes and cyclin D1 provide important insights into PKM2-promoted tumor progression and targets for treating human cancer.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institute of Health grants 2R01CA109035 (Z.L.), R01GM068566 (X.C.), and CA16672 (Cancer Center Support Grant) and a research grant (RP110252; Z. L.) from the Cancer Prevention and Research Institute of Texas (CPRIT).
Footnotes
CONTRIBUTIONS
This study was conceived by Z.L.; Z.L. and W.Y. designed the study; W.Y., Y.Z., Y.X., H.J., and C.A.L. performed experiments; K.A. provided pathology assistance; X.C., F.G., and L.C. provided reagents and conceptual advice; Z.L. wrote the paper with comments from all authors.
REFERENCES
- 1.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 3.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 4.Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Majumder PK, et al. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med. 2004;10:594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- 6.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clower CV, et al. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. 107:1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 9.Vander Heiden MG, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 329:1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo W, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Molecular cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Z. Nonmetabolic functions of pyruvate kinase isoform M2 in controlling cell cycle progression and tumorigenesis. Chinese journal of cancer. 2012;31:5–7. doi: 10.5732/cjc.011.10446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang W, et al. PKM2 Phosphorylates Histone H3 and Promotes Gene Transcription and Tumorigenesis. Cell. 2012;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Z, Xu S. ERK1/2 MAP kinases in cell survival and apoptosis. IUBMB Life. 2006;58:621–631. doi: 10.1080/15216540600957438. [DOI] [PubMed] [Google Scholar]
- 16.Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- 17.Zheng Y, et al. FAK phosphorylation by ERK primes ras-induced tyrosine dephosphorylation of FAK mediated by PIN1 and PTP-PEST. Molecular cell. 2009;35:11–25. doi: 10.1016/j.molcel.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lufei C, Cao X. Nuclear import of Pin1 is mediated by a novel sequence in the PPIase domain. FEBS Lett. 2009;583:271–276. doi: 10.1016/j.febslet.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Jans DA, Xiao CY, Lam MH. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays. 2000;22:532–544. doi: 10.1002/(SICI)1521-1878(200006)22:6<532::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 20.Mason DA, Stage DE, Goldfarb DS. Evolution of the metazoan-specific importin alpha gene family. J Mol Evol. 2009;68:351–365. doi: 10.1007/s00239-009-9215-8. [DOI] [PubMed] [Google Scholar]
- 21.Lu Z, Ghosh S, Wang Z, Hunter T. Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of beta-catenin, and enhanced tumor cell invasion. Cancer Cell. 2003;4:499–515. doi: 10.1016/s1535-6108(03)00304-0. [DOI] [PubMed] [Google Scholar]
- 22.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 24.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 25.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.