Abstract
Domestic dog breeds have undergone intense selection for a variety of morphologic features, including size. Among small-dog breeds, defined as those averaging less than ~15 in. at the withers, there remains still considerable variation in body size. Yet essentially all such dogs are fixed for the same allele at the insulin-like growth factor 1 gene, which we and others previously found to be a size locus of large effect. In this study we sought to identify additional genes that contribute to tiny size in dogs using an association scan with the single nucleotide polymorphism (SNP) dataset CanMap, in which 915 purebred dogs were genotyped at 60,968 SNP markers. Our strongest association for tiny size (defined as breed-average height not more than 10 in. at the withers) was on canine chromosome 3 (p = 1.9 × 10−70). Fine mapping revealed a nonsynonymous SNP at chr3:44,706,389 that changes a highly conserved arginine at amino acid 204 to histidine in the insulin-like growth factor 1 receptor (IGF1R). This mutation is predicted to prevent formation of several hydrogen bonds within the cysteine-rich domain of the receptor’s ligand-binding extracellular subunit. Nine of 13 tiny dog breeds carry the mutation and many dogs are homozygous for it. This work underscores the central importance of the IGF1 pathway in controlling the tremendous size diversity of dogs.
Introduction
Body size is a prototypical complex trait (Lettre 2011). Although size is easy to measure and is highly heritable, it has proved a challenging trait to study in humans (Aulchenko et al. 2009; Perola 2011). It is only relatively recently that large-scale genome-wide association scans (GWAS) have begun to identify loci associated with human height variation (Sanna et al. 2008; Weedon et al. 2007). Now, using dense SNP scans in large samples, 180 size-controlling loci have been identified in the human genome, but cumulatively they explain just ~12 % of the heritable variation in height and the effect size of each variant is very small (Hirschhorn and Gajdos 2011; Kim et al. 2010; Lango Allen et al. 2010; Liu et al. 2010; Okada et al. 2010; Soranzo et al. 2009; Weedon et al. 2008). Thus, the genetic control of human height is highly complex and studies to identify the genes involved require tremendous sample sizes to achieve sufficient statistical power.
Domestic dogs offer an important opportunity in this context: breeds are selected for specific body sizes and as a result purebred dogs vary fourfold in bone length measures (Sutter et al. 2008). The majority of purebred dog size variation is between breeds rather than within them, consistent with the intense selection for size applied by breeders. In stark contrast to the complexity of size genetics in humans, Chase et al. (2002) showed that a single locus on canine chromosome 15 was strongly associated with size and could explain 10 % of the variation in size in the Portuguese Water Dog breed, a result subsequently verified (Jones et al. 2008; Sutter et al. 2007; Vaysse et al. 2011). At this locus we found a single haplotype spanning the insulin-like growth factor 1 (IGF1) gene that had experienced a selective sweep to very high frequency in nearly all small breeds of dog (Sutter et al. 2007). Since the small-size-associated haplotype is widely shared among small breeds, developed for a variety of different reasons, the initial variant likely arose early in domestication. This haplotype appears to have originated in or near the Fertile Crescent (Gray et al. 2010), which may also be one of the sites of dog domestication (vonHoldt et al. 2010). Thus, via intense selection within inbred lines, an old haplotype rose to high frequency and today it has a very large effect on purebred dog size. This illustrates how the world’s ~400 dog breeds provide, in aggregate, a powerful tool for genetically dissecting complex traits (Parker et al. 2010). The power to map complex traits using dog breeds is especially high when the traits are under positive selection, as typified by size (Sutter et al. 2007), ear erectness (Vaysse et al. 2011), snout length and body mass (Boyko et al. 2010), brachycephaly (Bannasch et al. 2010), limb dwarfism (Parker et al. 2009), white spotting and hair ridge (Karlsson et al. 2007; Salmon Hillbertz et al. 2007), pelage (Cadieu et al. 2009), and skin wrinkling (Olsson et al. 2011).
Serum levels of IGF1 are correlated with both Poodle size (Eigenmann et al. 1984) and the selectively swept IGF1 haplotype (Sutter et al. 2007), but IGF1 is clearly not the only genetic variant controlling size. Jones et al. (2008) genotyped dogs from 148 breeds and identified the IGF1 locus as well as other loci significantly associated with breed-average height and weight traits, including five loci with strong candidate genes. Boyko et al. (2010) generated the CanMap dataset of 915 dogs from 80 breeds that were each genotyped at 60,968 SNPs and with these data identified numerous loci significantly associated with breed-average log(body weight) and morphometric measurements. A similar approach was recently used to study breed-average traits shared among 46 breeds using the Canine HD array of 172,000 SNPs (Vaysse et al. 2011).
Nearly all small breeds of dog are fixed for the IGF1 haplotype, yet within this group of breeds there is still considerable size variation. With the aim of identifying genes contributing to the tiniest dog sizes (i.e., dogs in breeds averaging no more than 10 in. at the withers), we utilized the CanMap genotypes to conduct a GWAS for tiny size. Following fine-mapping, we identified a nonsynonymous SNP in the IGF1 receptor (IGF1R) that is associated with dog size.
Materials and methods
Sample collection and DNA isolation
Purebred dog data and biomaterials were collected under approved animal care and use committee protocols at Cornell University and NHGRI/NIH. Blood samples were collected into EDTA anticoagulant at dog shows or by soliciting owners. Genomic DNA (gDNA) was extracted from white blood cells using standard proteinase K/phenol:chloroform isolation (Sambrook and Russell 2001). Pedigrees from samples used in the study were inspected to avoid including close relatives (usually meaning no dogs within any breed shared parents or grandparents with any other dog in that breed).
Phenotype assignment
Every dog within a given breed was assigned the same phenotypic value that represented an estimate of the typical size of dogs within that breed. For each breed we utilized web resources such as the published breed standards available at the American Kennel Club (AKC 1998) to estimate the average withers height for an adult male in the breed. This single value was then applied to all of our samples from that breed to represent that breed’s size.
SNP genotyping
We utilized the CanMap Affymetrix genotyping dataset collected by Boyko et al. (2010) and described in detail in their paper and elsewhere (Cadieu et al. 2009). We also genotyped SNPs using two other approaches: (1) we identified SNPs from capillary sequence traces collected for mutation scanning, and (2) by fine-mapping using the SNPlex custom genotyping platform (Applied Biosystems, Carlsbad, CA).
Capillary sequencing
To generate amplicons for capillary sequencing (list of primers in Supplementary Table 1), PCR primers were designed with Primer3 (Rozen and Skaletsky 2000). Between 1 and 15 ng of gDNA (usually 10 ng) was used as template in a 10-μl PCR with AmpliTaq Gold DNA polymerase in the manufacturer’s provided buffer (Applied Biosystems). PCR ran for 20 touchdown cycles (annealing temperature varying from 61 °C to 51 °C, dropping 0.5 °C each cycle) followed by 20 more cycles at 51 °C annealing temperature. Primers were removed from PCR amplicons using exonuclease I and shrimp alkaline phosphatase treatment at 37 °C for 30 min, followed by incubation for 15 min at 80 °C. Each PCR amplicon was sequenced in both directions using the PCR primer oligonucleotides to prime the sequencing reactions or, for a few amplicons, with internal sequencing primers. Sequencing reactions used the BigDye 3.1 Applied Biosystems master mix in a 5-μl reaction with 2 μl of the exo/SAP-cleaned PCR amplicon. Products from sequencing reactions were isolated by ethanol precipitation, suspended in 15 μl of water, and analyzed on an ABI3730 capillary sequencer.
SNPlex genotyping
Fine-mapping at the canine chromosome 3 locus utilized the SNPlex genotyping system from Applied Biosystems. Six pools of up to 48 SNPs each were designed to query a total of 287 SNPs. Genomic DNA samples (100–250 ng for each pool assay) were subjected to the SNPlex genotyping protocol, and raw genotype traces were collected on an ABI3730 following established procedures. SNP genotypes were called using GeneMapper (Applied Biosystems), and of the 287 attempted SNPs, 211 passed quality control and were found to be segregating two alleles in the sample population. These were included in the analyses.
Genetic analyses
PLINK (Purcell et al. 2007) and Haploview (Barrett et al. 2005) were used to test for significance of association and assess patterns of linkage disequilibrium (LD), respectively. PHASE (Stephens et al. 2001) was used to infer haplotypes spanning region 2 of the IGF1R gene. Using the phase_pairs output, we summed over the PHASE-assigned probability for each haplotype pair in each sample to accumulate the estimated chromosome counts for each haplotype for each breed.
IGF1R structure analysis and alignment
Crystallographic coordinates for domains 1–3 of the IGF1 receptor (Garrett et al. 1998) were downloaded from the RCSB protein databank (accession ID: 1IGR) and visualized using the PyMol Molecular Graphics System ver. 1.3 (Schroedinger, LLC). For the IGF1R and insulin receptor alignment, residues 192–213 of the predicted amino acid sequence for the dog IGF1R were used as a query in protein BLAST to search GenBank for sequences for the IGF1R and the insulin receptor from multiple species.
Results
We aimed to identify genetic factors that contribute to the body size phenotype of the tiniest dogs, defined as those with breed-average height ≤10 in. (25.4 cm) at the withers according to American Kennel Club resources (AKC 1998). To do this, we made use of the genotypes from a previously published GWAS, CanMap (Boyko et al. 2010), to identify size loci by scoring breeds either as “tiny” or “control.” We defined “tiny” breeds as those in which the average height is ≤10 in. (25.4 cm) at the withers. The nine breeds scored as tiny were the Brussels Griffon, Cairn Terrier, Chihuahua, Havanese, Norwich Terrier, Papillon, Pomeranian, Toy Poodle, and Yorkshire Terrier. These represent the smallest 10 % of breeds in the CanMap dataset (Supplementary Table 2) and are represented by 93 dogs. To avoid trait-confounding from limb-dwarfed breeds, which have very small withers heights despite their sometimes larger body sizes overall, we removed from the CanMap dataset 11 breeds that carry this trait (Parker et al. 2009). This left 676 dogs in 60 small, medium, and giant breeds to serve as non-tiny controls (Supplementary Table 2).
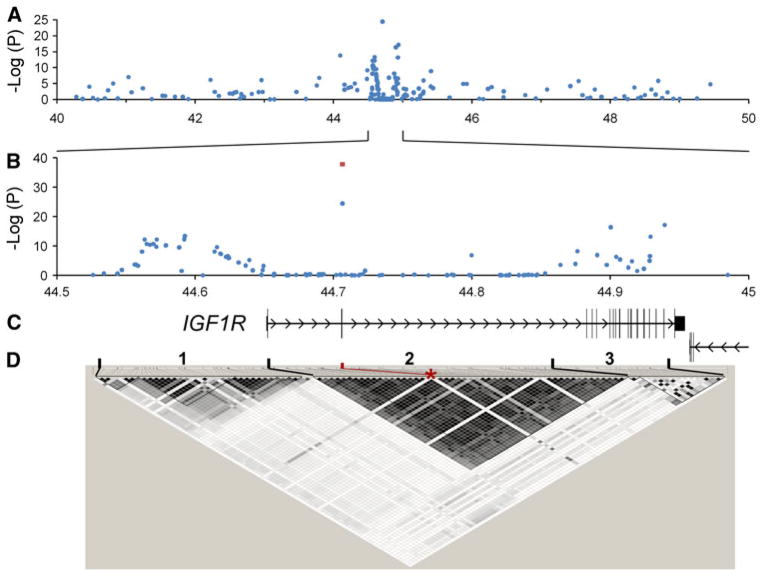
Comparing the nine tiny breeds to the remaining 60 breeds revealed a strong signal (p = 1.9 × 10−70) on canine chromosome 3 at 43,756,620 bp (Fig. 1; all genome positions are given in the CanFam2 dog genome reference assembly). Not surprisingly, the analysis also identified an association at IGF1, although this time, chromosome 15, where IGF1 lies, was not in the top tier of associations as it has been in other analyses for size (Boyko et al. 2010; Chase et al. 2002; Jones et al. 2008; Vaysse et al. 2011).
Fig. 1.
Genome-wide association scan (GWAS) for tiny size as a binary trait. Dogs from nine breeds (n = 93) with breed-average withers heights of 10 in. or smaller were coded “tiny,” while dogs from 60 small, medium, or large breeds were coded “control” (n = 676). Achondrodysplastic breeds were excluded from the analysis. The x axis shows each marker’s position in the genome
Additional size-associated loci were also observed in this GWAS; they are the focus of other follow-up studies. For chromosome 3, the strongest associated SNP was located in a large intron in the middle of the ADAM metallopeptidase with thrombospondin type 1 motif, 17 (ADAMTS17) gene. Interestingly, a SNP in ADAMTS17 has been previously associated with human height (Gudbjartsson et al. 2008), although the exact function of this secreted zinc metalloprotease is still unknown.
We scanned ADAMTS17 for putative variants controlling reduced size by collecting bidirectional capillary sequence from PCR amplicons generated in ten tiny and six large dogs (Supplementary Table 1). Our sequencing amplicons spanned the exons and exon/intron boundaries of the gene. Among the variants found (Supplementary Table 3), we identified three synonymous SNPs (chr3:43,472,198 C/T, chr3:43,472,288 C/T, and chr3:43,774,819 A/G) not reported in dbSNP build 131. We also found two synonymous (rs8575121 and rs8809065) and one nonsynonymous SNP (rs8809064), which are found in dbSNP build 131. The nonsynonymous SNP is at chr3:43,778,853 and causes a V966A substitution in exon 21; this amino acid is C-terminal to one of the thrombospondin domains of the protein. It is unlikely that the V966A substitution affects ADAMTS17 function: SIFT scores it as “tolerated,” and PolyPhen, which uses sequence-based and structure-based predictive features, labels it “benign,” with a score of 0.201 and a sensitivity of 0.92 (Adzhubei et al. 2010; Kumar et al. 2009). Furthermore, among available GenBank records, only the dog and horse ADAMTS17 protein sequences have valine at this position. Alanine, the variant amino acid generated by the nonsynonymous SNP, is actually more common in vertebrates and is found in the ADAMTS17 protein in humans, rats, and eight other vertebrates.
Upon analysis of LD patterns at this locus, we found that CanMap markers at ADAMTS17, including the size-associated SNP, were in LD with other markers on chromosome 3. Also, LD extended for longer distances in tiny versus other sized dogs (Supplementary Fig. 1). In particular, we found LD between the ADAMTS17 size-associated SNP and a set of seven contiguous SNPs that define a haplotype block spanning the insulin-like growth factor 1 receptor (IGF1R) gene, which is a highly plausible candidate for body-size variation.
Accordingly, we broadened our fine-mapping search by constructing and genotyping a custom panel of 211 SNPs in tiny and nontiny dogs. These SNPs cover a 10-Mb interval of canine chromosome 3, from 40 to 50 Mb, that is roughly centered on IGF1R, spanning 300 kb at chr3:44,650,000–44,950,000 bp. ADAMTS17 covers 330 kb at chr3:43,470,000–43,800,000 bp and is included in the fine-mapping scan by three SNPs within the gene plus a fourth SNP that is immediately upstream of the gene (Supplementary Table 4). We also discovered sequence variations in IGF1Rs exons and exon/intron boundaries by collecting bidirectional capillary sequences from PCR amplicons generated in ten tiny and six large dogs (Supplementary Table 2).
The fine-mapping SNPs were genotyped in 490 dogs representing 57 breeds, although the number of samples successfully genotyped at each SNP varied, partly due to the by-pool SNP-calling in the SNPlex genotyping platform (see Supplementary Table 4 for by-marker sample coverage). Of the 490 dogs, 132 were included in the GWAS and 358 were not. Because the overall number and breed number of this sample are smaller than those of the CanMap dataset used in the initial GWAS, and because the sample includes many small to medium-sized breeds, we tested for association with a quantitative size trait, i.e., we assigned a size value to every dog in a breed equal to the average withers height of an adult male in that breed (Supplementary Table 5). For this analysis we therefore ignored variation that occurs within each breed, instead focusing on the variation between breeds.
As shown in Fig. 2, we identified a strong association with breed-average height at the IGF1R gene, with the peak association occurring for a SNP at chr3:44,706,389 (p = 3.6 × 10−25). This is an A/G nonsynonymous SNP in the second coding exon of IGF1R that changes an arginine to histidine. None of the SNPs surrounding this one are associated, but additional association signals are obtained from SNPs immediately upstream of IGF1R and from several SNPs within the downstream third of the gene, where the majority of the gene’s exons lie (Fig. 2b, c). LD patterns over IGF1R show three independent regions of LD (Fig. 2d). The first spans the 100 kb upstream of IGF1R. The second spans the upstream two-thirds of the transcribed portion of the gene and includes the previously mentioned nonsynonymous SNP. This region contains the SNPs in LD with the SNP in ADAMTS17 that showed the highest association with size in the GWAS. The third region covers the downstream third of the gene and most of IGF1Rs exons. The nonsynonymous SNP, unlike other SNPs within region 2, is in LD with a large fraction of the SNPs in both regions 1 and 3 (Fig. 2d). Furthermore, it is the only SNP within region 2 that is associated with size.
Fig. 2.
Fine-mapping for dog size in the a chr3:40,000,000–50,000,000-Mb interval. At each marker, up to 475 dogs were genotyped and a statistical test was conducted for association with breed average height as a quantitative trait (blue-filled circles in a and b). Genotyping 298 additional dogs at the IGF1R nonsynonymous SNP (chr3:44,706,389) was used to confirm association. The filled square shows the p value resulting from the total set of sample genotypes for the nonsynonymous SNP. c Gene exons (vertical bars) and introns (lines with arrows in the direction of transcription) are shown to scale. d LD between SNP pairs, where darker diamonds indicate higher r2 values (Color figure online)
We questioned whether the LD between the nonsynonymous SNP and neighboring SNPs in regions 1 and 3 was responsible for the association signals we observe for those SNPs. We therefore repeated the association analysis, but this time it was conditioned on the nonsynonymous SNP by including the SNPs genotypes in the linear model as a covariate. The majority of the association signals previously observed in regions 1 and 3 are then lost (Supplementary Fig. 2), supporting the model that the size association at this locus is driven by the nonsynonymous SNP alone.
We tested the nonsynonymous SNPs association with size in a broader cross section of dogs by genotyping at this marker an additional 298 purebred dogs of variable breed-average height; the p value in the test for association with size decreased to 1.4 × 10−38 in the combined larger sample set of 619 samples (Fig. 2b).
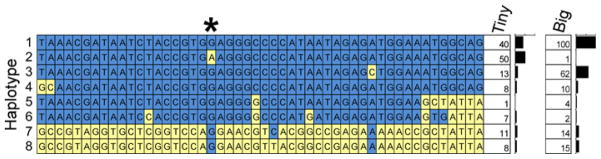
Although the nonsynonymous SNP in region 2 showed strong association with size, this was not true of the other SNPs in region 2. To investigate this, we inferred haplotypes with PHASE for the 50 SNPs of region 2, spanning chr3:44,656,749–44,852,968 (Fig. 3). Only haplotype #2 carries the “A” allele for the nonsynonymous SNP at chr3:44,706,389 and it differs from the very common haplotype #1 only at the nonsynonymous SNP. This explains the lack of size association at most markers in this interval. Based on the haplotype patterns in Fig. 3, we infer that the chr3:44,706,389 SNP “A” allele associated with tiny size is the derived allele, while the “G” allele is ancestral. Such an inference is also supported by the fact that four Golden Jackal (Canis aureus) chromosomes all carry a G at the site. This is consistent with a model in which haplotype #2 arose directly from haplotype #1 when the nonsynonymous SNPs “A” allele was created on a chromosome carrying haplotype #1 at IGF1Rs region 2.
Fig. 3.
Haplotypes from the linkage disequilibrium (LD) block that includes the nonsynonymous “A”/“G” SNP at chr3:44,706,389 (indicated by the asterisk). This haplotype block is indicated as “2” in Fig. 2 and spans the large first and second introns of IGF1R. The 50 SNPs are shown as columns and alleles are shaded. The bar graphs on the right show the total number of chromosomes carrying each haplotype for each of two groups of breeds: “Tiny” = 13 breeds in which the average adult male’s height at the withers is no more than 10 in., and “Big” = 15 breeds with average adult male’s height at the withers of at least 25 in. See also Supplementary Fig. 3 for breed-by-breed counts of chromosomes for each haplotype
Both large and tiny dog breeds carry haplotype #1, which we found in 23 breeds (Supplementary Fig. 3). In contrast, the nearly identical haplotype #2 is fairly common in tiny breeds but is essentially absent in large breeds (Supplementary Fig. 3). Interestingly, haplotype #2, which carries the nonsynonymous SNPs “A” allele, is present in the Dachshund and Poodle breeds, and in each breed it is found predominantly in the smaller-sized varieties of the breed. We assayed 13 Miniature Dachshunds for genotypes at the nonsynonymous SNP but detected only “AA” (Table 1). In contrast, the larger Standard-sized Dachshunds carry all three genotypes, “AA,” “AG,” and “GG,” and the difference in genotype frequencies between Miniatures and Standards is significant (p < 4.6 × 10−4, two-tailed Fisher’s exact test on three genotype classes between Miniature and Standard Dachshunds). In similar fashion, the small poodles (Toy and Miniature varieties) have a significantly different genotype frequency from that of the Standard-sized poodles; all but one of the 16 Standard Poodles assayed have the “GG” genotype (p < 6.0 × 10−6, two-tailed Fisher’s exact test on genotype classes, comparing Standard Poodles versus Toy and Miniature together).
Table 1.
Genotype counts and allele frequencies within Dachshund and Poodle size varieties for the IGF1R nonsynonymous SNP at chr3:44,706,389
| Dachshunds
|
Poodles
|
||||
|---|---|---|---|---|---|
| Miniature | Standard | Toy | Miniature | Standard | |
| Genotype | |||||
| AA | 13 | 4 | 4 | 3 | 0 |
| AG | 0 | 7 | 4 | 6 | 1 |
| GG | 0 | 2 | 2 | 1 | 15 |
| Allele frequency | |||||
| A | 1 | 0.58 | 0.6 | 0.6 | 0.03 |
| G | 0 | 0.42 | 0.4 | 0.4 | 0.97 |
We also addressed the question of how common this allele is in dogs of intermediate size by genotyping through sequencing dogs of different breeds. In looking at individual dogs from 21 additional breeds of diverse origin with average height at the withers >10 in. but <21 in., we found that samples from 13 of the 21 breeds carried no chromosomes with the tiny-size-associated “A” allele. Of the eight breeds in which the “A” allele was detected, most were quite small (Chinese Crested, Italian Greyhound, Toy Fox Terrier) or of variable size (Jack Russell Terrier, Shetland Sheepdog). We also found the “A” allele at low frequency in Miniature Schnauzers and Bulldogs and at a higher frequency in Sussex Spaniels. The only breeds larger than the Bulldog in which we detected the “A” allele were the Standard Poodle and the Irish Water Spaniel.
The SNP at chr3:44,706,389 changes amino acid 204 from an arginine to a histidine (R204H). IGF1R is a tyrosine kinase receptor with proteolytically derived α and β subunits (Forbes 2011). Amino acid 204 is found in the cysteine-rich portion of the extracellular ligand-binding domain of the receptor’s α subunit (Garrett et al. 1998), which contains some of the contact points made with IGF1 ligand when it binds (Keyhanfar et al. 2007). The arginine at position 204 is not only completely conserved in all of the sequenced IGF1R proteins (Fig. 4), but is one of just a few residues in this region of the polypeptide that is also completely conserved in the related molecule, insulin receptor. The high conservation of this amino acid, its location in the extracellular domain of the receptor, and the putative loss of hydrogen bonds in the R204H substitution (Fig. 5) all support the idea that this substitution affects receptor function. Furthermore, the SIFT score for the R204H substitution is “not tolerated” and PolyPhen scores it as “probably damaging,” with a sensitivity of 1.0 (Adzhubei et al. 2010; Kumar et al. 2009).
Fig. 4.
Amino acid alignment of the cysteine-rich domain from the IGF1 receptor and insulin receptor. Asterisks indicate invariant amino acids, and the arginine at position 204 is boxed
Fig. 5.
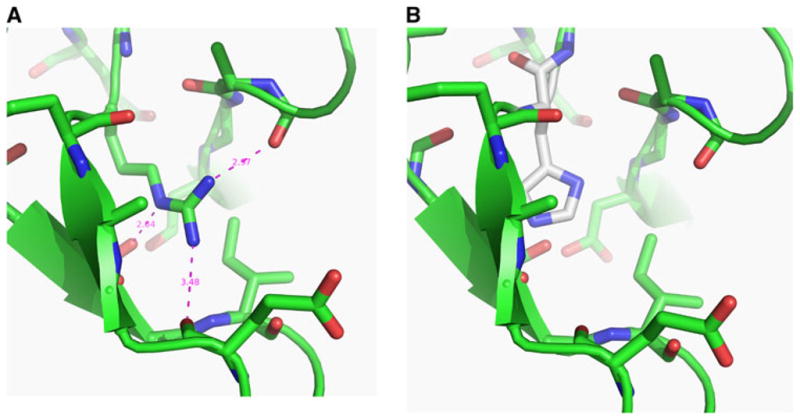
The R204H mutation in IGF1R is predicted to destroy three hydrogen bonds within a nonsolvent accessible region of the cysteine-rich domain. a The structure determined by Garrett et al. (1998) with arginine at 204. The arginine side chain is at the center of the image. b The structure with histidine at residue 204 where putative hydrogen bonds to S153 (carbonyl), E200 (side chain), and T195 (side chain) have been lost
Discussion
The domestic dog is a rich resource for studying the genetic control of body size. Dog breeds differ greatly from one another in body size, weight, bone lengths, and height at the withers such that many breeds represent semi-isolated populations, illustrating one possible outcome from intense selection on size. The population structure of purebred dogs is well suited to the difficult task of unraveling complex trait genetics (Karlsson et al. 2007). For 99 % of the history of the dog species, from ~15,000 to 100 years before the present, there was potential for gene flow between populations so that alleles under selection could move through subpopulations. In the late nineteenth century the newly formed kennel clubs initiated policies of strict reproductive isolation for each pure breed of dog. With few exceptions these policies continue to this day, so that each breed carries the particular set of alleles provided to it by its founding members a century ago. Under such conditions, the issues of founder effect, chronically small population sizes, genetic drift, and in some cases intense selection lead us to hypothesize that while there may be many size-controlling loci in the dog genome, perhaps any particular breed is likely fixed for a single allele at several such loci. This reduction in locus heterogeneity greatly simplifies mapping by limiting the number of segregating loci that must be simultaneously identified (Karlsson et al. 2007; Ostrander and Wayne 2005).
In this study we aimed to understand the genetic control of tiny size in the domestic dog. We first investigated the ADAMTS17 gene because it is directly within the top-ranked size-associated locus identified in our GWAS. However, our mutation scanning by capillary sequencing detected no obvious candidates likely to be causal for size. The only nonsynonymous SNP identified in the region represents a fairly conservative amino change from valine to alanine, and most vertebrates sequenced to date contain an alanine at this position. Furthermore, there are reports that have identified a severe truncating mutation in this gene as the cause of primary lens luxation in the dog (Farias et al. 2010), and as noted by the authors, none of their data or previous reports indicate a difference in size between affected and unaffected dogs. This mutation has been found in 14 of 30 breeds tested in a recent study (Gould et al. 2011), and although most of the breeds with the mutation are small, the more striking relationship common to most breeds with the mutation is that they are terriers or from breeds with putative terrier ancestry (Gould et al. 2011). Therefore, while our mapping does not formally exclude ADAMTS17 and, as proposed by Farias et al. (2010), a careful analysis of body size in breeds with the primary lens luxation mutation would be worthwhile, we consider the variation in ADAMTS17 an unlikely candidate for contributing to anything but perhaps subtle differences in dog body size.
The GWAS size-associated SNP within ADAMTS17 is in LD with several SNPs in a block spanning the IGF1R gene 900 kb away; LD often extends a megabase pair or more in purebred dogs (Lindblad-Toh et al. 2005; Sutter et al. 2004). Fine-mapping identified a nonsynonymous SNP within the second protein-coding exon of IGF1R that is associated with tiny size and is predicted to cause an amino acid change from arginine to histidine at residue 204. There are several lines of evidence supporting the idea that the alleles of this SNP functionally contribute to body size differences. First, inspection of the IGF1R protein structure (Garrett et al. 1998) suggests that the substitution to histidine may prevent formation of several hydrogen bonds that form between the arginine side group and neighboring atoms. Amino acid R204 is in a nonsolvent accessible region of the protein and, as would be predicted, there are potential acceptors in close proximity to form hydrogen bonds with the atoms of the side chain of arginine. The backbone carbonyl of S153, the carbonyl group of E200, and the side chain of T195 are all within 3.48 Å of atoms in R204s side chain. All of these residues are conserved in the predicted IGF1R protein in the dog. Replacement of the arginine at position 204 with a histidine residue eliminates the possibility of several of these hydrogen bonds and may also generate a steric clash with T195. The R204 amino acid is part of a loop found at the base of the extracellular domain, and the hydrogen bonds would be predicted to promote interaction of the loop with the rest of the domain. The arginine found at this position is highly conserved as it is invariant not only in IGF1R but also in the related insulin receptor proteins from mammals, birds, frogs, and fish.
In addition, we find the nonsynonymous SNP in a diverse set of tiny dog breeds, but almost no large breeds carry it. Furthermore, the trend within both Poodle and Dachshund size varieties is toward an increasing “A” allele frequency as size decreases (from the Standard variety down to the Miniature and Toy in the Poodle and from Standard to Miniature in the Dachshund). These results are also consistent with the identification of a signature of selection that spans the IGF1R gene (Akey et al. 2010; Vaysse et al. 2011). We hypothesize, therefore, that the IGF1R nonsynonymous SNP at chr3:44,706,389 is a causal mutation for tiny size in dogs. Future functional studies will be needed to determine the precise nature of this variant.
While we cannot exclude more complex scenarios, our results are consistent with the model that this SNP is the only causal mutation contributing to size variation at this locus. We did observe other association signals at IGF1R, both upstream of transcription and over the downstream coding exons, but these signals appear to be spuriously derived from the nonsynonymous SNP via LD. Mutations in the IGF1R gene in mice and humans that eliminate or decrease receptor levels or activity have been shown to affect body size. As noted earlier, homozygous knock outs of the IGF1R gene in mice is lethal perinatally, and pups are born at only 45 % of normal body mass (Liu et al. 1993). Mice heterozygous for gene knockouts (Bokov et al. 2011; Liu et al. 1993) or tissue-specific knockouts (Kappeler et al. 2008; Romero et al. 2010) also have reduced body size or mass. Several human families have been described in which heterozygous mutations in IGF1R have been linked to intrauterine and postnatal growth defects; some of these mutations have been shown to decrease IGF1 signaling through a variety of mechanisms (reviewed by Kawashima et al. 2012).
The results reported here, together with our previous findings, underscore the critical importance of the IGF1 pathway in controlling dog size. The IGF1 pathway affects a number of different tissues during development and postnatally, and complex interrelationships exist between this pathway and the growth hormone pathway (Netchine et al. 2011). The IGF1 small-size haplotype is correlated with a significant reduction in circulating levels of IGF1 hormone (Sutter et al. 2007). In this study we found that many of the tiniest dogs also carry an R204H mutation in their IGF1 receptor. Thus, these tiny dogs have reduced levels of IGF1 and, we hypothesize, may also have a reduced capacity to bind IGF1 at its receptor or transduce signals that should result from binding. We found many dogs that are homozygous for the R204H mutation, suggesting that the mutant IGF1R does have at least some function; R204H homozygous dogs do not die at birth as do mice homozygous for an IGF1R knockout (Liu et al. 1993).
IGF1 and IGF2 hormones promote cell growth, survival, and differentiation by binding the IGF1 receptor, which is a transmembrane receptor tyrosine kinase with a structure very similar to the insulin receptor: it contains two α and two β subunits (Forbes 2011). IGF1Rs extracellular α subunit binds ligands via contacts at several domains and there is evidence that the cysteine-rich domain is involved in the binding of IGF1 but not IGF2 (Forbes 2011; Keyhanfar et al. 2007). The dog R204H mutation is located in the cysteine-rich domain of the extracellular α subunit so it is possible that the mutation may affect IGF1 binding differently than IGF2 binding. A human mutation in the IGF1-C domain causes growth deficiency, just as other IGF1 mutations do, but it does so without also causing the severe development delay and deafness that result from other mutations in human IGF1 (Netchine et al. 2009, 2011). The IGF1-C domain makes contact with the IGF1R cysteine-rich domain; perhaps the dog R204H mutation in the cysteine-rich domain is able to confer a desirable small size without negative effects on hearing or developmental delay.
The IGF1R nonsynonymous SNP’s “A” allele is present in many, but not all, of the tiny breeds we studied. It may be that the pattern we previously found at the IGF1 gene, in which essentially all small dogs are fixed for a single allele, is the exception rather than the rule for many loci contributing to dog size. The IGF1 allele has a large effect on size, while the IGF1R mutation we report here appears to have a more modest effect: it contributes to very small size but dogs can be tiny without carrying it. Our data therefore support the idea that there exist different combinations of alleles that can produce tiny dog size. All tiny breeds are intensely selected for their diminutive sizes, but it seems they can arrive there by different genetic means. It appears that all such combinations of genetic factors do require the IGF1 allele, but small-size alleles at other loci, like IGF1R, may or may not be needed in a particular tiny breed.
Such a scenario is consistent with the known history of breed creation via tight population bottlenecks. Many alleles are likely to be lost in chronically small and highly inbred populations such as purebred dogs, and if a tiny-sized breed should ever become fixed for a large-size allele at a locus (via genetic drift despite selection for small size, for example), there are no means to ever recover the small-size allele except by new mutation. At the point when additional size loci have been identified in dogs, it will be interesting to learn whether any tiny breeds carry the small-size-associated alleles at every size locus. If none do, it will suggest that either the vagaries of drift and breed isolation prevented such a fortunate combination or that such a combination is not biologically compatible with life or health.
Supplementary Material
Acknowledgments
We thank members of the Ostrander and Sutter laboratories and the students of BCH. We thank Jeffrey Schoenebeck for helpful discussion and Roger Rowlett for assistance with IGF1R structure analysis. This research would not be possible without the generous support of dog owners who provided access to their animals. BCH carried out a majority of her portion of this work while on sabbatical in the Ostrander lab with support from the Colgate Research Council. This work was funded by the intramural program at NHGRI/NIH (EAO, MR, DL) and Cornell University internal funds (NBS).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00335-012-9417-z) contains supplementary material, which is available to authorized users.
Contributor Information
Barbara C. Hoopes, Department of Biology, Colgate University, 13 Oak Drive, Hamilton, NY 13346, USA
Maud Rimbault, National Human Genome Research Institute, Building 50, Room 5349, 50 South Drive MSC 8000, Bethesda, MD 20892, USA.
David Liebers, National Human Genome Research Institute, Building 50, Room 5349, 50 South Drive MSC 8000, Bethesda, MD 20892, USA.
Elaine A. Ostrander, National Human Genome Research Institute, Building 50, Room 5349, 50 South Drive MSC 8000, Bethesda, MD 20892, USA
Nathan B. Sutter, Email: nbs39@cornell.edu, Department of Clinical Sciences, College of Veterinary Medicine, Cornell University, Ithaca, NY 14853, USA. C3-179 Vet Medical Center, Cornell University, Ithaca, NY 14850, USA
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey JM, Ruhe AL, Akey DT, Wong AK, Connelly CF, Madeoy J, Nicholas TJ, Neff MW. Tracking footprints of artificial selection in the dog genome. Proc Natl Acad Sci USA. 2010;107:1160–1165. doi: 10.1073/pnas.0909918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Kennel Club (AKC) The complete dog book. 19. Howell Book House; New York: 1998. [Google Scholar]
- Aulchenko YS, Struchalin MV, Belonogova NM, Axenovich TI, Weedon MN, Hofman A, Uitterlinden AG, Kayser M, Oostra BA, van Duijn CM, Janssens AC, Borodin PM. Predicting human height by Victorian and genomic methods. Eur J Hum Genet. 2009;17:1070–1075. doi: 10.1038/ejhg.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannasch D, Young A, Myers J, Truve K, Dickinson P, Gregg J, Davis R, Bongcam-Rudloff E, Webster MT, Lindblad-Toh K, Pedersen N. Localization of canine brachycephaly using an across breed mapping approach. PLoS ONE. 2010;5:e9632. doi: 10.1371/journal.pone.0009632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bokov AF, Garg N, Ikeno Y, Thakur S, Musi N, DeFronzo RA, Zhang N, Erickson RC, Gelfond J, Hubbard GB, Adamo ML, Richardson A. Does reduced IGF1R signaling in Igf1r+/− mice alter aging? PLoS ONE. 2011;6(11):e26891. doi: 10.1371/journal.pone.0026891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Quignon P, Li L, Schoenebeck JJ, Degenhardt JD, Lohmueller KE, Zhao K, Brisbin A, Parker HG, vonHoldt BM, Cargill M, Auton A, Reynolds A, Elkahloun AG, Castelhano M, Mosher DS, Sutter NB, Johnson GS, Novembre J, Hubisz MJ, Siepel A, Wayne RK, Bustamante CD, Ostrander EA. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol. 2010;8:e1000451. doi: 10.1371/journal.pbio.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadieu E, Neff MW, Quignon P, Walsh K, Chase K, Parker HG, vonHoldt BM, Rhue A, Boyko A, Byers A, Wong A, Mosher DS, Elkahloun AG, Spady TC, Andre C, Lark KG, Cargill M, Bustamante CD, Wayne RK, Ostrander EA. Coat variation in the domestic dog is governed by variants in three genes. Science. 2009;326:150–153. doi: 10.1126/science.1177808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase K, Carrier DR, Adler FR, Jarvik T, Ostrander EA, Lorentzen TD, Lark KG. Genetic basis for systems of skeletal quantitative traits: principal component analysis of the canid skeleton. Proc Natl Acad Sci USA. 2002;99:9930–9935. doi: 10.1073/pnas.152333099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigenmann JE, Patterson DF, Froesch ER. Body size parallels insulin-like growth factor I levels but not growth hormone secretory capacity. Acta Endocrinol (Copenh) 1984;106:448–453. doi: 10.1530/acta.0.1060448. [DOI] [PubMed] [Google Scholar]
- Farias FH, Johnson GS, Taylor JF, Giuliano E, Katz ML, Sanders DN, Schnabel RD, McKay SD, Khan S, Gharahkhani P, O’Leary CA, Pettitt L, Forman OP, Boursnell M, McLaughlin B, Ahonen S, Lohi H, Hernandez-Merino E, Gould DJ, Sargan DR, Mellersh C. An ADAMTS17 splice donor site mutation in dogs with primary lens luxation. Investig Ophthalmol Vis Sci. 2010;51:4716–4721. doi: 10.1167/iovs.09-5142. [DOI] [PubMed] [Google Scholar]
- Forbes BE. Molecular mechanisms underlying insulin-like growth factor action: how mutations in the GH: IGF axis lead to short stature. Pediatr Endocrinol Rev. 2011;8:374–381. [PubMed] [Google Scholar]
- Garrett TP, McKern NM, Lou M, Frenkel MJ, Bentley JD, Lovrecz GO, Elleman TC, Cosgrove LJ, Ward CW. Crystal structure of the first three domains of the type-1 insulin-like growth factor receptor. Nature. 1998;394:395–399. doi: 10.1038/28668. [DOI] [PubMed] [Google Scholar]
- Gould D, Pettitt L, McLaughlin B, Holmes N, Forman O, Thomas A, Ahonen S, Lohi H, O’Leary C, Sargan D, Mellersh C. ADAMTS17 mutation associated with primary lens luxation is widespread among breeds. Vet Ophthalmol. 2011;14:378–384. doi: 10.1111/j.1463-5224.2011.00892.x. [DOI] [PubMed] [Google Scholar]
- Gray MM, Sutter NB, Ostrander EA, Wayne RK. The IGF1 small dog haplotype is derived from Middle Eastern grey wolves. BMC Biol. 2010;8:16. doi: 10.1186/1741-7007-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, Sulem P, Thorlacius S, Gylfason A, Steinberg S, Helgadottir A, Ingason A, Steinthorsdottir V, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Pedersen O, Aben KK, Witjes JA, Swinkels DW, den Heijer M, Franke B, Verbeek AL, Becker DM, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Gulcher J, Kiemeney LA, Kong A, Thorsteinsdottir U, Stefansson K. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Gajdos ZK. Genome-wide association studies: results from the first few years and potential implications for clinical medicine. Annu Rev Med. 2011;62:11–24. doi: 10.1146/annurev.med.091708.162036. [DOI] [PubMed] [Google Scholar]
- Jones P, Chase K, Martin A, Davern P, Ostrander EA, Lark KG. Single-nucleotide-polymorphism-based association mapping of dog stereotypes. Genetics. 2008;179:1033–1044. doi: 10.1534/genetics.108.087866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappeler L, De Magalhaes Filho C, Dupont J, Leneuve P, Cervera P, Perin L, Loudes C, Blaise A, Klein R, Epelbaum J, Le Bouc Y, Holzenberger M. Brain IGF-1 receptors control mammalian growth and lifespan through a neuroendocrine mechanism. PLoS Biol. 2008;6(10):e254. doi: 10.1371/journal.pbio.0060254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson EK, Baranowska I, Wade CM, Salmon Hillbertz NH, Zody MC, Anderson N, Biagi TM, Patterson N, Pielberg GR, Kulbokas EJ, III, Comstock KE, Keller ET, Mesirov JP, von Euler H, Kampe O, Hedhammar A, Lander ES, Andersson G, Andersson L, Lindblad-Toh K. Efficient mapping of Mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39:1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- Kawashima Y, Higaki K, Fukushima T, Hakuno F, Nagaishi J, Hanaki K, Nanba E, Takahashi S, Kanzaki S. Novel missense mutation in the IGF-1 receptor L2 domain results in intrauterine and postnatal growth retardation. Clin Endocrinol. 2012;77(2):246–254. doi: 10.1111/j.1365-2265.2012.04357.x. [DOI] [PubMed] [Google Scholar]
- Keyhanfar M, Booker GW, Whittaker J, Wallace JC, Forbes BE. Precise mapping of an IGF-I-binding site on the IGF-1R. Biochem J. 2007;401:269–277. doi: 10.1042/BJ20060890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Lee HI, Park T, Kim K, Lee JE, Cho NH, Shin C, Cho YS, Lee JY, Han BG, Yoo HW, Lee JK. Identification of 15 loci influencing height in a Korean population. J Hum Genet. 2010;55:27–31. doi: 10.1038/jhg.2009.116. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, Ferreira T, Wood AR, Weyant RJ, Segre AV, Speliotes EK, Wheeler E, Soranzo N, Park JH, Yang J, Gudbjartsson D, Heard-Costa NL, Randall JC, Qi L, Vernon Smith A, Magi R, Pastinen T, Liang L, Heid IM, Luan J, Thorleifsson G, Winkler TW, Goddard ME, Sin Lo K, Palmer C, Workalemahu T, Aulchenko YS, Johansson A, Zillikens MC, Feitosa MF, Esko T, Johnson T, Ketkar S, Kraft P, Mangino M, Prokopenko I, Absher D, Albrecht E, Ernst F, Glazer NL, Hayward C, Hottenga JJ, Jacobs KB, Knowles JW, Kutalik Z, Monda KL, Polasek O, Preuss M, Rayner NW, Robertson NR, Steinthorsdottir V, Tyrer JP, Voight BF, Wiklund F, Xu J, Zhao JH, Nyholt DR, Pellikka N, Perola M, Perry JR, Surakka I, Tammesoo ML, Altmaier EL, Amin N, Aspelund T, Bhangale T, Boucher G, Chasman DI, Chen C, Coin L, Cooper MN, Dixon AL, Gibson Q, Grundberg E, Hao K, Juhani Junttila M, Kaplan LM, Kettunen J, Konig IR, Kwan T, Lawrence RW, Levinson DF, Lorentzon M, McKnight B, Morris AP, Muller M, Suh Ngwa J, Purcell S, Rafelt S, Salem RM, Salvi E, Sanna S, Shi J, Sovio U, Thompson JR, Turchin MC, Vandenput L, Verlaan DJ, Vitart V, White CC, Ziegler A, Almgren P, Balmforth AJ, Campbell H, Citterio L, De Grandi A, Dominiczak A, Duan J, Elliott P, Elosua R, Eriksson JG, Freimer NB, Geus EJ, Glorioso N, Haiqing S, Hartikainen AL, Havulinna AS, Hicks AA, Hui J, Igl W, Illig T, Jula A, Kajantie E, Kilpelainen TO, Koiranen M, Kolcic I, Koskinen S, Kovacs P, Laitinen J, Liu J, Lokki ML, Marusic A, Maschio A, Meitinger T, Mulas A, Pare G, Parker AN, Peden JF, Petersmann A, Pichler I, Pietilainen KH, Pouta A, Ridderstrale M, Rotter JI, Sambrook JG, Sanders AR, Schmidt CO, Sinisalo J, Smit JH, Stringham HM, Bragi Walters G, Widen E, Wild SH, Willemsen G, Zagato L, Zgaga L, Zitting P, Alavere H, Farrall M, McArdle WL, Nelis M, Peters MJ, Ripatti S, van Meurs JB, Aben KK, Ardlie KG, Beckmann JS, Beilby JP, Bergman RN, Bergmann S, Collins FS, Cusi D, den Heijer M, Eiriksdottir G, Gejman PV, Hall AS, Hamsten A, Huikuri HV, Iribarren C, Kahonen M, Kaprio J, Kathiresan S, Kiemeney L, Kocher T, Launer LJ, Lehtimaki T, Melander O, Mosley TH, Jr, Musk AW, Nieminen MS, O’Donnell CJ, Ohlsson C, Oostra B, Palmer LJ, Raitakari O, Ridker PM, Rioux JD, Rissanen A, Rivolta C, Schunkert H, Shuldiner AR, Siscovick DS, Stumvoll M, Tonjes A, Tuomilehto J, van Ommen GJ, Viikari J, Heath AC, Martin NG, Montgomery GW, Province MA, Kayser M, Arnold AM, Atwood LD, Boerwinkle E, Chanock SJ, Deloukas P, Gieger C, Gronberg H, Hall P, Hattersley AT, Hengstenberg C, Hoffman W, Lathrop GM, Salomaa V, Schreiber S, Uda M, Waterworth D, Wright AF, Assimes TL, Barroso I, Hofman A, Mohlke KL, Boomsma DI, Caulfield MJ, Cupples LA, Erdmann J, Fox CS, Gudnason V, Gyllensten U, Harris TB, Hayes RB, Jarvelin MR, Mooser V, Munroe PB, Ouwehand WH, Penninx BW, Pramstaller PP, Quertermous T, Rudan I, Samani NJ, Spector TD, Volzke H, Watkins H, Wilson JF, Groop LC, Haritunians T, Hu FB, Kaplan RC, Metspalu A, North KE, Schlessinger D, Wareham NJ, Hunter DJ, O’Connell JR, Strachan DP, Wichmann HE, Borecki IB, van Duijn CM, Schadt EE, Thorsteinsdottir U, Peltonen L, Uitterlinden AG, Visscher PM, Chatterjee N, Loos RJ, Boehnke M, McCarthy MI, Ingelsson E, Lindgren CM, Abecasis GR, Stefansson K, Frayling TM, Hirschhorn JN. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G. Recent progress in the study of the genetics of height. Hum Genet. 2011;129:465–472. doi: 10.1007/s00439-011-0969-x. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, III, Zody MC, Mauceli E, Xie X, Breen M, Wayne RK, Ostrander EA, Ponting CP, Galibert F, Smith DR, DeJong PJ, Kirkness E, Alvarez P, Biagi T, Brockman W, Butler J, Chin CW, Cook A, Cuff J, Daly MJ, DeCaprio D, Gnerre S, Grabherr M, Kellis M, Kleber M, Bardeleben C, Goodstadt L, Heger A, Hitte C, Kim L, Koepfli KP, Parker HG, Pollinger JP, Searle SM, Sutter NB, Thomas R, Webber C, Baldwin J, Abebe A, Abouelleil A, Aftuck L, Ait-Zahra M, Aldredge T, Allen N, An P, Anderson S, Antoine C, Arachchi H, Aslam A, Ayotte L, Bachantsang P, Barry A, Bayul T, Benamara M, Berlin A, Bessette D, Blitshteyn B, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Brown A, Cahill P, Calixte N, Camarata J, Cheshatsang Y, Chu J, Citroen M, Collymore A, Cooke P, Dawoe T, Daza R, Decktor K, DeGray S, Dhargay N, Dooley K, Dorje P, Dorjee K, Dorris L, Duffey N, Dupes A, Egbiremolen O, Elong R, Falk J, Farina A, Faro S, Ferguson D, Ferreira P, Fisher S, FitzGerald M, Foley K, Foley C, Franke A, Friedrich D, Gage D, Garber M, Gearin G, Giannoukos G, Goode T, Goyette A, Graham J, Grandbois E, Gyaltsen K, Hafez N, Hagopian D, Hagos B, Hall J, Healy C, Hegarty R, Honan T, Horn A, Houde N, Hughes L, Hunnicutt L, Husby M, Jester B, Jones C, Kamat A, Kanga B, Kells C, Khazanovich D, Kieu AC, Kisner P, Kumar M, Lance K, Landers T, Lara M, Lee W, Leger JP, Lennon N, Leuper L, LeVine S, Liu J, Liu X, Lokyitsang Y, Lokyitsang T, Lui A, Macdonald J, Major J, Marabella R, Maru K, Matthews C, McDonough S, Mehta T, Meldrim J, Melnikov A, Meneus L, Mihalev A, Mihova T, Miller K, Mittelman R, Mlenga V, Mulrain L, Munson G, Navidi A, Naylor J, Nguyen T, Nguyen N, Nguyen C, Nicol R, Norbu N, Norbu C, Novod N, Nyima T, Olandt P, O’Neill B, O’Neill K, Osman S, Oyono L, Patti C, Perrin D, Phunkhang P, Pierre F, Priest M, Rachupka A, Raghuraman S, Rameau R, Ray V, Raymond C, Rege F, Rise C, Rogers J, Rogov P, Sahalie J, Settipalli S, Sharpe T, Shea T, Sheehan M, Sherpa N, Shi J, Shih D, Sloan J, Smith C, Sparrow T, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Stone S, Sykes S, Tchuinga P, Tenzing P, Tesfaye S, Thoulutsang D, Thoulutsang Y, Topham K, Topping I, Tsamla T, Vassiliev H, Venkataraman V, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Yang S, Yang X, Young G, Yu Q, Zainoun J, Zembek L, Zimmer A, Lander ES. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- Liu JZ, Medland SE, Wright MJ, Henders AK, Heath AC, Madden PA, Duncan A, Montgomery GW, Martin NG, McRae AF. Genome-wide association study of height and body mass index in Australian twin families. Twin Res Hum Genet. 2010;13:179–193. doi: 10.1375/twin.13.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netchine I, Azzi S, Houang M, Seurin D, Perin L, Ricort JM, Daubas C, Legay C, Mester J, Herich R, Godeau F, Le Bouc Y. Partial primary deficiency of insulin-like growth factor (IGF)-I activity associated with IGF1 mutation demonstrates its critical role in growth and brain development. J Clin Endocrinol Metab. 2009;94:3913–3921. doi: 10.1210/jc.2009-0452. [DOI] [PubMed] [Google Scholar]
- Netchine I, Azzi S, Le Bouc Y, Savage MO. IGF1 molecular anomalies demonstrate its critical role in fetal, postnatal growth and brain development. Best Pract Res Clin Endocrinol Metab. 2011;25:181–190. doi: 10.1016/j.beem.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Okada Y, Kamatani Y, Takahashi A, Matsuda K, Hosono N, Ohmiya H, Daigo Y, Yamamoto K, Kubo M, Nakamura Y, Kamatani N. A genome-wide association study in 19,633 Japanese subjects identified LHX3-QSOX2 and IGF1 as adult height loci. Hum Mol Genet. 2010;19:2303–2312. doi: 10.1093/hmg/ddq091. [DOI] [PubMed] [Google Scholar]
- Olsson M, Meadows JR, Truve K, Rosengren Pielberg G, Puppo F, Mauceli E, Quilez J, Tonomura N, Zanna G, Docampo MJ, Bassols A, Avery AC, Karlsson EK, Thomas A, Kastner DL, Bongcam-Rudloff E, Webster MT, Sanchez A, Hedhammar A, Remmers EF, Andersson L, Ferrer L, Tintle L, Lindblad-Toh K. A novel unstable duplication upstream of HAS2 predisposes to a breed-defining skin phenotype and a periodic fever syndrome in Chinese Shar-Pei dogs. PLoS Genet. 2011;7:e1001332. doi: 10.1371/journal.pgen.1001332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander EA, Wayne RK. The canine genome. Genome Res. 2005;15:1706–1716. doi: 10.1101/gr.3736605. [DOI] [PubMed] [Google Scholar]
- Parker HG, vonHoldt BM, Quignon P, Margulies EH, Shao S, Mosher DS, Spady TC, Elkahloun A, Cargill M, Jones PG, Maslen CL, Acland GM, Sutter NB, Kuroki K, Bustamante CD, Wayne RK, Ostrander EA. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science. 2009;325:995–998. doi: 10.1126/science.1173275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, Shearin AL, Ostrander EA. Man’s best friend becomes biology’s best in show: genome analyses in the domestic dog. Annu Rev Genet. 2010;44:309–336. doi: 10.1146/annurev-genet-102808-115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perola M. Genome-wide association approaches for identifying loci for human height genes. Best Pract Res Clin Endocrinol Metab. 2011;25:19–23. doi: 10.1016/j.beem.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero CJ, Ng Y, Luque RM, Kineman RD, Koch L, Bruning JC, Radovick S. Targeted deletion of somatotroph insulin-like growth factor-I signaling in a cell-specific knockout mouse model. Mol Endocrinol. 2010;24(5):1077–1089. doi: 10.1210/me.2009-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Salmon Hillbertz NH, Isaksson M, Karlsson EK, Hellmen E, Pielberg GR, Savolainen P, Wade CM, von Euler H, Gustafson U, Hedhammar A, Nilsson M, Lindblad-Toh K, Andersson L, Andersson G. Duplication of FGF3, FGF4, FGF19 and ORAOV1 causes hair ridge and predisposition to dermoid sinus in Ridgeback dogs. Nat Genet. 2007;39:1318–1320. doi: 10.1038/ng.2007.4. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning, a laboratory manual. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2001. [Google Scholar]
- Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, Bonnycastle LL, Shen H, Timpson N, Lettre G, Usala G, Chines PS, Stringham HM, Scott LJ, Dei M, Lai S, Albai G, Crisponi L, Naitza S, Doheny KF, Pugh EW, Ben-Shlomo Y, Ebrahim S, Lawlor DA, Bergman RN, Watanabe RM, Uda M, Tuomilehto J, Coresh J, Hirschhorn JN, Shuldiner AR, Schlessinger D, Collins FS, Davey Smith G, Boerwinkle E, Cao A, Boehnke M, Abecasis GR, Mohlke KL. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soranzo N, Rivadeneira F, Chinappen-Horsley U, Malkina I, Richards JB, Hammond N, Stolk L, Nica A, Inouye M, Hofman A, Stephens J, Wheeler E, Arp P, Gwilliam R, Jhamai PM, Potter S, Chaney A, Ghori MJ, Ravindrarajah R, Ermakov S, Estrada K, Pols HA, Williams FM, McArdle WL, van Meurs JB, Loos RJ, Dermitzakis ET, Ahmadi KR, Hart DJ, Ouwehand WH, Wareham NJ, Barroso I, Sandhu MS, Strachan DP, Livshits G, Spector TD, Uitterlinden AG, Deloukas P. Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5:e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter NB, Eberle MA, Parker HG, Pullar BJ, Kirkness EF, Kruglyak L, Ostrander EA. Extensive and breed-specific linkage disequilibrium in Canis familiaris. Genome Res. 2004;14:2388–2396. doi: 10.1101/gr.3147604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, Zhu L, Padhukasahasram B, Karlins E, Davis S, Jones PG, Quignon P, Johnson GS, Parker HG, Fretwell N, Mosher DS, Lawler DF, Satyaraj E, Nordborg M, Lark KG, Wayne RK, Ostrander EA. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter NB, Mosher DS, Gray MM, Ostrander EA. Morphometrics within dog breeds are highly reproducible and dispute Rensch’s rule. Mamm Genome. 2008;19:713–723. doi: 10.1007/s00335-008-9153-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaysse A, Ratnakumar A, Derrien T, Axelsson E, Rosengren Pielberg G, Sigurdsson S, Fall T, Seppala EH, Hansen MS, Lawley CT, Karlsson EK, Bannasch D, Vila C, Lohi H, Galibert F, Fredholm M, Haggstrom J, Hedhammar A, Andre C, Lindblad-Toh K, Hitte C, Webster MT The Lupa Consortium. Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet. 2011;7:e1002316. doi: 10.1371/journal.pgen.1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vonHoldt BM, Pollinger JP, Lohmueller KE, Han E, Parker HG, Quignon P, Degenhardt JD, Boyko AR, Earl DA, Auton A, Reynolds A, Bryc K, Brisbin A, Knowles JC, Mosher DS, Spady TC, Elkahloun A, Geffen E, Pilot M, Jedrzejewski W, Greco C, Randi E, Bannasch D, Wilton A, Shearman J, Musiani M, Cargill M, Jones PG, Qian Z, Huang W, Ding ZL, Zhang YP, Bustamante CD, Ostrander EA, Novembre J, Wayne RK. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464:898–902. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, Perry JR, Elliott KS, Hackett R, Guiducci C, Shields B, Zeggini E, Lango H, Lyssenko V, Timpson NJ, Burtt NP, Rayner NW, Saxena R, Ardlie K, Tobias JH, Ness AR, Ring SM, Palmer CN, Morris AD, Peltonen L, Salomaa V, Davey Smith G, Groop LC, Hattersley AT, McCarthy MI, Hirschhorn JN, Frayling TM The Diabetes Genetics Initiative, The Wellcome Trust Case Control Consortium. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, Samani NJ, Shields B, Prokopenko I, Farrall M, Dominiczak A, Johnson T, Bergmann S, Beckmann JS, Vollenweider P, Waterworth DM, Mooser V, Palmer CN, Morris AD, Ouwehand WH, Caulfield M, Munroe PB, Hattersley AT, McCarthy MI, Frayling TM The Diabetes Genetics Initiative, Wellcome Trust Case Control Consortium, Cambridge GEM Consortium. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.