Abstract
Quantitative neuropathologic methods provide information that is important for both research and clinical applications. The technological advancement of digital pathology and image analysis offers new solutions to enable valid quantification of pathological severity that is reproducible between raters regardless of experience. Using an Aperio ScanScope XT and its accompanying image analysis software, we designed algorithms for quantitation of amyloid and tau pathologies on 65 β-amyloid (6F/3D antibody) and 48 phospho-tau (PHF-1)-immunostained sections of human temporal neocortex. Quantitative digital pathologic data were compared with manual pathology counts. There were excellent correlations between manually counted and digitally analyzed neuropathologic parameters (R2 values 0.56-0.72). Data were highly reproducible among 3 participants with varying degrees of expertise in neuropathology (Intra-class correlation coefficient values >0.910). Digital quantification also provided additional parameters, including average plaque area, which show statistically significant differences when samples are stratified according to APOE allele status (average plaque area 380.9 μm2 in ApoE ε4 carriers vs. 274.4 μm2 for non-carriers, p < 0.001). Thus, digital pathology offers a rigorous and reproducible method for quantifying AD neuropathologic changes and may provide additional insight into morphologic characteristics that were previously more challenging to assess due to technical limitations.
Keywords: Alzheimer disease, Autopsy, Digital pathology, Image analysis, Neuropathology
INTRODUCTION
The association between the severity of Alzheimer disease neuropathologic changes (ADNCs) and cognitive impairment has long been established (1-19). The burden of neurofibrillary tangles (NFTs) and neuritic Aβ plaques (NPs), as well as their distribution throughout the cortex, are the bases for the Braak and Consortium to Establish a Registry for Alzheimer’s Disease (CERAD) staging schemes, respectively (1, 20, 21). Previous studies from the University of Kentucky Alzheimer’s Disease Center (UK-ADC) have demonstrated the utility of quantitative assessment of ADNCs. Manual quantitation of AD pathologies has been performed at the UK-ADC for more than 20 years. The quantitation of ADNCs helped to demonstrate the association of the different lesion type densities with cognitive impairment across the spectrum of disease from preclinical AD (11, 22) and mild cognitive impairment (5) to end-stage disease (23). Using the quantitative manual counts, the severity of ADNCs can also be correlated with possible risk factors such as diabetes (24) and to related diseases such as tangle-only pathology lacking amyloid plaques (25); they can also highlight differences in the anatomical distribution of ADNCs (26). Further, having quantitative neuropathologic assessment enabled us to develop a model of how each pathologic subtype contributes to cognitive impairment (27). In these studies, we previously found evidence of a “leveling off” of amyloid plaques in late-stage disease (18), but our confidence in these results was limited due to the technical drawbacks of the manual counting method (i.e. with caps on amyloid plaque counts, as described [23, 25, 28]). Work from other centers has also showed the benefits of quantitative assessment of AD neuropathologic changes (29-33). A common thread in these published studies is that the benefits of quantitative pathology can best be obtained if they are used in combination with detailed data about the patients’ non-AD pathological findings, medical histories, and cognitive status.
Although insights can be gained by quantitative assessment of ADNCs, manual methods as performed at UK-ADC are painstaking efforts that suffer from certain drawbacks. This process, which involves manual inspection and quantitation of 8 different areas of the brain processed via the modified Bielschowsky silver impregnation protocol, is extremely time-consuming. Also, despite our longstanding expertise in this area there is still suboptimal inter-rater reliability, which limits the ability to generalize our findings to those of other institutions. A final drawback includes the practice of limiting the counted area to 5 microscopic fields and implementing a cap value to plaques. While this is necessary to allow manual counting to be manageable, it introduces a source of error to our quantitation results.
The advent of digital slide scanners and software analysis packages is revolutionizing the practice of pathology and the research questions that can be addressed using pathological material. Digital slide scanners create digitized images of glass slides that can be viewed from a computer at varying magnifications and stored indefinitely. Such files can then be used for high-throughput, sophisticated image analyses, which offer quantitation capabilities that far exceed those that can be achieved manually in terms of reproducibility, unbiased image recognition algorithms and sheer volume. Previous studies have demonstrated some of the advantages of digital image analysis within AD research; either through montaged still images (34) or a variety of digital slide systems (35, 36). One such software package is Aperio’s Genie Histologic Pattern Recognition Tool (37). The Genie software has the ability to “learn” objects of our specification, in this case NFTs and NPs, based on a set of representative slides. Once this “teaching” is complete, the software has the ability to identify NFTs and NPs on any PHF-1-stained section in a very reproducible manner. This pattern recognition software has been successfully used in a variety of pathologic conditions, ranging from kidney failure to breast carcinoma (38, 39).
We took advantage of the strengths of digital pathology to update our quantitation protocol for ADNCs. In addition to parameters similar to those that we also obtain manually, we anticipated finding additional changes within plaque morphology that were impossible to quantify rigorously by manual methods. Additional data provided routinely by the algorithms include average plaque area and varying staining intensities. Such variables applied over many cases may reveal new insights into the pathological expression of the disease that may never have been possible through manual methods alone.
MATERIALS AND METHODS
Case Selection and Scanning
Slides and paraffin-embedded tissue from the superior and middle temporal gyri were analyzed from which manual quantitation of diffuse Aβ plaques (DPs), NPs, and NFTs had already been performed on modified Bielschowsky impregnations of near-serial sections via methods previously described in detail (23, 25, 28). Briefly, paraffin-embedded tissue was processed and 8-μm-thick sections were cut and processed using a modified Bielschowsky method (34). DPs were counted through a 10x objective (field size 2.35 mm2) in the 5 most involved fields, with an arbitrary “cap” at 250 plaques over all 5 fields. NPs were also counted in a similar manner but without a cap. NFTs were counted in the 5 most severely affected fields through a 20x objective (field size, 0.586 mm2). An arithmetic mean was then calculated for each parameter, resulting in a DP, NP, and NFT count/field.
For digital amyloid quantitation, a convenience sample of 65 cases was chosen based on the manual DP counts. Cases were selected with the goal of ensuring an adequate distribution of DP counts, ranging from 0 and 50 DP/field, i.e. not “anchored” in very high or very low DP counts. For each case, Aβ immunohistochemical (IHC) staining had already been performed via previously described methods (28), with a monoclonal NCL-β-amyloid 6F/3D antibody (Novocastra, Newcastle, UK). Further analyses of amyloid plaque size distributions were performed on those cases with known Apolipoprotein E (APOE) status (APOE ε4, n = 19; APOE non-ε4, n = 41), as well as a subset of these with a final Mini-Mental State Examination (MMSE) score of 27 to 30 (APOE ε4: n = 6; APOE non-ε4: n = 24).
For digital quantification of tau (neurofibrillary) pathologies, a convenience sample of 48 cases were chosen based on the Braak stage to ensure a full range of tau levels (4 cases Braak 0, 5 cases Braak I, 4 cases Braak II, 3 cases Braak III, 4 cases Braak IV, 14 cases Braak V, and 14 cases Braak VI). PHF-1 (a kind gift from Dr. Peter Davies, Bronx, NY) IHC was performed on the superior and middle temporal gyri. Additional analyses to compare digital amyloid and tau pathologies were performed on a subset of the cases in which both PHF-1 and AβIHC stains were performed. Cases with confounding pathologies (e.g. neocortical Lewy bodies, hippocampal sclerosis, vascular disease, frontotemporal dementias, and progressive supranuclear palsy) were excluded from this analysis, leaving a total of 24 cases across the full spectrum of ADNC severity. Slides were then loaded into an Aperio ScanScope XT and scanned at 40x magnification (0.25 μm/pixel) via the semi-automated method and then stored on a dedicated server. Slides were checked for image quality by using an Aperio ‘quality factor >90’ and visual inspection.
Analysis Region Selection
Whole slide analysis was impractical because it was overly time-consuming and severely biased by differential distribution of gray/white matter in different cases. To enable a more focused analysis that could be reproducible between users, a square analysis region (4 mm2) was created. For each case, that box was placed within the gray matter at the site of highest concentration of pathological findings. Subsequent boxes were then placed as far from the existing boxes as possible without overlapping other analysis regions. Areas of poor stain quality were avoided. To minimize analysis alterations caused by portions of folded tissue or foreign material, these were eliminated from the analysis boxes with the negative pen tool. In the amyloid analyses, meningeal vessels were also excluded by the negative pen tool, so as to limit the burden and plaque analyses to parenchymal content. Parenchymal vessels that were involved by amyloid angiopathy were included in the burden analysis; these vessels were not picked up by the plaque density algorithm.
Analysis Workflow
Once the analysis regions were selected, the next step was to identify the appropriate quantitation algorithms to use. For cases in which there was no need to subtype pathologies within a given immunostain, for example, with the AβIHC stain, the pattern recognition software adds an unnecessary and time-consuming step. Therefore, analysis of either amyloid plaque density or overall amyloid burden could be performed by simple modification of the pre-existing algorithm templates. In contrast, within a single PHF-1 immunostain we wanted to quantitate NFTs and NPs separately. The pattern recognition software allows each of these structures to be highlighted in isolation. Once identified, they could then be quantitated by a variety of mechanisms. A representative schematic of the digital workflow is in Figure 1.
Figure 1.
(A, B) Analytic workflow for high-throughput quantitation of Alzheimer Disease-type neuropathologic changes (A), with a more detailed explanation of the image analysis steps (B). IHC, immunohistochemistry; NFTs, neurofibrillary tangles; NP, neuritic Aβ plaques; PHF-1, phospho-tau immunostain.
β-Amyloid Quantitation
Two different parameters were calculated for amyloid pathologies: an overall amyloid burden and an amyloid plaque density. For the overall amyloid burden, the Aperio Image Analysis Toolbox Positive Pixel Count (PPC), version 9.1, was used. Modifications were made to the input parameters of the algorithm to optimize for our staining protocol based on comparison of sample analysis markups with manual counting results by visual inspection in multiple areas upon 3 representative slides (Table, Supplemental Digital Content 1, http://links.lww.com/NEN/A398). The modified PPC was then run on the 10 4-mm2 boxes selected on each slide as described above. The amyloid burden was calculated by adding up all of the weak (1+, yellow), positive (2+, orange), and strong (3+, red) pixels from the data and dividing by the overall analysis area (40 mm2). For the amyloid plaque density, the Aperio Image Analysis Toolbox Nuclear algorithm, version 9.1, was used with subsequent modifications, derived in a similar manner to the amyloid burden (Table ,Supplemental Digital Content 2, http://links.lww.com/NEN/A399). This modified algorithm was then run on the same 10 boxes chosen for the amyloid burden analysis. The amyloid plaque density was calculated by adding up all the weakly (1+, yellow), moderately (2+, orange), and strongly (3+, red) positive ‘nuclei’ and dividing by the overall analysis area (40 mm2).
Tau Quantitation
Three different parameters were quantified for tau pathologies: NFT density, NP burden, and overall tau burden. For the NFT and NP quantitation, a single Genie algorithm was first developed to separate NFTs and NPs from the background. Approximately 10 to 15 representative samples for each of the 3 classes (NFTs, NPs, and background) were highlighted using a digital pen tool. In selecting sample structure, care was taken to make sure the range of possible morphologies was covered for any given pathology (e.g. making sure that multiple areas with white matter, vasculature, or meningeal tissue were included for the ‘background’ training). Then the Genie training algorithm, consisting of 2000 iterations, was run with the apparent 5X magnification. The algorithm was then evaluated by visual inspection in an unmarked area to assess its accuracy. This process was repeated until the algorithm, or “classifier” could identify both structures to our satisfaction. The NFT/NP Genie algorithm was then used in subsequent analyses for quantification.
The NFT density was calculated via a modified nuclear algorithm, version 9.1, with our NFT/NP Genie classifier limited to NFTs. An input parameter that was important to address was the minimum nuclear cutoff size (μm2). To find the nuclear size that would both maximize the number of NFTs and minimize the number of non-NFT particles counted (i.e. oligodendroglia and glial inclusions), the algorithm was run multiple times on a subset of 10 cases using a series of ‘minimum nuclear size’ cutoffs ranging from 35 to 100 μm2. The NFT density for each cutoff was calculated and then compared to both the manual counts and the final MMSE scores. Using this method, it was found that a minimum nuclear size of 40 μm2 provided the most accurate result. The algorithm input parameters are listed in Table, Supplemental Digital Content 3, http://links.lww.com/NEN/A400. The modified nuclear algorithm was run on 10 4-mm2 boxes selected by the method stated above within the gray matter in the PHF-1-immunostained sections. The NFT density was calculated by dividing the total number of nuclei counted by the overall analysis area (40 mm2).
For the NP burden, the Positive Pixel Count (PPC), version 9.1, was used with our NFT/NP Genie classifier limited to NPs and modified for our staining protocol via visual inspection. The algorithm input parameters are listed in Table, Supplemental Digital Content 4, http://links.lww.com/NEN/A401. The modified PPC was then run on the same boxes selected for NFT analysis. The NP burden was calculated by adding up all the weak (1+, yellow), positive (2+, orange), and strong (3+, red) pixels from the data and dividing by the overall analysis area (40 mm2).
The overall tau burden was determined using a PPC algorithm, version 9.1, similar to the amyloid burden quantitation, but with stain-specific modifications based on visual inspection. The algorithm input parameters are listed in Table, Supplemental Digital Content 5, http://links.lww.com/NEN/A402. The modified PPC was then run on the same 10 4-mm2 boxes selected for the other tau analyses. The tau burden was calculated by dividing the “strong” (3+, red) pixels by the overall analysis area (40 mm2).
Inter-observer Variability
To test the reproducibility of the algorithms, a subset of 30 Aβ- and 30 PHF-1-stained sections were given to 2 additional individuals with varying degrees of neuropathology expertise, i.e. the brain bank coordinator involved in the manual counting of these pathologies and an undergraduate student with no neuropathology experience. Each individual was given the same set of digital slides, along with instructions for analyses as described above, with no pre-existing analysis boxes selected. This method was only performed a single time. All 5 digital analyses were then run on these selected regions and compared to the original data received from the fields selected by a neuropathologist.
Statistical Analysis
Ordinary least-squares regression was conducted to assess the relationship between manual and digital counts. Group comparisons involved paired t-tests, with graphed results of amyloid plaque morphology shown as arithmetic mean ± SEM, using MS Excel. Intra-class correlation coefficients were calculated to assess inter-rater reliability for the 3 users. One-way analysis of variance tables (with autopsy case as the factor) were constructed for each measure using PROC GLM in SAS/STAT 9.3, and estimated Intra-class correlation coefficients and 95% confidence intervals were calculated based on the mean squared errors for within and between subjects.
RESULTS
After scanning, the next step was to identify the regions for analysis. Although initial analyses were run on the entire slide, it was found to be inefficient, taking hours to identify one parameter when the Genie software was used. Limiting the analysis area to a preset region of known size and shape led to far faster analyses. In order to balance accuracy and efficiency, a series of 30 4-mm2 annotation boxes were placed in the manner described above (Fig. 2A) on 15 representative Aβ-stained slides. Amyloid quantitation via the modified nuclear algorithm was then performed in each of the square regions. By adding the plaque numbers within each box together in a sequential manner and then dividing by the total area analyzed (e.g. Box 1 plaque #/4 mm2, Box 1+2 plaque #/8 mm2, Box 1+2+3 plaque #/12 mm2, etc.), an additive amyloid plaque density was calculated. When these additive densities were graphed with respect to box number, it became clear that the density approached an asymptote (Fig. 2B). In all cases tested this asymptote was approached by 8 to 10 analysis boxes. To ensure that the box selection procedure was not significantly biased, we also performed the same analyses on a subset of 5 cases, choosing the first square in a region with the least involvement of pathology. In all cases, the analyses approached the same asymptote as was reached in the original selection. A second algorithm, the overall amyloid burden, was also tested and showed similar results. Because of these findings, we used 10 4-mm2 boxes for each slide to quantitate amyloid and tau pathologies. This dropped the analysis time from an average of 1 and 6.5 hours to 5 and 45 minutes for each amyloid and tau algorithm, respectively.
Figure 2.
Determination of appropriate analysis area. (A) A 4-mm2-square annotation was initially placed in the gray matter region with highest density of pathology (*). Subsequent regions were then randomly selected, up to 30 regions. The plaque density was then calculated for each individual box and then sequentially averaged together in a step-wise fashion. (B) The average density approached an asymptote by 10 boxes in all test cases studied. Scale bar = 2 mm.
Amyloid Quantitation
Both the amyloid burden and amyloid plaque density (Fig. 3) correlated strongly with manual DP counts, with R2 values of 0.62 and 0.72, respectively (Table 1, Fig. 4A). By altering our quantitation protocol from the modified Bielschowsky to amyloid IHC, the ability to distinguish DPs from NPs during amyloid quantitation was lost. Because the amyloid IHC should pick up both plaque types, the digital parameters were correlated to the sum of the manually derived DP and NP counts. This resulted in similar correlations between our methods (R2 values 0.60 and 0.67 for amyloid burden and plaque density, respectively) (Table 1, Fig. 4B). An additional confounding issue involved the artifactual capping of manual counts at 50 plaques/field. When a similar cap was employed on the digitally derived numbers, purely for the sake of correlation, there was marked improvement of correlation between the amyloid plaque density and manual DP counts, with an R2 value of 0.82, as well as with the sum of DP and NP manual counts (R2 = 0.79). Finally, the 2 separate digital amyloid parameters were correlated to each other to confirm concordance, which was found to be the case (R2 = 0.76, Fig. 4C).
Figure 3.
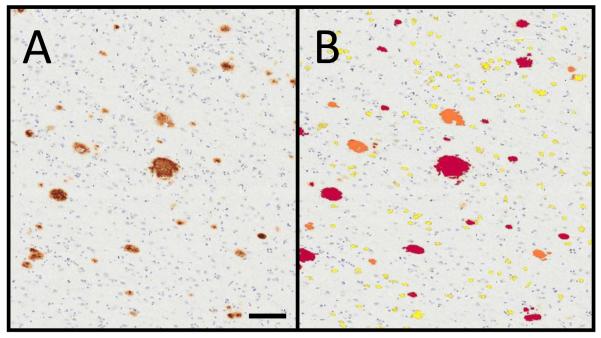
Amyloid quantification. (A) β-amyloid immunostain was performed on the superior and middle temporal gyri (SMTG) of all cases. (B) Digital analysis of amyloid plaque density in number of diffuse Aβ plaques (DP #)/mm2 via a modified nuclear algorithm. In addition to quantitation of the amyloid plaques, the digital analysis also separated the plaques by varying degrees of intensity (0: blue, 1+: yellow; 2+: orange; 3+: red). Scale bar = 50 μm.
Table 1. Ordinary Least Squares Regression R2 Correlation Coefficients for Comparisons Between Manual Counts and Digital Parameters.
| Digital Amyloid (n=65) | Correlation with Manual DPs R2 (p) values |
Correlation with Manual DPs + NPs R2 (p) values |
|---|---|---|
| Amyloid Burden | 0.62 (<0.001) | 0.60 (<0.001) |
| Amyloid Plaque Density |
0.72 (0.001) | 0.67 (0.09) |
| Digital Tau (n=48) | Manual NFTs R2 values |
Manual NPs R2 values |
| NFT Density | 0.57 (0.04) | -- |
| Tau Burden | 0.56 (NA) | 0.27 (NA) |
| NP Burden | -- | 0.31 (<0.001) |
DPs, diffuse plaques; NPs, neuritic plaques; NFTs, neurofibrillary tangles; NA, not applicable.
Figure 4.
Amyloid quantification correlations (n = 65). (A) There was good correlation between the digital quantification via digital amyloid plaque density and the manually derived diffuse Aβ plaque (DP) counts. (B) Inclusion of the manually counted neuritic Aβ plaques (NPs), which were also picked up by the amyloid immunohistochemistry, also showed excellent correlation. (C) Correlation of the digital plaque density to digital amyloid burden.
Tau Quantitation
Correlations between tau pathologies were more variable (Table 1). Representative images of the Genie NFT/NP recognition and the subsequent tau algorithms are shown in Figures 5 and 6, respectively. Digital NFT density correlated well with the manual NFT counts (R2 = 0.57, Fig. 7). Whereas there was no official capping policy of tau pathologies, the manual NFTs were never more than 50 NFTs/field, despite the Aperio density often being significantly larger. We hypothesize that a bias due to counting fatigue likely affected the manual data. For correlation purposes, the Aperio data were capped at 50 NFTs/mm2, as was done on the amyloid plaque density. This resulted in a significant improvement in the correlation, with an R2 value of 0.7369. Overall tau burden showed good correlation with manual NFT counts (R2 = 0.56) and the digitally derived NP burden (R2 = 0.73) and NFT density (R2 = 0.86), but weaker correlation with the manual NP counts (R2 = 0.27). NP burden showed the weakest correlation with manual counts, with an R2 value of 0.31. Even implementing a cap (as with the NFT and amyloid plaque densities), there was only a mild improvement in correlation (R2 = 0.37) (Fig. 8). By visual inspection, the NP burden algorithm appeared to appropriately highlight NPs. Further visual comparison between the original silver impregnation preparations (on which the manual counts had been obtained) and the new PHF-1 immunostain highlighted the markedly increased sensitivity of the PHF-1, which identified more NPs than were seen on with the Bielschowsky method (Fig. 8B, C).
Figure 5.
Use of Genie Histopathology Pattern Recognition Tool to identify tau pathologies on phospho-tau (PHF-1) immunohistochemistry. (A) An area selected for Genie analysis contained both neuritic Aβ plaques (NPs) (arrowheads) and neurofibrillary tangles (NFTs) (arrows). (B) The crafted algorithm highlighted these structures automatically (NFTs green, NPs gray). Scale bar = 25 μm.
Figure 6.
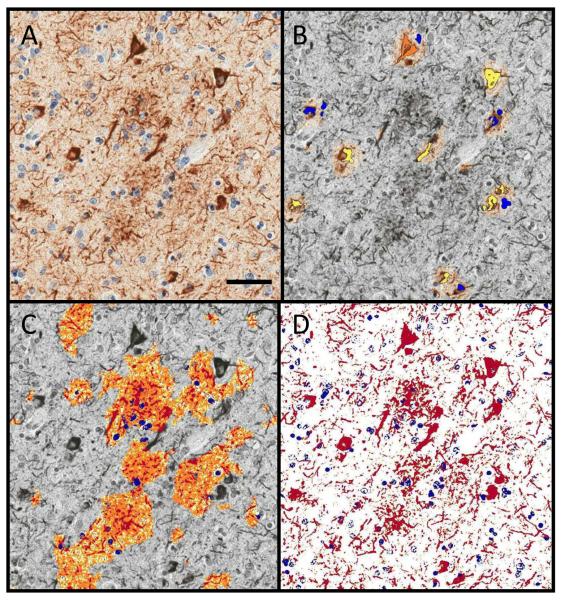
Digital quantification of tau pathologies. (A) Analyses were performed on phospho-tau (PHF-1)-immunostained sections. (B) After using the crafted Genie NFT/NP algorithm to isolate the neurofibrillary tangles (NFTs), the NFT density (NFTs/mm2) was determined by a modified nuclear algorithm, with NFTs ranging in staining intensities from 0+ (blue), 1+ (yellow), 2+ (orange), and 3+ (red). (C) In a similar manner, the neuritic Aβ plaque (NP) burden was calculated by first using the same crafted Genie algorithm to isolate the NPs and then running a modified positive pixel count to highlight each pixel based on a similar staining intensity as above. (D) An overall tau burden was also calculated (red: positive immunohistochemical (IHC) staining; blue: negative IHC staining). Scale bar = 25 μm.
Figure 7.
Neurofibrillary tangle (NFT) quantification correlations (n = 48). The correlation between digital NFT density and the manual NFT counts are shown.
Figure 8.
Neuritic Aβ plaque (NP) quantitation correlation (n = 48). (A) Even with a virtual cap, the correlation between the digital NP burden and the manual NP counts was below that of the neurofibrillary tangles (NFTs). Upon review of the cases with high digital NP burden and low manual NP counts, much of this was thought to be due to the increased sensitivity of the phospho-tau (PHF-1) immunostain when compared to the modified Bielschowsky method. (B, C) A representative 10x field in the superior and middle temporal gyri (SMTG) of the case highlighted with an arrow in (A). Those that were likely counted as diffuse Aβ plaques (DPs) on the silver stain (B) were actually NPs by PHF-1 IHC (C). Scale bar = 50 μm.
Inter-observer-Variation
The observed agreement among the 3 users was very good for all measures assessed (Table 2). Intra-class correlation coefficients ranged from 0.910 (95% confidence interval 0.855, 0.945) for the NFT density to 0.986 (95% confidence interval 0.977, 0.992) for the tau burden. R2 correlation coefficients were also calculated via least squares regression and found to range from 0.93 to 0.98 between the individual users.
Table 2. Intra-Class Correlation Coefficients with 95% Confidence Intervals to Assess Inter-Rater Reliability for 3 Users.
| Measure | Estimated ICC (95% Confidence Interval) |
|---|---|
| Amyloid Burden | 0.973 (0.956, 0.984) |
| Amyloid Plaque Density | 0.974 (0.957, 0.984) |
| Tau Burden | 0.986 (0.977, 0.992) |
| NFT Density | 0.910 (0.855, 0.945) |
| NP Density | 0.970 (0.951, 0.982) |
ICC, intra-class correlation coefficient; NP, neuritic plaque; NFT, neurofibrillary tangles
Additional Analyses
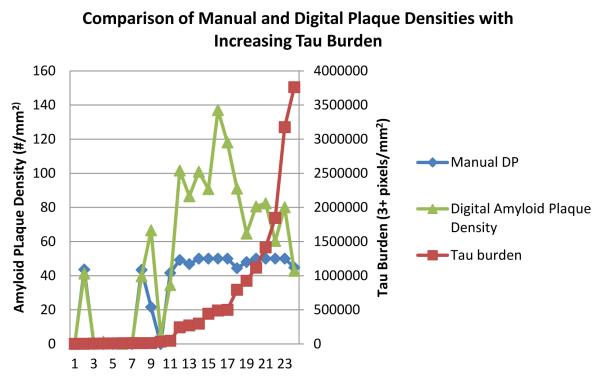
By examining subsets of the above cases, additional comparisons were made using information gathered from the above algorithms. The first subset of cases was limited to those with only ADNCs and had both PHF-1 and Aβ immunostaining performed (n = 24). Digital amyloid plaque densities (as well as the manual sum of DPs and NPs) were examined as a function of increasing tau burden. The results showed the digital amyloid plaque density began decreasing when the tau burden approached 500,000 3+pixels/mm2 (Fig. 9); this trend was not identified through the manual counts due to the capping procedure.
Figure 9.
Comparison of varying amyloid plaque densities as overall tau burden increases (n = 24). With the increased counting capabilities offered by digital quantitation, it was clear that plaque number tended to decrease as tau burden increased, a trend that could not be seen using our manual data alone. DP = diffuse Aβ plaques.
The second additional analysis involved the comparison of amyloid plaque area for cases with known APOE allele status. When separated, 2 two cohorts based on the presence (n = 7) or absence (n = 41) of the APOE ε4 allele, those cases with at least one APOE ε4 allele had statistically larger plaque sizes (mean = 380.9 μm2) vs. those lacking the APOE ε4 allele (mean = 274.4 μm2, p < 0.001) (Fig. 10A). This finding maintained significance even when the analysis was limited to those cases with a final MMSE score of 27 to 30. Those with APOE ε4 (n = 5) had an average plaque size of 333.20 μm2, whereas those without APOE ε4 alleles (n = 25) had a significantly smaller plaque size (233.48 μm2. p = 0.023) (Fig. 10B).
Figure 10.
Examination of amyloid plaque area in patients with known APOE allele status (APOE ε4 n = 7; non-APOE ε4 = 41). (A) Patients with APOE ε4 alleles had a significantly larger average plaque area vs. those without an APOE ε4 allele. (B) This finding holds true even when the degree of cognitive impairment is controlled by limiting the final Mini-Mental State Examination (MMSE) scores to 27 to 30 (APOE ε4 n= 5; non-APOE ε4 = 25) (B). Graphs are mean ± SE; *p < 0.001, **p < 0.023.
DISCUSSION
The goals of this study were to establish a set of computer algorithms to replace manual quantitation at our center and to highlight the possible benefits that digital pathology may provide over the traditional counting methods at other research centers. The results indicate that amyloid plaque parameters, NFT density, and overall tau burden correlate strongly with the manual count data that have been used for years and in dozens of published studies at the UK-ADC. We also were able to elucidate new features of plaque morphology that would be practically impossible to evaluate using most other methods.
Digital pathology offers multiple benefits that surpass both semiquantitative methods and manual counts. Digital algorithms offer superior reproducibility and higher throughput performance that enables a far more standardized approach to the assessment of ADNCs. If individual centers begin to use a standard algorithm for quantitation, results could be used across institutions, thereby exponentially increasing the statistical power available to all centers involved. The digital approach is relatively efficient when it comes to manpower. While it does take additional time to scan the slide and set up the analysis windows (approximately 45 minutes to prepare and scan at 40x via the semi-automated method and an additional 5 to 10 minutes to select the analysis windows per slide), the bulk of the analysis work is done by the server alone. These analyses can be set up during the day and then allowed to run overnight without interruption. In addition, neuropathologic expertise is not a requirement for this method. Anyone at any level of expertise can be taught to select analysis areas in a few minutes. Because the analysis algorithms are held constant, regardless of who sets up the windows, the data will be consistent. This could be expanded to involve algorithm sharing between institutions and thus improve the inter-rater reliability between the different research centers to help standardize the field of quantitative ADNCs.
Prior work showed that digital pathology could help elucidate different subtypes of AD cases based on quantifiable patterns of ADNCs (29). We confirmed that digital pathology can be used to discover new and interesting trends that we were not able to identify before at our center despite decades of work in quantitative assessment of ADNCs. While our manually quantified ADNC numbers suggested that amyloid plaque burden leveled off with increasing pathology, we failed to identify that it actually decreases with increasing tau burden by our manual methods alone. Intuitively, it makes sense that individuals with ApoE ε4 alleles would have more amyloid plaque pathology (40-43), and this might correspond with having larger amyloid plaques; however, using manual counts, we could not demonstrate this trend reliably. These data may enable other new insights into the pathologic changes seen in AD.
Despite the benefits of the digital pathology methods, they also entail potential drawbacks. The up-front cost of digital pathology could be problematic for some centers and hospitals; the system described here cost almost $300,000 in 2010. Whole slide analysis was the theoretical goal; however, the massive amount of analysis time this required made this impractical. Our protocol still quantitates far more area than was previously examined manually. The most problematic pathology to quantitate were the NPs, as indicated by the weaker correlations between digitally and manually counted numbers. Due to the heterogeneous nature of the NPs, single plaques could not be counted individually, as they could by manual methods. Thus, we had to convert to an NP burden, which gives a picture of overall NP surface area, rather than individual plaque number. We also switched our staining protocol from the modified Bielschowsky histochemical method to the PHF-1 IHC stain. This stain has a better sensitivity for tau-related abnormalities (44-47), and thus highlighted many more NPs than were seen with the silver method. It is likely this combination of variables led to the weaker correlation seen between our NP parameters.
Digital pathology offers a valuable resource for quantitative pathology in neurodegenerative disease. With these algorithms, more AD-type pathology can be counted faster and more reproducibly than by manual inspection alone. In addition, more parameters can be rigorously examined, from staining intensity to plaque size and more. As our use of this technology advances, it will open up new understanding of the pathologies in human brain aging.
Supplementary Material
ACKNOWLEDGMENT
We are deeply grateful to all of the participants in our longitudinal aging study and to the patients with Alzheimer disease in our Alzheimer’s Disease Center’s research clinic.
This study was supported by Grants P30 AG028383 and S10RR026489 from the National Institutes of Health, Bethesda, MD.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Braak H, Braak E. Neuropathological staging of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–59. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 2.Mayeux R, Hyslop PS. Alzheimer’s disease: Advances in trafficking. Lancet Neurol. 2008;7:2–3. doi: 10.1016/S1474-4422(07)70298-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg L, McKeel DW, Jr, Miller JP, et al. Clinicopathologic studies in cognitively healthy aging and Alzheimer’s disease: Relation of histologic markers to dementia severity, age, sex, and apolipoprotein E genotype. Arch Neurol. 1998;55:326–35. doi: 10.1001/archneur.55.3.326. [DOI] [PubMed] [Google Scholar]
- 4.Braak H, Braak E. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol Aging. 1995;16:271–8. doi: 10.1016/0197-4580(95)00021-6. [DOI] [PubMed] [Google Scholar]
- 5.Markesbery WR, Schmitt FA, Kryscio RJ, et al. Neuropathologic substrate of mild cognitive impairment. Arch Neurol. 2006;63:38–46. doi: 10.1001/archneur.63.1.38. [DOI] [PubMed] [Google Scholar]
- 6.Green MS, Kaye JA, Ball MJ. The Oregon brain aging study: Neuropathology accompanying healthy aging in the oldest old. Neurology. 2000;54:105–13. doi: 10.1212/wnl.54.1.105. [DOI] [PubMed] [Google Scholar]
- 7.Haroutunian V, Purohit DP, Perl DP, et al. Neurofibrillary tangles in nondemented elderly subjects and mild Alzheimer disease. Arch Neurol. 1999;56:713–18. doi: 10.1001/archneur.56.6.713. [DOI] [PubMed] [Google Scholar]
- 8.Sonnen JA, Larson EB, Crane PK, et al. Pathological correlates of dementia in a longitudinal, population-based sample of aging. Ann Neurol. 2007;62:406–13. doi: 10.1002/ana.21208. [DOI] [PubMed] [Google Scholar]
- 9.Arriagada PV, Marzloff K, Hyman BT. Distribution of Alzheimer-type pathologic changes in nondemented elderly individuals matches the pattern in Alzheimer’s disease. Neurology. 1992;42:1681–88. doi: 10.1212/wnl.42.9.1681. [DOI] [PubMed] [Google Scholar]
- 10.Bancher C, Jellinger K, Lassmann H, et al. Correlations between mental state and quantitative neuropathology in the Vienna longitudinal study on dementia. Eur Arch Psychiatry Clin Neurosci. 1996;246:137–46. doi: 10.1007/BF02189115. [DOI] [PubMed] [Google Scholar]
- 11.Nelson PT, Jicha GA, Schmitt FA, et al. Clinicopathologic correlations in a large Alzheimer disease center autopsy cohort: Neuritic plaques and neurofibrillary tangles do count when staging disease severity. J Neuropathol Exp Neurol. 2007;66:1136–46. doi: 10.1097/nen.0b013e31815c5efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guillozet AL, Weintraub S, Mash DC, et al. Neurofibrillary tangles, amyloid, and memory in aging and mild cognitive impairment. Arch Neurol. 2003;60:729–36. doi: 10.1001/archneur.60.5.729. [DOI] [PubMed] [Google Scholar]
- 13.Imhof A, Kovari E, von Gunten A, et al. Morphological substrates of cognitive decline in nonagenarians and centenarians: A new paradigm? J Neurol Sci. 2007;257:72–9. doi: 10.1016/j.jns.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 14.Cummings BJ, Pike CJ, Shankle R, et al. Beta-amyloid deposition and other measures of neuropathology predict cognitive status in Alzheimer’s disease. Neurobiol Aging. 1996;17:921–33. doi: 10.1016/s0197-4580(96)00170-4. [DOI] [PubMed] [Google Scholar]
- 15.Davis PC, Gearing M, Gray L, et al. The CERAD experience, Part VIII: Neuroimaging-neuropathology correlates of temporal lobe changes in Alzheimer’s disease. Neurology. 1995;45:178–9. doi: 10.1212/wnl.45.1.178. [DOI] [PubMed] [Google Scholar]
- 16.Koepsell TD, Kurland BF, Harel O, et al. Education, cognitive function, and severity of neuropathology in Alzheimer disease. Neurology. 2008;70:1732–39. doi: 10.1212/01.wnl.0000284603.85621.aa. [DOI] [PubMed] [Google Scholar]
- 17.Braak H, Braak E, Bohl J. Staging of Alzheimer-related cortical destruction. Eur Neurol. 1993;33:403–8. doi: 10.1159/000116984. [DOI] [PubMed] [Google Scholar]
- 18.Nelson PT, Braak H, Markesbery WM. Neuropathology and cognitive impairment in Alzheimer disease: A complex but coherent relationship. J Neuropathol Exp Neurol. 2008;68:1–14. doi: 10.1097/NEN.0b013e3181919a48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jicha GA, Carr SA. Conceptual Evolution in Alzheimer’s disease: Implications for Understanding the clinical phenotype of progressive neurodegenerative disease. J Alz Disease. 2010;19:253–72. doi: 10.3233/JAD-2010-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirra SS, Heyman A, McKeel D, et al. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD) II. Standardization of the neuropathological assessment of Alzheimer’s disease. Neurol. 1991;41:479–86. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- 21.Mirra SS. The CERAD neuropathology protocol and consensus recommendations for the postmortem diagnosis of Alzheimer’s disease: A commentary. Neurobiol Aging. 1997;18:S91–94. doi: 10.1016/s0197-4580(97)00058-4. [DOI] [PubMed] [Google Scholar]
- 22.icha GA, Abner EL, Schmitt FA, et al. Preclinical AD Workgroup staging: pathological correlates and potential challenges. Neurobiol Aging. 2012;33:622.e621–622.e616. doi: 10.1016/j.neurobiolaging.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abner EL, Kryscio RJ, Schmitt FA, et al. “End-stage” neurofibrillary tangle pathology in preclinical Alzheimer’s disease: fact or fiction? J Alzheimers Dis. 2011;25:445–53. doi: 10.3233/JAD-2011-101980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson PT, Smith CD, Abner EA, et al. Human cerebral neuropathology of Type 2 diabetes mellitus. Biochim Biophys Acta. 2009;1792:454–69. doi: 10.1016/j.bbadis.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson PT, Abner EL, Schmitt FA, et al. Brains with medial temporal lobe neurofibrillary tangles but no neuritic amyloid plaques are a diagnostic dilemma but may have pathogenetic aspects distinct from Alzheimer disease. J Neuropathol Exp Neurol. 2009;68:774–84. doi: 10.1097/NEN.0b013e3181aacbe9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson PT, Abner EL, Scheff SW, et al. Alzheimer’s-type neuropathology in the precuneus is not increased relative to other areas of neocortex across a range of cognitive impairment. Neurosci Lett. 2009;450:336–9. doi: 10.1016/j.neulet.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson PT, Abner EL, Schmitt FA, et al. Modeling the association between 43 different clinical and pathological variables and the severity of cognitive impairment in a large autopsy cohort of elderly persons. Brain Pathol. 2010;20:66–79. doi: 10.1111/j.1750-3639.2008.00244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson PT, Jicha GA, Schmitt FA, et al. Clinicopathologic correlations in a large Alzheimer disease center autopsy cohort: neuritic plaques and neurofibrillary tangles “do count” when staging disease severity. J Neuropathol Exp Neurol. 2007;66:1136–1146. doi: 10.1097/nen.0b013e31815c5efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray ME, Graff-Radford NR, Ross OA, et al. Neuropathologically defined subtypes of Alzheimer’s disease with distinct clinical characteristics: a retrospective study. Lancet Neurol. 2010;10:785–96. doi: 10.1016/S1474-4422(11)70156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chibnik LB, Shulman JM, Leurgans SE, et al. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann Neurol. 2011;69:560–9. doi: 10.1002/ana.22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennett DA, Wilson RS, Schneider JA, et al. Apolipoprotein E epsilon4 allele, AD pathology, and the clinical expression of Alzheimer’s disease. Neurology. 2003;60:246–52. doi: 10.1212/01.wnl.0000042478.08543.f7. [DOI] [PubMed] [Google Scholar]
- 32.Robinson JL, Geser F, Corrada MM, et al. Neocortical and hippocampal amyloid-beta and tau measures associate with dementia in the oldest-old. Brain. 2011;134:3708–15. doi: 10.1093/brain/awr308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Armstrong RA, Cairns NJ. Size frequency distribution of the beta-amyloid (abeta) deposits in dementia with Lewy bodies with associated Alzheimer’s disease pathology. Neurol Sci. 2009;30:471–7. doi: 10.1007/s10072-009-0135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Byrne UT, Ross JM, Faull RL, et al. High-throughput quantification of Alzheimer’s disease pathological markers in the post-mortem human brain. J Neurosci Methods. 2009;176:298–309. doi: 10.1016/j.jneumeth.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Samaroo HD, Opsahl AC, Schreiber J, et al. High throughput object-based image analysis of beta-amyloid plaques in human and transgenic mouse brain. J Neurosci Methods. 2012;204:179–88. doi: 10.1016/j.jneumeth.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Chubb C, Inagaki Y, Sheu P, et al. BioVision: an application for the automated image analysis of histological sections. Neurobiol Aging. 2006;27:1462–76. doi: 10.1016/j.neurobiolaging.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 37.Staniszewski W. Virtual microscopy, data management and image analysis in Aperio ScanScope system. Folia Histochem Cytobiol. 2009;47:699–701. doi: 10.2478/v10042-009-0115-y. [DOI] [PubMed] [Google Scholar]
- 38.Klapczynski M, Gagne GD, Morgan SJ, et al. Computer-assisted imaging algorithms facilitate histomorphometric quantification of kidney damage in rodent renal failure models. J Pathol Inform. 2012;3:20. doi: 10.4103/2153-3539.95456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laurinavicius A, Laurinaviciene A, Ostapenko V, et al. Immunohistochemistry profiles of breast ductal carcinoma: factor analysis of digital image analysis data. Diagn Pathol. 2012;7:27. doi: 10.1186/1746-1596-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grimmer T, Tholen S, Yousefi BH, et al. Progression of cerebral amyloid load is associated with the apolipoprotein E epsilon4 genotype in Alzheimer’s disease. Biol Psychiatry. 2010;68:79–84. doi: 10.1016/j.biopsych.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caselli RJ, Walker D, Sue L, et al. Amyloid load in nondemented brains correlates with APOE e4. Neurosci Lett. 2010;473:168–71. doi: 10.1016/j.neulet.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Josephs KA, Tsuboi Y, Cookson N, et al. Apolipoprotein E epsilon 4 is a determinant for Alzheimer-type pathologic features in tauopathies, synucleinopathies, and frontotemporal degeneration. Arch Neurol. 2004;61:1579–84. doi: 10.1001/archneur.61.10.1579. [DOI] [PubMed] [Google Scholar]
- 43.Schmechel DE, Saunders AM, Strittmatter WJ, et al. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc Natl Acad Sci USA. 1993;90:9649–53. doi: 10.1073/pnas.90.20.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alafuzoff I, Arzberger T, Al-Sarraj S, et al. Staging of neurofibrillary pathology in Alzheimer’s disease: a study of the BrainNet Europe Consortium. Brain Pathol. 2008;18:484–96. doi: 10.1111/j.1750-3639.2008.00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alafuzoff I, Pikkarainen M, Arzberger T, et al. Inter-laboratory comparison of neuropathological assessments of beta-amyloid protein: a study of the BrainNet Europe consortium. Acta Neuropathol. 2008;115:533–46. doi: 10.1007/s00401-008-0358-2. [DOI] [PubMed] [Google Scholar]
- 46.Alafuzoff I, Pikkarainen M, Al-Sarraj S, et al. Interlaboratory comparison of assessments of Alzheimer disease-related lesions: a study of the BrainNet Europe Consortium. J Neuropathol Exp Neurol. 2006;65:740–57. doi: 10.1097/01.jnen.0000229986.17548.27. [DOI] [PubMed] [Google Scholar]
- 47.Greenberg SG, Davies P. A preparation of Alzheimer paired helical filaments that displays distinct tau proteins by polyacrylamide gel electrophoresis. Proc Natl Acad Sci USA. 1990;87:5827–31. doi: 10.1073/pnas.87.15.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.