Abstract
Ethnopharmacological relevance
Eclipta alba, traditionally known as bhringraj, has been used in Ayurvedic medicine for more than 1000 years in India. It is used for the treatment of infective hepatitis, liver cirrhosis, liver enlargement and other ailments of liver and gall bladder in India.
Aim of the study
To evaluate anti-hepatitis C virus activity present in the Eclipta alba extract, perform bioassay based fractionation and identify anti-HCV phytochemicals from the active fractions.
Materials and methods
Identification of active compounds was performed by bio-activity guided fractionation approach. Active isolates were separated by the combination of silica gel chromatography and preparative scale reverse phase HPLC. Eclipta alba extract and its isolates were examined for their ability to inhibit HCV replicase (HCV NS5B) activity in vitro and HCV replication in a cell culture system carrying replicating HCV subgenomic RNA replicon. The purified isolates were also examined for their binding affinity to HCV replicase by fluorescence quenching and their cytotoxicity by MTT assay.
Results
Eclipta alba extract strongly inhibited RNA dependent RNA polymerase (RdRp) activity of HCV replicase in vitro. In cell culture system, it effectively inhibited HCV replication which resulted in reduced HCV RNA titer and translation level of viral proteins. Bioassay-based fractionations of the extracts and purification of anti-HCV phytochemicals present in the active fractions have identified three compounds, wedelolactone, luteolin, and apigenin. These compounds exhibited dose dependent inhibition of HCV replicase in vitro, and anti-HCV replication activity in the cell culture system
Conclusion
Eclipta alba extract and phytochemicals isolated from active fractions display anti-HCV activity in vitro and in cell culture system. The standardized Eclipta alba extract or its isolates can be used as an effective alternative and complementary treatment against HCV.
Keywords: Hepatitis C virus, RNA dependent RNA polymerase, NS5B, Fluorescence quenching, Wedelolactone, Luteolin, Apigenin
1. Introduction
Approximately 170 million people worldwide are chronically infected with HCV (Manns et al., 2001). HCV infection often may leads to the development of a chronic condition that frequently progresses to liver cirrhosis and hepatocellular carcinoma (Raimondi et al., 2009). Current therapy for HCV infection includes the prolonged administration of a combination of ribavirin and pegylated interferon-α, which cures 40-50% of patients (Palumbo, 2011). Recently, the FDA approved the use of two protease inhibitors, Boceprevir and Telaprevir in combination with ribavirin and pegylated interferon-α which have a cure rate of 60- 80%, but exhibited some serious side effects in clinical trials (Poordad et al., 2011; Zeuzem et al., 2011). Consequently, there is urgent need to develop new remedies for the treatment of illness caused by HCV infection.
HCV is a positive single-stranded RNA virus of approximately 9.6 KB in length that encodes a large polyprotein of approximately 3,010 amino acids in a single reading frame (Moradpour et al., 2007). In infected cells, this poly protein is cleaved at multiple sites by a combination of cellular and viral proteases to generate structural and nonstructural (NS) viral proteins: C, E1, E2, P7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B. As with other positive-stranded RNA viruses, the replication of HCV proceeds through initial synthesis of a complementary negative strand of RNA, which serves as a template for the production of new positive strand genomic RNA molecules. One of the NS proteins is HCV replicase (NS5B), an RNA-dependent RNA polymerase (RdRp) that is responsible for replication of the viral RNA genome (Butcher et al., 2001). The NS5B has, therefore, been the primary target for drug development. Many small molecules targeting this enzyme have demonstrated antiviral efficacy in clinical trials (Beaulieu, 2007).
More than 40% of all currently prescribed drugs are derived from chemicals that have initially been identified in plants. Many phytochemicals have been identified that display considerable inhibition of HCV at some stage of the life cycle (Hu et al., 2007). In particular, silymarin, from milk thistle, has been evaluated (Polyak et al., 2010). We found that aqueous extract of Eclipta alba exhibited potent anti-HCV activity both in vitro and in a cell-culture system. Eclipta alba belongs to the Asteraceae family and is known for its numerous biological activities. It is used to treat infective hepatitis in India (Singh et al., 2001), hair loss (Datta et al., 2009), snake venom poisoning, and other viral infections (Diogo et al., 2009). In Ayurveda, the Indian traditional system of medicine, Eclipta alba has been shown to have hepato-protective activity and used to restore liver function. Therefore, it has frequently been used to treat many liver ailments as well as hepatitis. It is also used as general tonic for longevity and rejuvenation. Ethanolic extract of Eclipta alba has been shown to have hepatoprotective effects on damaged livers in rat and mice (Singh et al., 1993). To the best of our knowledge, the Eclipta alba extract or its purified isolates have not been evaluated for their anti-HCV activity. Here we describe bio-assay based fractionation of Eclipta alba extract, purification and identification of phytochemicals from the active fractions and evaluation of their anti-HCV activity in vitro and in cell culture system.
2. MATERIALS AND METHODS
Silica gel chromatography was done using silica gel (70-230 mesh, 60Å,) obtained from Sigma-Aldrich. Reverse-phase analytical HPLC analyses were done on HP1040 HPLC equipped with an autosampler and diode array detector (DAD), applying a Bio-Sil ODS-5S (Bio-Rad) 150 mm × 4 mm column and water (0.1% TFA)-acetonitrile gradient as the solvent system. Absorbance at 220 nM and 254 nM were recorded on DAD. Preparative scale purification was carried out using a Discovery® C18 reverse-phase preparative scale column (25 cm × 10 mm, 5 μm, Sigma-Aldrich) and a Varian HPLC 9010 equipped with a 9060 polychrome detector. Silica gel 60 F254 TLC plates obtained from Merck were used for TLC. Fluorescence quenching assay was performed on Varian Cary Eclipse fluorescence spectrophotometer (Varian Inc.). All compounds were made soluble in DMSO for the RdRp assay, fluorescence quenching assay and cell culture experiments. DMEM, antibiotics (penicillin-streptomycin) and L-Glutamine were purchased from Mediatech. Dulbecco's phosphate-buffered saline (DPBS), and other cell culture reagents were purchased from Invitrogen Life Technologies (Carlsbad, CA). Primary and secondary antibody for NS5A, NS5B were purchased from Santa Cruz Biotechnology. β-Actin antibody was purchased from Calbiochem. All gel images (Western blotting and RT-PCR) were quantified by QuantityOne software (version 4.4.1; Bio-Rad). IC50 values were calculated using CalcuSyn (Biosoft) software and graphs were drawn using GraphPad Prism 4.0 (GraphPad software) (Chou, 1977).
2.1 Plant material
The plant was identified and collected by Ganesh Ayurvedic Pharmacy at Varanasi, India. The voucher specimen (No. 2011-05) of collected plants was deposited at pharmacy. The aqueous extract of the plant was prepared and concentrated at Ganesh Pharmacy in India.
2.2 Extraction and Purification
The freshly collected whole Eclipta alba plants (40 kg) were thoroughly washed, cut into pieces, and crushed into pulp in deionized water. The pulp was then diluted 4 fold and boiled for 1 h. The suspension was filtered through 4 layers of cheesecloth and concentrated by evaporation. The 250 g of concentrated aqueous extract was homogenized in methanol/water (30:70, v/v) (0.6 L × 4) and incubated overnight at room temperature with stirring. This mixture was extracted by ethyl acetate (0.5 L × 4) and dichloromethane (0.5 L X 3). After removing water from the organic layers over sodium sulfate, it was dried to obtain 9 g dark-brown viscous material. The solvent-extracted material was mixed with silica gel, dried under vacuum, and loaded on the top of a silica-gel (50 g) gravity column. Elution was carried out in dichloromethane solvent with a gradient of methanol from 0% to 50%. While collecting the fractions, the eluants were monitored by TLC (methanol: dichloromethane, 10:90). Based on TLC purity, a total of 18 fractions (F1 to F18) were collected. Fractions with similar purity were pooled and examined for inhibition of RdRp activity. Preliminary in vitro screening showed the presence of inhibitory activity in many fractions, with maximum being in fractions F4-F8. The active fractions with similar purity were pooled and further purified. The fractions F14 and F15 were pooled and purified by silica gel flash column chromatography (methanol: dichloromethane, 5:95 to 50:50) to yield compound 1 (15.4 mg). Fractions F11-F13 were combined and purified by reverse phase HPLC, (0-15% acetonitrile in 0.1% TFA in 100 min; 2 ml/min) to yield compound 2 (3.5 mg). Fractions F4 to F8 were individually subjected to normal-phase chromatography (30 g silica gel column; 0-10% methanol in dichloromethane) produced three compounds, 3, 4, and 5. with modest purity which were further purified individually on a reverse-phase preparative HPLC (20-50% acetonitrile in 0.1% TFA in 180 min; 2 ml / min) to yield 4.0 mg, 6.7 mg and 3.0 mg of purified compounds, 3, 4, and 5, respectively.
2.3 Expression and purification of HCV NS5B
The plasmid pET-21d-5BΔ21 expressing His-tagged HCV NS5B carrying a deletion of 21 amino acids from the C terminal was obtained from Dr. Sergey Kochetkov laboratory (Ivanov et al., 2005). The plasmid was transformed in Escherichia coli Rosetta (DE3) and His-tagged HCV NS5B was expressed and purified by immobilized metal affinity chromatography (IMAC) using Ni-NTA (nickel-nitrilotriacetic acid) column, as described previously (Ivanov et al, 2005). The IMAC purified factions were pooled and further purified on a pre-packed 1 ml Hi-Trap Heparin column (Pharmacia) applying on the Pharmacia FPLC (fast protein liquid chromatography). In brief, the Ni-NTA eluted fractions with similar purity were pooled and diluted 3 fold with buffer A (Tris HCl (pH 7.0), 5% glycerol, 0.5% triton X-100 and 1 mM 2-mercaptoethanol) and loaded on the Hi-Trap Heparin column. The column was washed with 2 column volumes of buffer A, and eluted with a linear gradient (0% to 80%) of 1 M KCl in the same buffer in 20 min (1 ml/min). Eluted fractions showing more that 98% purity on 8% SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) were pooled, dialyzed overnight against a buffer containing 50 mM Tris HCl (pH 7.8), 5 mM DTT (dithiothreitol), 100 mM NaCl and 50% glycerol and stored at -80°C.
2.4 RdRp assay
RdRp assay was carried out in a final volume of 25 μl. The reaction mixture contained 20 mM Tris HCl (pH 7.8), 100 mM NaCl, 100 mM sodium glutamate, 0.5 mM DTT, 0.01% BSA (bovine serum albumin), 0.01% tween-20, 5% glycerol, 20 U/ml RNase Out, 25 μM cold UTP, 1μCi/assay 3H UTP, 0.5 mM MnCl2,, and 588 nM (1 μg) HCV NS5B. The enzyme was preincubated in the reaction buffer along with test compounds or DMSO (control) at room temperature for 20 min, after which polymerase reactions were initiated by the addition of preannealed 500 nM PolyrA/dT18. After 1h at 37 °C, reactions were terminated by addition of ice-cold 5% TCA and kept on ice for 30 min. Acid-insoluble materials were filtered on a glass fiber filter (GF/B) and the filters were washed successively with 5% TCA, water, and ethanol. Filters were air dried and placed in a vial containing 5 ml EcoLite scintillation fluid and counted for radioactivity using a Peckard 2200-CA Tri-Carb scintillation counter.
2.5 Fluorescence quenching assay
Fluorescence quenching based binding assay was performed in a final volume of 100 μl per well in a 96 well plate (Fluotrac 200). The HCV NS5B (CΔ21) was diluted to a final concentration of 1 μM in a buffer containing 50 mM Tris HCl (pH 7.8), 50 mM NaCl, 10% glycerol, 10% DMSO and 0.5 mM MnCl2. The test compounds were mixed and incubated for 20 min at room temperature. The fluorescence emission of enzyme-ligand complex was monitored at 300 to 420 nm with an excitation wavelength at 280 nm. The emission fluorescence intensity of NS5B at 330 nm was maximum in the presence or absence of the test compounds and, therefore excitation and emission intensities at 280 nm and 330 nm, respectively, were used to determine the dissociation constant (Kd) value for the compounds (ligand). The excitation and emission wavelengths were set at monochromator bandwidths of 20 and 10 nm, respectively. The background emission was eliminated by buffer containing equal amount of test compound or DMSO. The Kd value was determined using the equation, ΔF/ΔFmax = [ligand] total / (Kd + [ligand] total), where ΔF is the difference in emission fluorescence intensity of NS5B in the absence and presence of compound. ΔFmax is the difference in emission fluorescence intensity of NS5B in the absence of compound and in the presence of infinite concentration of compound and Kd is the dissociation constant (Bougie et al., 2003). The data were also analyzed by non-linear and double-reciprocal representation of (ΔF/F0 × 100) vs [compound] where F0 is the emission fluorescence intensity of NS5B in absence of compound.
2.6 Western blotting
MH14 cells, carrying the stably replicating HCV subgenomic replicon (Murata et al., 2005) was used to evaluate anti-HCV activity of Eclipta alba extract and its purified isolates. MH14 cells were grown at 37° C with 5% CO2 in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FBS (fetal bovine serum), 1% penicillin/streptomycin, 1% glutamine, and 300 μg/ml of G418 (Calbiochem). After 48- h, cells were washed twice with Dulbecco's phosphate-buffered saline and lysed in a buffer containing 10 mM Tris HCl (pH 6.8), 1 mM DTT, 1% SDS, and 10% glycerol. The cell lysates were boiled for 5 min, cooled to room temperature, and centrifuged at 10,000×gat 4 °C. The supernatant was saved and total protein concentration was determined (DC protein assay, Bio-Rad). An aliquot of the supernatant equivalent to 15 μg protein was resolved by electrophoresis on 8% SDS-polyacrylamide gel and then transferred to nitrocellulose membrane (Schleicher and Schuell Bioscience). The membrane was first blocked with 5% nonfat dry milk in PBS containing 0.5% Tween 20 for 1 h at room temperature. The membrane was washed 4-5 times with the same buffer and treated with 1:500 diluted primary antibody of selected protein target ( NS5B, NS5A, or actin) overnight at 4°C, The membrane was washed again several times with the same buffer, then treated with 1:5,000 dilution of horseradish-peroxidase conjugated secondary antibody. After 1 h incubation at room temperature, membranes were washed 5 times with the same buffer and incubated with horseradish peroxidase substrate (Western lightening chemiluminescence reagent; PerkinElmer Life Sciences) for 10 sec to 1 min, exposed on Kodak X-Omat AR X-ray film and developed. Housekeeping β-actin served as the loading control.
2.7 Quantitation of HCV RNA by RT-PCR
Total RNA from MH14 cells was isolated by TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The isolated RNA was dissolved in DEPC-treated water, quantified by NanoDrop (ND1000, NanoDrop Technologies), divided into aliquots, and stored at -80°C. For RT-PCR, one microgram of total RNA was reverse transcribed by Superscript II reverse transcriptase (Invitrogen) as per the manufacturer's protocol in a final volume of 20 μl. Two microliters of synthesized cDNA was subjected to PCR in a total volume of 50 μl using ChoiceTaq DNA polymerase (Denville Scientific). PCR was done for 25 cycles consisting 1 min at 94 °C, 1 min at 52 °C, and 1 min at 72 °C. For HCV RNA, the forward PCR primer was 5'-CGGGAGAGCCATAGTGG-3'; the reverse primer for reverse transcription and PCR was 5'-AGTACCACAAGGCCTTTCG -3'. The reverse transcription of GAPDH mRNA was done as an internal control. For GADPH mRNA, the forward primer was PCR 5'-CTCTGCTCCTCCTGTTCGAC-3' and the reverse primer for reverse transcription and PCR was 5'-ATGGGTGGAATCATATTGGAAC-3'. The products were analyzed by electrophoresis on 2% agarose gel. The image was captured using Bio-Rad imager.
2.8 Endogenous HCV replication assay in cell-free replication lysate of MH 14 cells
The replicative cytoplasmic fractions from the MH14 cells were prepared following the protocol described previously (Ali et al., 2002). Cells were washed with cold buffer containing 30 mM HEPES (pH 7.4), 150 mM sucrose, 33 mM ammonium chloride, 7 mM KCl, and 4.5 mM magnesium acetate. The cells were then treated using lysolecithin solution (250 μg/ml) in the washing buffer for 1 min and then added 3 ml of washing buffer to each dish. The buffer were discarded and cells were collected by scraping them into 120 μl of replication buffer containing 100 mM HEPES (pH 7.4); 50 mM ammonium chloride; 7 mM KCl; 1 mM spermidine; 1 mM (each) of ATP, GTP, CTP, and UTP; 1 mM DTT; and 10% glycerol. The cells were lysed gently by pipetting up and down several times, and then centrifuged the lysed cells at 1,600 rpm for 5 min at 4°C. The replicative lysate (supernatant) was divided into aliquots and stored at -80°C until further use. For the cell-free HCV replication assay, we incubated 50 μl of replicative lysate for 0 h or 3 h at 30°C in the presence and absence of Eclipta alba extract. The replication was then terminated by adding 0.5% SDS in STE buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 150 mM NaCl) and extracted total RNA twice with phenol-chloroform-isoamyl alcohol (25:24:1) and twice with water-saturated ether. We precipitated the RNA by ethanol and dissolved it in water. The total RNA underwent subsequent RT-PCR analysis (SuperScript II reverse transcriptase (Invitrogen) and ChoiceTaq DNA polymerase (Denville Scientific) for HCV RNA and GAPDH mRNA. The PCR products were visualized by 2% agarose gel electrophoresis. The experiments were performed in duplicate.
2.9 MTT assay
The MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay was carried out for determining the toxicity of Eclipta alba extract and its purified isolates according to published protocol (Kodama et al., 1996). MH14 cells were plated on a 96 well plate at 1-1.5 × 104 cells per well. Cells were allowed to grow overnight at 37 °C. The cells were washed with PBS and incubated with the increasing concentrations of Eclipta alba extract or its purified isolates in a final volume of 200 μl of the culture medium. The cells were grown for 48 h and then supplemented with 10 μl MTT (4 mg/ml dissolved in PBS) to each well. The MTT is selectively and rapidly reduced by living cells to form a purple formazan precipitate. After 4 h incubation at 37 °C, medium was carefully aspirated, and formazan crystals were dissolved in 100 μl DMSO. Absorbance at 570 nm was measured using a Biotek Synergy HT Spectrophotometer, with a reference filter set at 620 nm. Cells treated with equal amounts of DMSO were kept as control.
3. RESULTS
3.1 Eclipta alba extract inhibited NS5B RdRp activity in vitro
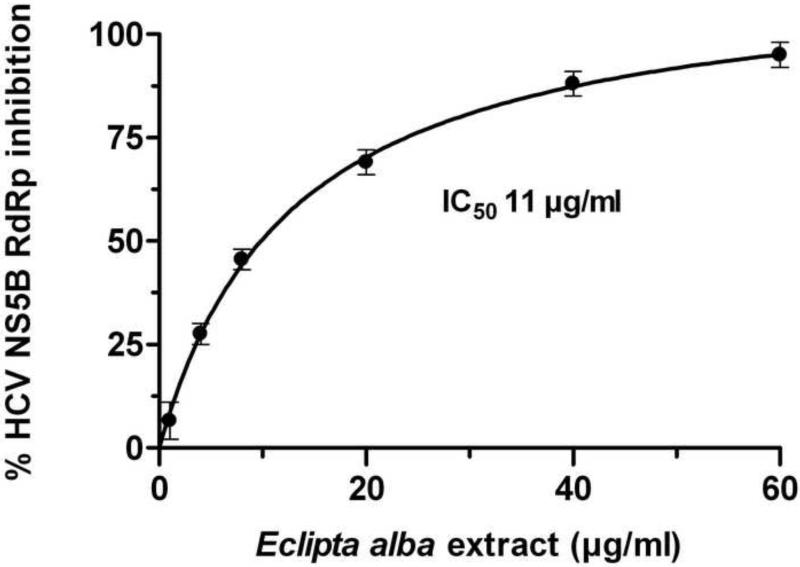
In our preliminary screening of plant extracts for inhibitory activity on HCV replicase (NS5B), we noted that Eclipta alba extract strongly inhibited the RdRp activity of NS5B in vitro. We therefore determined the IC50 of the extract against NS5B activity. The Eclipta alba extract was then lyophilized and a small amount of the dried material was dissolved in DMSO and examined for RdRp inhibition as a function of increasing concentration of the extract. The inhibitory constant (IC50) of the extract was found to be 11μg/ml (Fig. 1).
Figure 1. Inhibition of HCV NS5B RdRp activity by Eclipta alba extract.
The inhibition of RdRp activity of HCV NS5B by Eclipta alba extract is shown as percent inhibition with respect to control. The results depicted are an average of three independent set of experiments.
3.2 Eclipta alba extract blocked viral replication and HCV protein expression in cell culture system
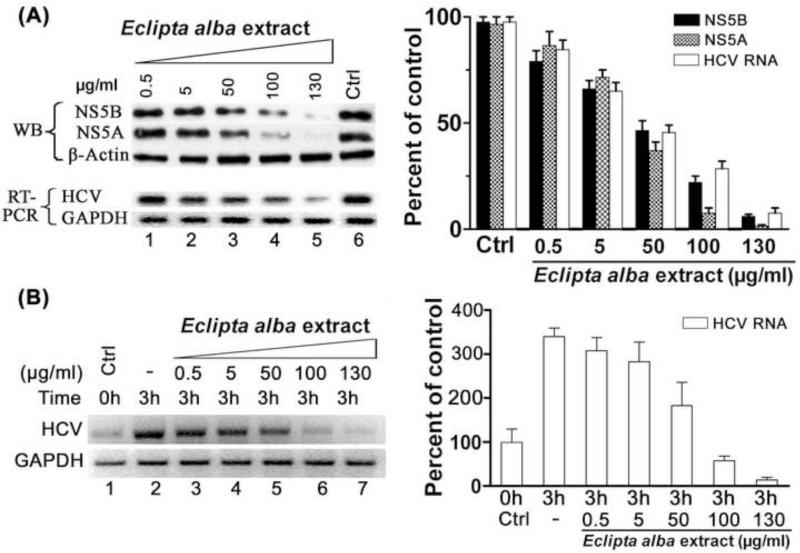
Since the solvent-extracted fraction of Eclipta alba displayed significant inhibition of RdRp activity of HCV NS5B in vitro, we examined whether the extract also has anti-HCV activity in cell culture. We used MH14 cells carrying stably replicating HCV subgenomic replicons. The MH 14 cells were incubated with increasing amounts of the extract and after 48 h, cells were harvested, lysed, and analyzed for viral proteins (NS5B and NS5A) by Western blotting and viral RNA by RT PCR. The expression of both NS5B and NS5A was significantly decreased as compared to controls (Fig. 2A). More than 95% inhibition was noted at concentration of 130 μg/ml of extract within 48 h (Fig. 2A, lane 5). We also noted a significant dose-dependent decrease in the viral RNA level, which was consistent with the decrease in viral protein expression. At 130 μg/ml concentration of the extract a greater than 90% reduction in the viral RNA level (Figure 2A, lane 5) was observed, suggesting that the extract contained anti-HCV phytochemicals inhibiting HCV replication in the cell-culture system.
Figure 2. Effect of Eclipta alba extract on HCV replication.
(A) MH14 cells carrying subgenomic HCV replicon were grown in the presence of indicated concentration of Eclipta alba extract. After 48 h, cells were harvested, lysed and subjected to SDS-PAGE and Western blotting (WB) for HCV NS5B and HCV NS5A. The β-actin served as the loading control. In the second set of the experiment, MH14 cells were grown for 60 h in the presence and absence of Eclipta alba extract. Cells were harvested and total RNA was isolated and analyzed for HCV RNA by RT-PCR. The RT-PCR of GAPDH mRNA was also carried out as an internal control. Quantitation of WB and RT-PCR results are shown on right panel. (B) The inhibition of endogenous HCV replication in cell free replicative lysate of MH14 cells by Eclipta alba extract. An aliquot of cell free replicative lysate was incubated in the absence (-) and presence of indicated concentration of Eclipta alba extract. At the end of incubation, RT-PCR of HCV RNA and GAPDH RNA was carried out. Right panel shows the quantitation of RT-PCR results of two independent experiments with error bars.
Having found the anti-HCV activity of Eclipta alba extract in MH14 cells, we postulated this could be due to the direct inhibition of NS5B in vitro. To test this hypothesis, we used the cell-free replicative lysate from MH14 cells and applied it to determine endogenous HCV replication activity as described previously (Zhang et al., 2008). An aliquot of the cell free replication lysate prepared from MH 14 cells was incubated in absence and presence of different concentrations of Eclipta alba extract at 30 °C for 0h and 3h. Following incubation, the total RNA was isolated and level of HCV RNA was determined by RTPCR, which reflected the activity of HCV replicative complexes in the replicative lysate (Figure 2B). As shown in the figure, the level of HCV RNA in control after 3 h of incubation was 3-4 fold higher than at 0 h control (lanes 1 and 2). However, in the presence of increasing concentration of Eclipta alba extract, significant inhibition of endogenous HCV replication was noted Fig. 2B, lanes 3-7). This result clearly indicates that the inhibition of HCV replication observed in cell culture system is due to its inhibitory effect on endogenous HCV replication via HCV replicase.
3.3 Identification of active compounds from the extract and its effect on HCV replication
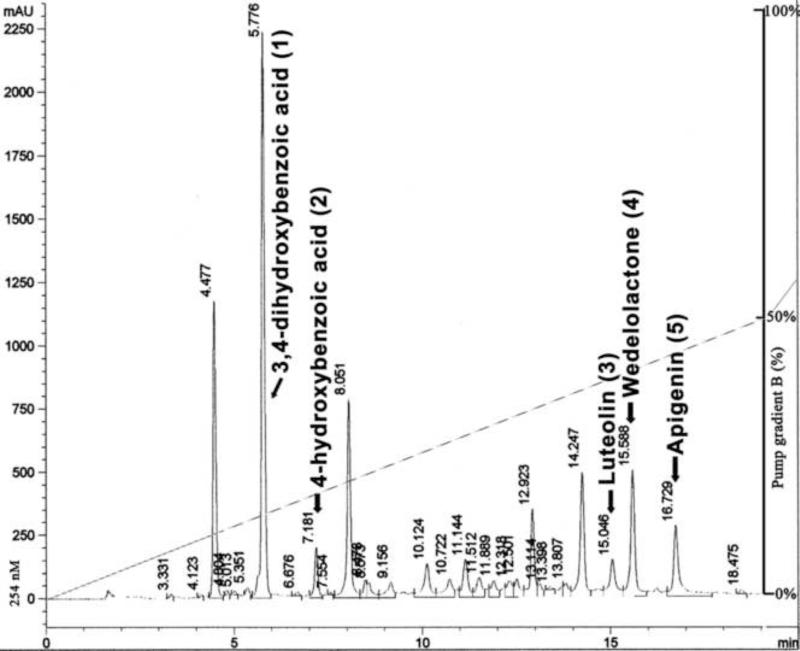
Since Eclipta alba extract strongly inhibited RdRp activity of NS5B in vitro and demonstrated anti-HCV activity in cell culture, we did further bio-assay-based fractionation of the concentrated extract to isolate and identify the constituents that are responsible for anti-HCV activity. An analytical HPLC method was developed to obtain a pattern of phytochemicals present in the solvent-extracted fraction using a reverse-phase C18 column (Fig. 3).
Figure 3. HPLC profile of solvent-extracted Eclipta alba.
HPLC separation was done on a C18 reverse-phase column, Bio-Sil ODS-5S using a gradient method: 0-50% B in 20 min, 50-95% B in 25 min. (A= water with 0.1% TFA; B = acetonitrile).
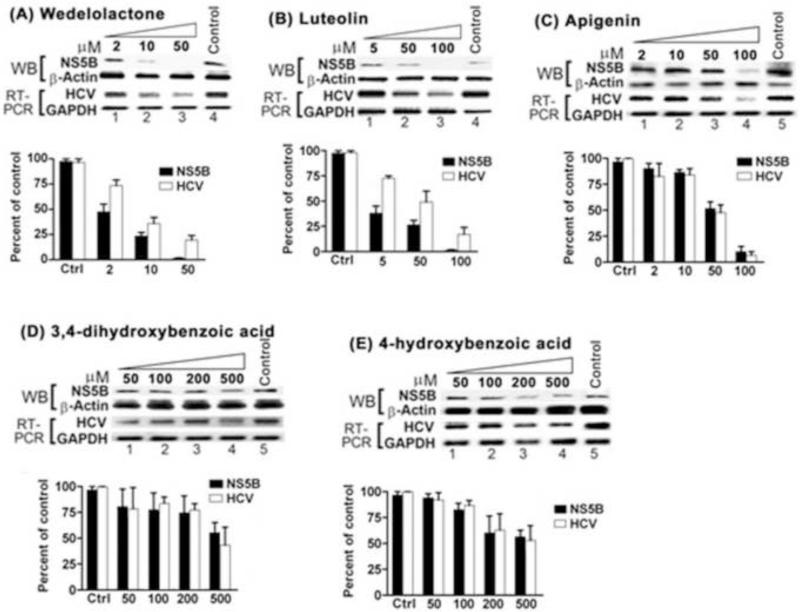
To identify the bioactive fraction, the Eclipta alba extract was further fractionated on silica gel, from which 18 fractions were collected. Fractions 4-8 exhibited highest inhibition activity against HCV NS5B. These fractions were pooled and applied again on to the silica-gel column for further separation with a shallow gradient of methanol in dichloromethane. Analytical HPLC profiles of these purified fractions showed the presence of minor impurities. The fractions underwent additional purification by preparative C18 chromatography. Preparative HPLC separation yielded compound 3-5. Based on spectroscopic data, compound 4 was elucidated as wedelolactone. It was also confirmed by the co-analysis with HPLC reference standard (sigma). We found that our isolated wedelolactone (4) inhibited NS5B RdRp activity in vitro with IC50 of 7.7 μM. In a cell culture system, wedelolactone at 10 μM inhibited > 80% of NS5B expression and at 50 μM concentration, we could not detect the expression of NS5B by Western blot analysis (Fig. 4A, lane 3). As expected, wedelolactone also inhibited HCV replication in cell culture system; 30% and 80% of HCV replication was inhibited at 10 μM and 50 μM concentration, respectively, as judged by the HCV RNA level (Fig. 4A, lane 2, 3).
Figure 4. Anti-HCV activity of Eclipta alba isolates.
MH14 cells were grown in the absence (control) or presence of indicated concentration of Wedelolactone (A), Luteolin (B), Apigenin (C), 3,4-dihydroxybenzoic acid (D) or 4-hydroxybenzoic acid (E). After 48 h, cells were harvested, lysed and Western blotted for NS5B and β-actin. In another set, cells were grown for 60 h and levels of HCV RNA and GADPH mRNA were determined by RT-PCR. The Y axis expresses the percentage of NS5B expression and HCV RNA level with respect to the control. Quantitation represents the results of two independent experiments with standard deviation.
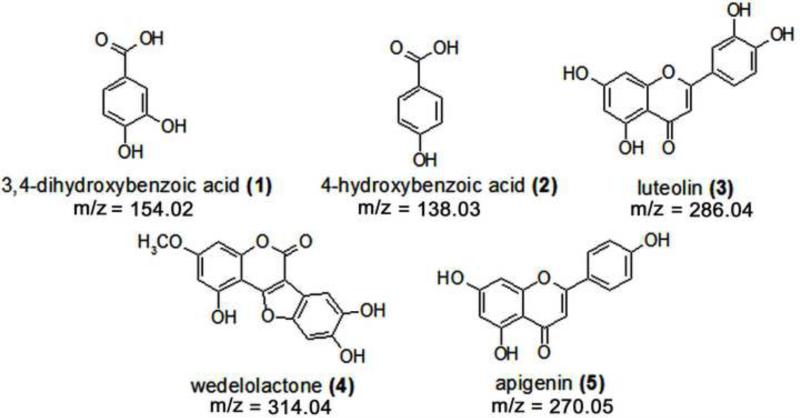
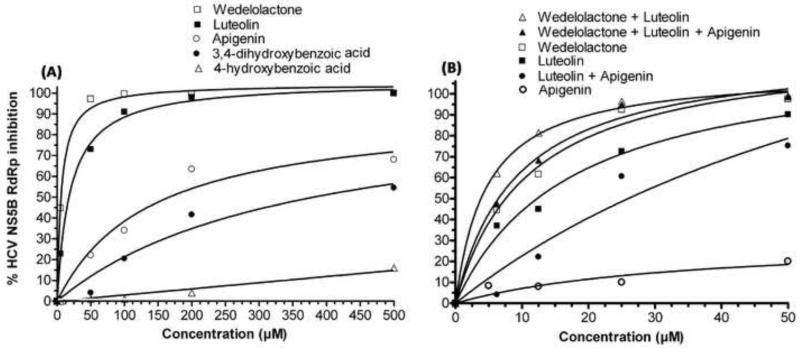
Compounds 3 and 5 were found to have almost similar structure. They were identified as luteolin (3) and apigenin (5), respectively (Fig. 5). 1H NMR data (DMSO-d6) of 5, as compared to 3, showed one additional proton with δ 6.91, (d, J = 9Hz). The NMR data confirmed the 4-hydroxyphenyl subunit in apigenin and 3,4-dihydroxyphenyl subunit in luteolin (3). The 1H NMR data of 5 explained two identical doublets (δ 6.92, d, J = 9Hz), (δ 7.91, d, J = 8.5Hz), clarified the identical environment of protons in the 4-OH-phenyl subunit in apigenin (5). The HPLC reference standards of luteolin (sigma) and apigenin (sigma) were also co-analyzed in HPLC with our isolated compound 3 and 5, confirmed their identical retention times at Rt 15.0 and Rt 16.7, respectively (Fig. 3). Biological evaluation of 3 and 5 indicated that both compounds inhibited the RdRp activity of HCV NS5B with respective IC50 values of 11.3 μM and 175.5 μM (Fig. 6). The antiviral activity of 3 and 5 in a cell culture system was also determined at different concentrations. We found that the inhibition of viral replication was reduced approximately by 50% at 50 μM concentration of 3 and 5 At 100 μM concentration of 3 and 5, greater than 90% inhibition of NS5B was noted (Fig. 4 B, C). Both the compounds 3 & 5 also inhibited 50% HCV replication in cell culture system at 50 μM concentrations. At 100 μM of compounds 3 and 5, HCV replication was reduced by approximately 80 % and 90%, respectively (Fig. 4 B, C).
Figure 5. Chemical structures of the compounds isolated from Eclipta alba.
m/z value of each compounds are shown below the name.
Figure 6. Dose-dependent inhibition of HCV NS5B by Eclipta alba isolates.
Each point represents the mean value of two independent set of experiments with standard deviations.
Compounds 1 and 2 were identified as 3,4-dihydroxybenzoic acid (1), 4-hydroxybenzoic acid (2) from 1H NMR and ESI-MS analysis. Compound 1 also inhibited RdRp activity of NS5B in vitro (IC50 400 μM), whereas 2 did not inhibit NS5B activity (Fig. 6A). The antiviral efficacy evaluation of isolate 1 and 2 in a cell culture system indicated that compound 1 and 2 inhibited HCV RNA replication only at very high concentrations at 500 μM (Fig. 4 D, E).
3.4 A combination of active isolates has synergistic effects
We found that wedelolactone (4) and luteolin (3) in 1:1 combination increased inhibitory effect on NS5B RdRp activity with a combined IC50 value of 4.5 μM as compared to their individual IC50 values (Fig. 6B). Also, at equimolar concentrations, wedelolactone (4), luteolin (3) and apigenin (5) yielded a combined IC50 value of 7.6 μM, Equimolar concentration of luteolin (3) and apigenin (4) in 1:1 combination also showed improved IC50 value of 24.6 μM, indicating their synergistic inhibitory effect on the enzyme. This may be due to their different binding sites on the enzyme molecule. We did not observe any synergistic effect in combinations of compound 1 and 2.
3.5 Fluorescence quenching of HCV polymerase by active isolates
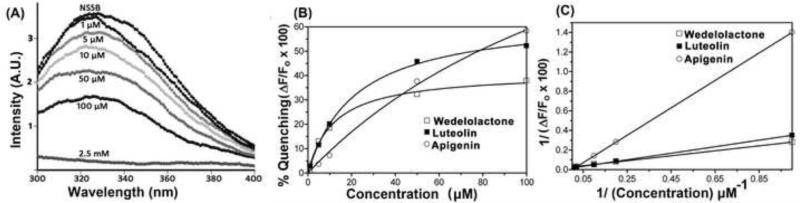
The HCV NS5B has significant intrinsic fluorescence emission at 330 nm when excited at 280 nm. The intrinsic fluorescence is reduced upon binding of ligand molecules such as nucleotide substrate or inhibitor (Flowers et al., 2003). We therefore examined whether Eclipta alba isolates showing inhibitory activity against HCV NS5B can quench the intrinsic fluorescence upon binding to the enzyme. We found that compounds 3, 4 and 5 rapidly quenched the fluorescence emission of the HCV NS5B indicating their ability to bind with the enzyme. We therefore, determine their binding affinity (Kd value) by titrating a fixed concentration of enzyme with increasing concentration of individual compounds, 3, 4 and 5. A typical fluorescence emission spectrum of NS5B in presence of increasing concentration of luteolin (3) is shown in Fig. 7A. The fluorescence quenching data showed that these compounds bind with the enzyme in a concentration-dependent manner and, follow a typical drug binding hyperbolic curve (Fig. 7B). The double-reciprocal plot of these data is shown in Fig. 7C. Kd values for wedelolactone (4), luteolin (3) and apigenin (5) were calculated to be 24.3 μM, 31.5 μM and 122 μM, respectively.
Figure 7. Fluorescence quenching of NS5B by wedelolactone, luteolin and apigenin.
The fixed concentration of NS5B (1 μM) was incubated with increasing concentration of the test compound. The change of intrinsic protein fluorescence was monitored at the emission spectrum of 300 to 400 nm, with excitation at 280 nm. (A) Fluorescence emission pattern of NS5B in the presence of increasing concentration of luteolin. (B), the fluorescence quenching of NS5B in the presence of increasing concentration of wedelolactone (□), luteolin (■), and apigenin (○). The % fluorescence quenching (ΔF/Fo × 100) was calculated relative to the fluorescence of NS5B in the absence of test compound. (C) Double reciprocal plot of data shown in B.
3.6 Cytotoxicity of Eclipta alba extract and its isolates
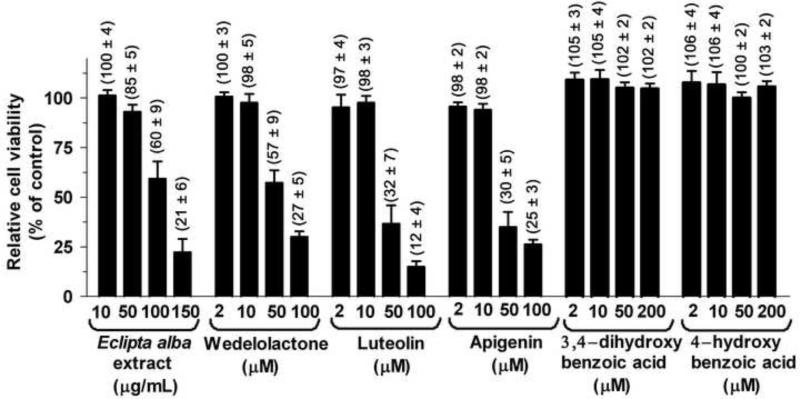
MTT is soluble in the aqueous medium and metabolized to purple formazan crystals by the active live cells. The amount of formazan produced is proportional to the number of viable cells. We performed MTT assay to determine the toxicity of Eclipta alba extract and its purified isolates in MH14 cells. The cells were grown in the absence and presence of increasing concentrations of Eclipta alba extract or its purified isolates. After 48 h, the cells were assayed for the possible cytotoxicity. As illustrated in Fig. 8, Eclipta alba extract did not display any cytotoxicity up to 50 μg/ml. However, it was marginally toxic at 100 μg/ml showing 60% cell viability. Wedelolactone (4), luteolin (3) and apigenin (5) at 2 μM and 10 μM, respectively, had no influence on cell viability, but were toxic at 50 μM and 100 μM concentrations. In contrast, 3,4-dihydroxybenzoic acid (1) and 4-hydroxybenzoic acid (2) were found to beneficial for cell growth as they enhanced cellular proliferation. These results suggest that Eclipta alba contains a cocktail of phytochemicals, some of which are moderately cytotoxic at higher concentration while others are endowed with cytoprotective activity.
Figure 8. Effect of Eclipta alba extract and its purified compounds on cell viability.
Cells were grown in the presence and absence of Eclipta alba extract or its purified isolates. After 48 h, cell viability was evaluated by MTT assay. The results expressed as relative cell viability in compare to control. Results shown are the averages of three sets of independent experiments with error bars.
4. DISCUSSION
Conventional drug development is not only expensive, but slow, taking many years to develop a new drug (DiMasi et al., 2003). Moreover, the final products are often unaffordable in poor countries. Traditional medicinal plants and their successful use in treatment have made it possible to do reverse pharmacology by identifying biologically active compounds present in plants. Examples of plant-derived drugs are aspirin, derived from the bark of the willow tree; the antimalarial drug artemisinin, isolated from the sweet wormwood plant, Artemesia annua; and quinine from the bark of cinchona trees (Cinchona officialinis) (Duke and Paul, 1993; Klayman, 1985; Mahdi et al., 2006; Woodward and Doering, 1944).
Eclipta alba has been shown to have hepato-protective properties (Singh et al., 2001; Singh et al., 1993). The extract, prepared from whole plant, has traditionally been used in India to treat liver cirrhosis, infective hepatitis, liver enlargement, jaundice, and other ailments of the liver and gallbladder. Given the known therapeutic properties of Eclipta alba, we carried out reverse pharmacology involving bio-assay based fractionation of Eclipta alba extract to identify anti-HCV phytochemicals present in the extract. We first examined the effect of Eclipta alba extract on HCV replicase in vitro, finding that Millipore-filtered extract had potently inhibited the polymerase activity of HCV replicase (NS5B). We then examined the effect of crude extract on HCV replication in MH14 cell culture system that carries stably replicating HCV subgenomic replicons. We found that the extract of Eclipta alba strongly inhibited HCV replication (Fig. 1 and. 2) with an IC50 value of 11 μg/ml. These results were in contrast to results reported for silymarine (a standardized crude extract from the milk thistle Silybum marianum), which has been shown to possess anti-HCV activity only at very high concentration with IC50 of 300-600 μM (Polyak et al., 2010).
These screening results indicated the presence of phytochemicals of clinical significance, having potent antiviral activity against HCV, in the extract of Eclipta alba. The bio-assay- based fractionation of crude extract identified five different compounds with anti-HCV activity. These are 3,4-dihydroxybenzoic acid (1), 4-hydroxybenzoic acid (2), luteolin (3), wedelolactone (4) and apigenin (5). Wedelolactone, luteolin and apigenin were the highly active anti-HCV compounds exhibiting IC50 of 7.7 μM, 11.3 μM and 175.5 μM, respectively. These compounds from Eclipta alba were significantly remarkable as compared to the purified flavonoids of silymarine, which had a fairly high IC50 (300-600 μM) for RdRp activity (Polyak et al., 2010). In addition, wedelolactone (4), luteolin (3) and apigenin (5) reached rapid binding equilibrium with NS5B, as within 1 minute of incubation of these compound with NS5B, the emission fluorescence were quenched and reached to its equilibrium (data not shown). Kd values determined by emission fluorescence quenching were found to be within 1-3 fold of their IC50 values obtained by the inhibition of RdRp activity of HCV NS5B.
Wedelolactone has been shown to have diverse bioactivities, inhibiting HCV NS5B RdRp (Kaushik-Basu et al., 2008), topoisomerase 2α (Benes et al., 2011), phospholipase A2 (Diogo et al., 2009), IKK kinase (Kobori et al., 2004), and Na+, K+-ATPase activity (Pocas et al., 2006). The IC50 values of isolated wedelolactone and commercially available wedelolactone were indistinguishable. Testing of the antiviral efficacy of our purified wedelolactone (4) in an HCV subgenomic replicon system exhibited potent antiviral activity in a cell culture system. In that system, wedelolactone inhibited HCV replication resulting in reduced HCV RNA titer and subsequent translation of viral proteins (Fig. 4A).
Luteolin and apigenin are the natural flavones, which have been shown to inhibit cyclindependant kinase 9 (CDK9) and block phosphorylation of the carboxy-terminal domain of RNA polymerase II in cancer cells (Polier et al., 2011). It also rapidly down-regulates anti-apoptotic protein, Mcl 1 (myeloid cell leukemia 1) and promotes apoptosis in cancerous cells. Recently, these flavones have been shown as an adjuvant for TRAIL (tumor necrosis factor-related apoptosis-inducing ligand) mediated anti-cancer therapy (Ding et al., 2012). The anti-HCV evaluation of luteolin and apigenin showed that the two compounds have minor difference in their structure but have significant difference in their ability to inhibit the RdRp activity of HCV NS5B in vitro, as well as to inhibit HCV replication in cell culture system (Fig. 4B, C). This could be due to a substructural requirement in binding with NS5B active sites. Both, luteolin and apigenin showed comparable high cytotoxicity in MH14 cell which is derived from nonneoplastic human hepatocytes immortalized by SV40 large T antigen. We also found that both luteolin and apigenin strongly up-regulate tumor suppressor protein, p53 while they have no effect on p53 expression in peripheral blood mononuclear cells (PBMC) (data will be communicated elsewhere). In view of these findings, the traditional use of Eclipta alba extract in treatment of infective hepatitis and related liver ailments could also be beneficial for treatment of liver cancer associated with chronic hepatitis C.
It has been argued that herbal medicines may acts synergistically rather than in isolated form (Wagner and Ulrich-Merzenich, 2009). This may be due to the binding of more than one compound at different sites on the same target or receptor. HCV NS5B has been shown to have five different binding sites for small molecules that might bind and block enzyme activity (Hang et al., 2009). Our results showed that these compounds have a synergistic inhibitory effect on RdRp activity of HCV NS5B also confirm different binding sites for these compounds on HCV NS5B molecule. We found that, combined in a 1:1 ratio, wedelolactone (4) and luteolin (3) synergistically inhibited the RdRp activity of HCV NS5B with IC90 of the combination reduced to 21 μM, as opposed to their respective individual IC90 values of 32 μM and 52 μM (Fig. 6B).
The observed synergistic or additive effect of these compounds against HCV replication suggests that these compounds are more beneficial when used together in the crude extract. Although the additive effect may be lost in the purified form, it was important to isolate and identify these active molecules and evaluate their antiviral activity in the cell culture system. We report for the first time the identification of anti-HCV phytochemicals from Eclipta alba that may have therapeutic potential in treatment of HCV infection. This study provides a strong basis for further optimization and standardization of Eclipta alba extract as well as its anti-HCV isolates for development of anti-HCV therapy.
5. CONCLUSION
This study suggests that the Eclipta alba extract and its isolates (Wedelolactone, luteolin, and apigenin) exhibit anti-HCV activity in vitro and in cell culture system. Using endogenous HCV replication system, we further established that the observed inhibition of HCV replication in cell culture system was via inhibition of HCV replicase activity in the cell. The standardized Eclipta alba extract or its isolates can be used as an effective alternative and complementary treatment against HCV.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Makoto Hijikata of Koyoto University, Japan, for providing MH14 cells and pMH14 plasmid, and Dr. Sergey Kochetkov, of the Russian Academy of Sciences, Moscow, for HCV NS5B expression clone. We also appreciate Dr. Tanaji Talele for MS analysis of the isolated compounds. This research was partly supported by grants from the NIH/NIAID and NIH/NIDDK (AI073703 and DK083560 to VNP)
ABBREVIATIONS
- HCV
Hepatitis C virus
- NS5B
non-structural protein 5B of hepatitis C virus
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- Poly (rA).(dT)18
polyriboadenylic acid annealed with (oligodeoxythymidylic acid)18
- RdRp
RNA-dependant RNA polymerase
- Ni-NTA
nickel-nitrilotriacetic acid
- FPLC
fast protein liquid chromatography
- HPLC
high pressure liquid chromatography
- TLC
thin layer chromatography
- DTT
dithiothreitol
- BSA
Bovine serum albumin
- FBS
fetal bovine serum
- DMEM
Dulbecco's modified eagle medium
- DPBS
Dulbecco's phosphate buffered saline
- TFA
trifluoroacetic acid
- DMSO
dimethylsulfoxide
- IC50
half-maximal inhibitory concentration
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPORTING INFORMATION:
Spectroscopic data (1H NMR, ESI-MS) as well as HPLC analysis of the isolated compounds are attached.
REFERENCES
- Ali N, Tardif KD, Siddiqui A. Cell-free replication of the hepatitis C virus subgenomic replicon. Journal of Virology. 2002;76:12001–12007. doi: 10.1128/JVI.76.23.12001-12007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu PL. Non-nucleoside inhibitors of the HCV NS5B polymerase: progress in the discovery and development of novel agents for the treatment of HCV infections. Current Opinion In Investigational Drugs. 2007;8:614–634. [PubMed] [Google Scholar]
- Benes P, Knopfova L, Trcka F, Nemajerova A, Pinheiro D, Soucek K, Fojta M, Smarda J. Inhibition of topoisomerase IIalpha: novel function of wedelolactone. Cancer Letters. 2011;303:29–38. doi: 10.1016/j.canlet.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Bougie I, Charpentier S, Bisaillon M. Characterization of the metal ion binding properties of the hepatitis C virus RNA polymerase. The Journal of Biological Chemistry. 2003;278:3868–3875. doi: 10.1074/jbc.M209785200. [DOI] [PubMed] [Google Scholar]
- Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- Chou TC. On the determination of availability of ligand binding sites in steady-state systems. Journal of Theoretical Biology. 1977;65:345–356. doi: 10.1016/0022-5193(77)90329-0. [DOI] [PubMed] [Google Scholar]
- Datta K, Singh AT, Mukherjee A, Bhat B, Ramesh B, Burman AC. Eclipta alba extract with potential for hair growth promoting activity. J Ethnopharmacology. 2009;124:450–456. doi: 10.1016/j.jep.2009.05.023. [DOI] [PubMed] [Google Scholar]
- DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: new estimates of drug development costs. Journal of Health Economics. 2003;22:151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- Ding J, Polier G, Köhler R, Giaisi M, Krammer PH, Li-Weber M. Wogonin and Related Natural Flavones Overcome Tumor Necrosis Factor-related Apoptosis-inducing Ligand (TRAIL) Protein Resistance of Tumors by Down-regulation of c-FLIP Protein and Up-regulation of TRAIL Receptor 2 Expression. Journal of Biological Chemistry. 2012;287:641–649. doi: 10.1074/jbc.M111.286526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diogo LC, Fernandes RS, Marcussi S, Menaldo DL, Roberto PG, Matrangulo PV, Pereira PS, Franca SC, Giuliatti S, Soares AM, Lourenco MV. Inhibition of snake venoms and phospholipases A(2) by extracts from native and genetically modified Eclipta alba: isolation of active coumestans. Basic & Clinical Pharmacology & Toxicology. 2009;104:293–299. doi: 10.1111/j.1742-7843.2008.00350.x. [DOI] [PubMed] [Google Scholar]
- Duke SO, Paul RN. Development and Fine Structure of the Glandular Trichomes of Artemisia annua L. International Journal of Plant Sciences. 1993;154:107–118. [Google Scholar]
- Flowers S, Biswas EE, Biswas SB. Conformational dynamics of DnaB helicase upon DNA and nucleotide binding: analysis by intrinsic tryptophan fluorescence quenching. Biochemistry. 2003;42:1910–1921. doi: 10.1021/bi025992v. [DOI] [PubMed] [Google Scholar]
- Hu JF, Patel R, Li B, Garo E, Hough GW, Goering MG, Yoo HD, O'Neil-Johnson M, Eldridge GR. Anti-HCV bioactivity of pseudoguaianolides from Parthenium hispitum. Journal of Natural Products. 2007;70:604–607. doi: 10.1021/np060567e. [DOI] [PubMed] [Google Scholar]
- Ivanov AV, Korovina AN, Tunitskaya VL, Kostyuk DA, Rechinsky VO, Kukhanova MK, Kochetkov SN. Development of the system ensuring a high-level expression of hepatitis C virus nonstructural NS5B and NS5A proteins. Protein Expression and Purification. 2006;48:14–23. doi: 10.1016/j.pep.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Kaushik-Basu N, Bopda-Waffo A, Talele TT, Basu A, Costa PR, da Silva AJ, Sarafianos SG, Noel F. Identification and characterization of coumestans as novel HCV NS5B polymerase inhibitors. Nucleic Acids Research. 2008;36:1482–1496. doi: 10.1093/nar/gkm1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klayman DL. Qinghaosu (artemisinin): an antimalarial drug from China. Science. 1985;228:1049–1055. doi: 10.1126/science.3887571. [DOI] [PubMed] [Google Scholar]
- Kobori M, Yang Z, Gong D, Heissmeyer V, Zhu H, Jung YK, Gakidis MA, Rao A, Sekine T, Ikegami F, Yuan C, Yuan J. Wedelolactone suppresses LPS-induced caspase-11 expression by directly inhibiting the IKK complex. Cell Death and Differentiation. 2004;11:123–130. doi: 10.1038/sj.cdd.4401325. [DOI] [PubMed] [Google Scholar]
- Kodama E, Shigeta S, Suzuki T, De Clercq E. Application of a gastric cancer cell line (MKN-28) for anti-adenovirus screening using the MTT method. Antiviral Res. 1996;31:159–164. doi: 10.1016/0166-3542(96)06966-5. [DOI] [PubMed] [Google Scholar]
- Mahdi JG, Mahdi AJ, Bowen ID. The historical analysis of aspirin discovery, its relation to the willow tree and antiproliferative and anticancer potential. Cell Proliferation. 2006;39:147–155. doi: 10.1111/j.1365-2184.2006.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nature Review in Microbiology. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- Murata T, Ohshima T, Yamaji M, Hosaka M, Miyanari Y, Hijikata M, Shimotohno K. Suppression of hepatitis C virus replicon by TGF-beta. Virology. 2005;331:407–417. doi: 10.1016/j.virol.2004.10.036. [DOI] [PubMed] [Google Scholar]
- Palumbo E. Pegylated interferon and ribavirin treatment for hepatitis C virus infection. Therapeutic Advances in Chronic Disease. 2011;2:39–45. doi: 10.1177/2040622310384308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocas ES, Lopes DV, da Silva AJ, Pimenta PH, Leitao FB, Netto CD, Buarque CD, Brito FV, Costa PR, Noel F. Structure-activity relationship of wedelolactone analogues: structural requirements for inhibition of Na+, K+ -ATPase and binding to the central benzodiazepine receptor. Bioorganic and Medicinal Chemistry. 2006;14:7962–7966. doi: 10.1016/j.bmc.2006.07.053. [DOI] [PubMed] [Google Scholar]
- Polier G, Ding J, Konkimalla BV, Eick D, Ribeiro N, Kohler R, Giaisi M, Efferth T, Desaubry L, Krammer PH, Li-Weber M. Wogonin and related natural flavones are inhibitors of CDK9 that induce apoptosis in cancer cells by transcriptional suppression of Mcl-1. Cell Death & Disease. 2011;2:e182. doi: 10.1038/cddis.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak SJ, Morishima C, Lohmann V, Pal S, Lee DY, Liu Y, Graf TN, Oberlies NH. Identification of hepatoprotective flavonolignans from silymarin. Proceedings of Nattional Academy of Sciences of the United States of America. 2010;107:5995–5999. doi: 10.1073/pnas.0914009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poordad F, McCone J, Jr., Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N, DiNubile MJ, Sniukiene V, Brass CA, Albrecht JK, Bronowicki JP. Boceprevir for untreated chronic HCV genotype 1 infection. New England Journal of Medicine. 2011;364:1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi S, Bruno S, Mondelli MU, Maisonneuve P. Hepatitis C virus genotype 1b as a risk factor for hepatocellular carcinoma development: A meta-analysis. Journal of Hepatology. 2009;50:1142–1154. doi: 10.1016/j.jhep.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Singh B, Saxena AK, Chandan BK, Agarwal SG, Anand KK. In vivo hepatoprotective activity of active fraction from ethanolic extract of Eclipta alba leaves. Indian J Physiological Pharmacology. 2001;45:435–441. [PubMed] [Google Scholar]
- Singh B, Saxena AK, Chandan BK, Agarwal SG, Bhatia MS, Anand KK. Hepatoprotective effect of ethanolic extract of Eclipta alba on experimental liver damage in rats and mice. Phytotherapy Research. 1993;7:154–158. [Google Scholar]
- Wagner H, Ulrich-Merzenich G. Synergy research: approaching a new generation of phytopharmaceuticals. Phytomedicine. 2009;16:97–110. doi: 10.1016/j.phymed.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Woodward RB, Doering WE. The total synthesis of quinine1. Journal of the American Chemical Society. 1944;66:849–849. [Google Scholar]
- Zeuzem S, Andreone P, Pol S, Lawitz E, Diago M, Roberts S, Focaccia R, Younossi Z, Foster GR, Horban A, Ferenci P, Nevens F, Mullhaupt B, Pockros P, Terg R, Shouval D, van Hoek B, Weiland O, Van Heeswijk R, De Meyer S, Luo D, Boogaerts G, Polo R, Picchio G, Beumont M. Telaprevir for retreatment of HCV infection. New England Journal of Medicine. 2011;364:2417–2428. doi: 10.1056/NEJMoa1013086. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Harris D, Pandey VN. The FUSE binding protein is a cellular factor required for efficient replication of hepatitis C virus. Journal of Virology. 2008;82:5761–5773. doi: 10.1128/JVI.00064-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.