Summary
Factor IXa (FIXa) is a vitamin K-dependent coagulation serine protease which binds to factor VIIIa (FVIIIa) on negatively charged phospholipid vesicles (PCPS) to catalyze the activation of factor X (FX) to factor Xa (FXa) in the intrinsic pathway. Fluorescence resonance energy transfer (FRET) studies have indicated that the Gla-domain-dependent interaction of FIXa and FX with PCPS in the presence of FVIIIa positions the active-site of the protease at an appropriate height above the membrane surface to optimize the catalytic reaction. In this study, we investigated the contribution of the NH2-terminal EGF-domain (EGF1) of FIXa to the recognition specificity of intrinsic Tenase by constructing an EGF1 deletion mutant of FIXa (FIXa-desEGF1) and characterizing the properties of the mutant in kinetic, direct binding and FRET assays. The results of direct binding and kinetic studies demonstrated that the binding affinity of the mutant for interaction with FVIIIa on PCPS has been impaired greater than 10- fold and the catalytic efficiency of the mutant protease-FVIIIa-PCPS complex in the activation of FX has been decreased ~100-fold. By contrast, the mutant protease exhibited a normal activity toward FX in the absence of the protein cofactor. FRET measurements revealed that the distance of the active-site of the mutant FIXa relative to PCPS vesicles has been decreased 10 Å from 75 ±2 Å for FIXa to 65 ±2 Å for FIXa-desEGF1 independent of FVIIIa. These results suggest that the NH2-terminal EGF-domain of FIXa provides a binding-site for FVIIIa and plays an essential spacer function in the intrinsic Tenase complex.
Keywords: Factor IXa, factor VIIIa, intrinsic Tenase, FRET, Gla-domain, EGF-domains
Introduction
Factor IX (FIX)1 is a vitamin K-dependent coagulation serine protease zymogen that upon activation to factor IXa (FIXa) by either one of its physiological activators, tissue factor-factor VIIa (TF-FVIIa) or factor XIa, binds to factor VIIIa (FVIIIa) on negatively charged membrane surfaces in the presence of Ca2+ (intrinsic Tenase) to activate factor X (FX) to factor Xa (FXa) during the blood coagulation process (1–5). The complex formation with FVIIIa in intrinsic Tenase improves the catalytic efficiency of FIXa toward FX by more than 4–5 orders of magnitude by a not completely understood mechanism (1–5). Unlike its dramatic rate accelerating effect on FX activation, FVIIIa has a minimal cofactor effect on the activity of FIXa toward small synthetic substrates (6,7). This and other observations have lead to the hypothesis that FVIIIa-mediated exosite interactions with FX, involving sites distant from the active-site pocket of FIXa, play dominant roles in determining the catalytic specificity of the protease in the intrinsic Tenase complex (6–8).
Following activation by either TF-FVIIa or FXIa, the single chain zymogen FIX is converted to a two-chain serine protease consisting of a light and a heavy chain held together by a single disulfide bond (9). The N-terminal light chain of FIXa contains the non-catalytic Gla and two epidermal growth factor (EGF)-like domains, while the C-terminal heavy chain contains the trypsin-like catalytic domain of the protease (9). Similar to other vitamin K-dependent coagulation proteases, the Gla domain of FIXa binds to negatively charged phospholipids in the presence of Ca2+ to facilitate the interaction of the protease with its substrate FX on the membrane surface (9). While an interactive-site for FVIIIa has been identified in the catalytic domain of FIXa (10), conflicting results have been reported as to whether or not the Gla and/or EGF1 domain of the protease directly interact with the cofactor (11). One study, using synthetic Gla domain-derived peptides, identified a binding site for the Gla domain of FIXa in FVIIIa (12). Other studies using chimeric molecules in which the EGF1 domain of FIX was replaced with the same domain of the homologous vitamin K-dependent proteins, FVII, FX and protein C, have reported a role for EGF1 of FIX in the activation of the zymogen by the FVIIa-TF complex but not in its activation by FXIa (11). However, the protease activity of some of the EGF1 chimeric mutants was either markedly improved or impaired in the intrinsic Tenase complex (13,14), thus rendering it difficult to unambiguously determine what specific role the EGF1 domain plays in the activation complex.
To address this question, we took a different mutagenesis approach. Thus, instead of replacing EGF1 of FIXa with the homologous domain of other vitamin K-dependent coagulation proteases, we deleted this domain and then characterized the biochemical and biophysical properties of the mutant in the absence and presence of FVIIIa by kinetic, direct binding and FRET assays. The results demonstrate that EGF1 of FIXa is required for the high affinity interaction of the protease with FVIIIa in the intrinsic Tenase complex. Furthermore, FRET analysis indicates that EGF1 plays an essential spacer function to maintain the active-site pocket of FIXa above the membrane surface. Thus its deletion led to lowering the height of the active-site pocket of FIXa by ~10 Å relative to the membrane surface, thereby leading to the inability of the protease to cleave the scissile bond of the substrate FX in the intrinsic Tenase complex. Finally our results support the previous observation that EGF1 of the zymogen FIX is dispensable for its activation by FXIa, however, this domain is required for the substrate activation by the TF-FVIIa complex (11).
Materials and methods
Construction, expression and purification of recombinant FIX mutants
The EGF1 deletion mutant of FIX (FIX-desEGF1) was constructed by the PCR mutagenesis approach and expressed in human embryonic kidney (HEK-293) cells as described (15). A FIX mutant in which the catalytic residue Ser195 (chymotrypsinogen numbering) (15) was substituted with a Cys (FIX-Cys195) was constructed by the same methods and expressed in the same expression/purification vector system as described (16). The construction and expression of FIX in both full-length and Gla-domainless (GD-FIX) forms have been described (16,17). Recombinant soluble tissue-factor (sTF), and TF lacking the cytoplasmic domain (dcTF) were prepared as described (18,19).
Human plasma-derived proteins FIXa, FXa and FX, and RVV-X and fluorescein-labeled Phe-Pro-Arg-ck (Fl-FPR) were purchased from (Haematologic Technologies, Essex Junction, VT). The fluorescent dyes; fluorescein-5-maleimide, octadecylrhodamine (OR) and Oregon Green488 (OG488) were purchased from Invitrogen (Carlsbad, CA). Dioleoylphosphatidylcholine (PC) and dioleoylphosphatidylserine (PS) were purchased from Avanti Polar Lipids (Alabaster, AL). The Nα-[(acetylthio) acetyl] derivative of EGR-ck (ATA-EGR-ck) was a generous gift from Dr. Paul Bock (Vanderbilt University, Nashville, TN). Human recombinant FVIIIa was a generous gift from Dr. Philip Fay (University of Rochester, Rochester, NY). Heparin-Sepharose was purchased from Amersham Pharmacia (Piscataway, NJ). The chromogenic substrates, Spectrozyme FXa (SpFXa) was purchased from American Diagnostica (Greenwich, CT) and CBS 31.39 (LGR-pNA) was purchased from Midwest Bio-Tech. Inc. (Fishers, IN).
Activation of FIX derivatives by RVV-X
All FIX derivatives were converted to active forms by RVV-X, separated from venom and active-site titrated as described (16,17).
Activation by the Factor VIIa-TF Complex
The initial rate of recombinant FIX and FIX-desEGF1 activation was studied in the presence of both soluble TF (sTF) and dcTF incorporated into PCPS vesicles (15 μM) as described (18,19). Briefly, the initial rate of the activation of wild-type and mutant FIX (1 μM) by FVIIa (50 nM in the presence of sTF and 1 nM in the presence of dcTF) was monitored in the presence of a saturating concentration of sTF (200 nM) and relipidated dcTF (5 nM) in 0.02 M NaCl, 0.02 M Tris-HCl, pH 7.4 (TBS) containing 0.1 mg/mL BSA, 0.1% polyethylene glycol (PEG) 8000 and 5 mM Ca2+ (TBS/Ca2+) at room temperature in 30 μL reactions in 96-well assay plates. The activation reactions were terminated by an addition of 20 μL EDTA to obtain a final concentration of 20 mM. The concentration of FIXa generated was determined from standard curves by a chromogenic substrate assay using CBS 31.39 as described (16,17).
Phospholipid vesicles preparation
Phospholipid vesicles at a molar ratio of PC to PS of 4:1 containing or lacking different amounts of the acceptor dye OR were prepared by extrusion method as described (20). The OR containing vesicles received various amounts of the dye in ethyl acetate prior to lyophilization and extrusion. The % yield of phospholipids recovery was determined by comparing the amount of choline from small samples before and after centrifugation step by a colorimetric assay using Wako phospholipid B kit (Wako Chemicals USA, Inc., Richmond, VA) as described (20). The concentration of OR in the acceptor-containing samples was determined from absorbance at 564 nm using a molar extinction coefficient of 95,400 M−1 cm−1 as described (21,22). The acceptor density of OR (σ, in OR/Å2) was calculated using molecular weights 786.1 and 810.0 Da for PC and PS, respectively, assuming that acceptors were distributed randomly at the phospholipid surface, and assuming that each phospholipid molecule occupies 70 Å2 of the surface area as described (21,23).
FX activation by FIXa derivatives
The initial rate of FX activation by FIXa and FIXa-desEGF1 was measured on PCPS vesicles both in the absence and presence of FVIIIa in TBS containing 0.1 mg/mL BSA, 0.1% polyethylene glycol (PEG) 8000 and 5 mM Ca2+ (TBS/Ca2+) at room temperature as described (16). The apparent Kd for the interaction of FIXa derivatives with FVIIIa and the kinetic constants Km and kcat for the activation of FX by the FIXa derivatives in intrinsic Tenase parameters were determined as described (16,17).
Fluorescein labeling
Both wild-type and FIXa-desEGF1 (~0.5 mg), in the Mono Q elution buffer (~0.45 M NaCl, 0.02 M Tris-HCl, pH 7.4) were incubated with 10-fold molar excess of Fl-FPR for 2 h at room temperature in the dark. The extent of active-site labeling was monitored by the loss of the enzymatic activity using CBS 31.39. Incubation was continued until more than 99.9% of the activity was inhibited. For the Cys195 labeling of both the FIXa-Cys195 enzyme and the FIX-Cys195 zymogen, the same amount of the mutant proteins were incubated with fluorescein-5-malimide (200 μM) in TBS containing 25 μM dithiothreitol to ensure reduction of the free cysteine for 2h at room temperature in the dark. This level of reducing agent did not influence the activity of FIXa. The free inhibitor or dye was separated from the labeled proteins by gel filtration on the PD-10 column followed by their extensive dialysis in 0.1 M NaCl and 0.05 M Hepes, pH 7.4 containing 5 mM Ca2+ at 4 °C in the dark. An extinction coefficient of 84,000 M−1 cm−1 at 498 nm was used to calculate the fluorescein concentration and the ratio ε280nm/ε498nm = 0.19 was used to correct for the contribution of the dye to 280-nm absorbance of the proteins as described (24).
Oregon Green488 labeling
The methods developed by Bock were used to conjugate OG488 to the active sites of the ATA-EGR-ck inhibited FIXa derivatives as described (24).
Spectral measurements
Steady-state anisotropy and fluorescence measurements were made using an Aminco-Bowman series 2 Spectrophotometer (Spectronic Unicam, Rochester, NY) equipped with an automatic polarizer interfaced to a PC computer for data acquisition and analysis as described (20). The equilibrium dissociation constants (KD) for the interaction of OG488-EGR labeled FIXa derivatives with FVIIIa on PCPS vesicles were determined from the increase in the anisotropy of OG488 in the active-sites of FIXa derivatives upon interaction with the cofactor as described (25). This was done by titrating increasing concentrations of FVIIIa (0.1–800 nM) with a fixed concentration of each labeled protease (20–50 nM) on PCPS vesicles (50 μM) in TBS/Ca2+. The KD for the interaction of the labeled FIXa with FVIIIa was calculated by nonlinear least-squares computer fitting of the data by the quadratic binding equation as described (25).
The anisotropy of OR labeled PCPS vesicles containing a limiting concentration of the acceptor (σ = 0.9 × 10−4 OR/Å2) was measured as described above except that the excitation and emission wavelengths were set to 521 nm and 586 nm, respectively. Absorbance measurements were made using a Beckman Coulter DU 800 spectrophotometer. The values for the quantum yield (Q), spectral overlap between donor (fluorescein) and acceptor (OR) dyes (JDA), and the distance between donor and acceptor dyes at the 50% FRET efficiency (R0) were calculated as described previously (20–22). The values of Q and JDA were separately measured for each fluorescein-labeled protein both in the absence and presence of PCPS vesicles and FVIIIa.
FRET measurements
All FRET measurements were performed exactly as described (20–22). Briefly, the donor (fluorescein) containing (cuvette D) and both the donor and acceptor (OR) containing cuvettes (cuvette DA) each received 50 nM fluorescein-labeled FIXa derivatives in 0.1 M NaCl, 0.05 M Hepes, pH 7.4, and 5 mM Ca2+ while blanks (cuvette B) and acceptor-containing cuvettes (cuvette A) received 50 nM of unlabeled FIXa derivatives in the same buffer. The same procedures were used for the measurements in the presence of FVIIIa (50 nM) with the exception that the concentrations of the labeled FIXa derivatives were reduced to 20 nM. The net initial emission intensities were obtained by subtracting the initial intensities of A from DA (FDA-FA)o a)nd B from D (FD-FB)o. Samples D and B were then titrated with PCPS vesicles lacking the acceptor OR, while samples DA and A were titrated with PCPS vesicles containing the acceptor. Similarly, the intensities of A and B were subtracted from DA and D, respectively and the values were then corrected for dilutions (less than 4% at the end of titration). Following 5 min incubation emission intensities were measured. The ratio of the donor quantum yields (Q) in the D and DA samples based on their fluorescence emission intensities (F) is given by equation 1 as described (21):
| (1) |
at the end of experiments, the fluorescent labeled proteins were released from the membrane surface by the addition of EDTA to 10 mM to ensure that energy transfer is reversible. For calculating the distance of the closest approach by equation 2 below, the QD/QDA value was calculated by dividing the value before EDTA by the value after addition of EDTA. This normalization procedure corrects for the contribution of the acceptor inner filter effects and a potential membrane-binding-independent energy transfer as described (21).
Calculation of the distance of closest approach
Assuming that both donor and acceptor dyes are randomly and uniformly distributed (κ2 = 2/3), the distance of closest approach (L) between the plane of the donor dye attached to the active-site of the proteins and the plane of the acceptor dyes at the surface of PCPS vesicles can be determined using equation 2 as described (21,22):
| (2) |
where π is 3.14, σ is the acceptor density at the membrane surface in OR/Å2 and Ro is the distance at which the singlet-singlet energy transfer from the donor dye to the acceptor dye is 50% efficient. The net QD/ODA values were plotted as a function of product between Ro2 and the acceptor density (σ) for at least 5–16 different energy transfer experiments to obtain the average value of L as described (20–22).
Results
Expression, purification and characterization of mutant proteins
FIX derivatives were all expressed in HEK-293 cells and purified to homogeneity by a combination of immunoaffinity and ion exchange chromatography using the HPC4 monoclonal antibody immobilized on Affi-gel 10 and FPLC Mono Q column, respectively, as described (16). The purity of all mutants was verified by the SDS-PAGE. The FIX derivatives were converted to their active forms by RVV-X as described under “Materials and methods” and in refs. 16 and 17. We have characterized the kinetic properties of the Gla-domainless FIXa (GD-FIXa) in a previous study and noted a normal amidolytic activity for the mutant protease (17). The FIXa-desEGF1 mutant also exhibited a normal amidolytic activity toward hydrolysis of CBS 31.39 yielding Km and kcat values of 6.9 ±0.4 mM and 9.5 ±0.3 s−1, respectively, which are similar to the corresponding values observed for wild-type FIXa (6.6 ±0.3 mM and 10.4 ±0.2 s−1). These results suggest that the deletion of EGF1 domain did not adversely affect the folding and the reactivity of the catalytic triad of the mutant protease.
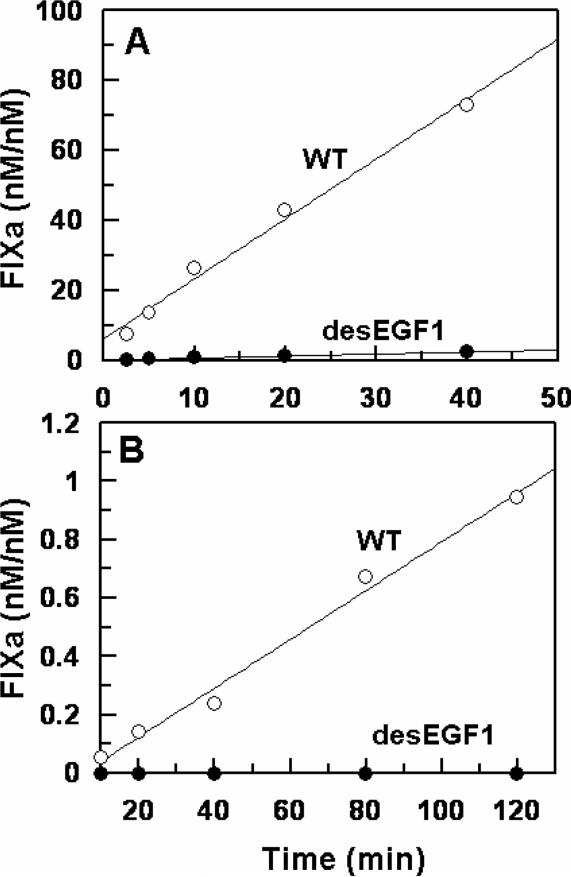
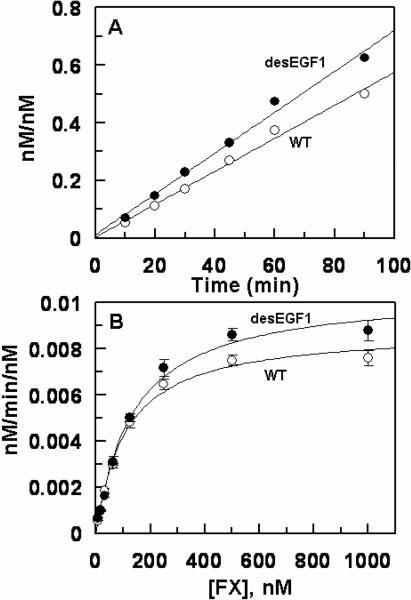
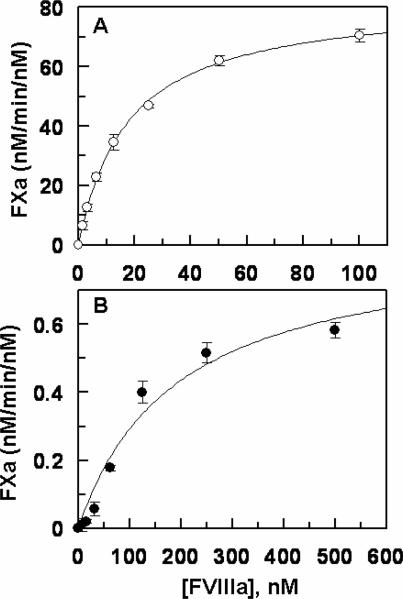
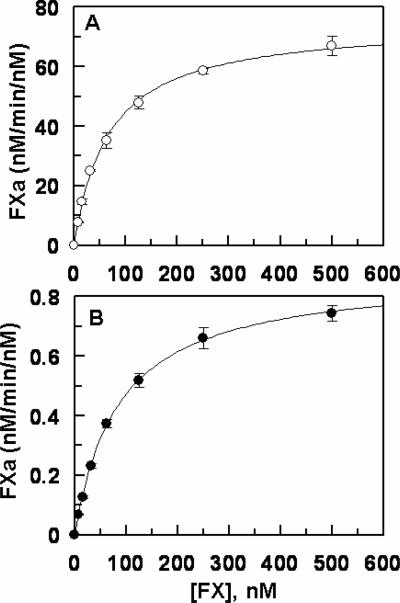
Next, the activation of FIX-desEGF1 was monitored by the two physiological activators of FIX, the TF-FVIIa complex and FXIa. The results indicated that while FXIa activates the mutant zymogen at a near normal rate (data not shown), its activation by TF-FVIIa on PCPS vesicles has been dramatically impaired. As presented in Fig. 1A, relative to activation of recombinant wild-type FIX, the initial rate of FIX-desEGF1 activation by the TF-FVIIa complex on PCPS vesicles was impaired greater than 30-fold. In the absence of PCPS, the extent of the activation of the deletion mutant by FVIIa in complex with soluble TF in solution was also severely impaired since no significant amount of the activated form of the deletion mutant could be detected by the time course analysis for up to 120 min (Fig. 1B). In agreement with previous data in the literature (11), these results suggest that EGF1 domain of FIX is required for its activation by TF-FVIIa but not by FXIa. The activity of FIXa-desEGF1 toward FX was then compared to that of wild-type FIXa in the absence and presence of FVIIIa on PCPS vesicles. Comparisons of the time course and concentration-dependence of FX activation suggested that the FIXa-desEGF1 mutant activates FX with a similar or improved rate on PCPS vesicles in the absence of FVIIIa (Fig. 2, Table 1). However, the FVIIIa concentration-dependence of FX activation revealed that the affinity of the mutant protease for interaction with FVIIIa has been markedly decreased (Fig. 3). Thus, in contrast to an apparent Kd of 17.6 nM for wild-type FIXa interaction with FVIIIa, the corresponding value was increased to ~190 nM for FIXa-desEGF1 (Fig. 3, Table 1). Further kinetic studies in the presence of saturating concentrations of FVIIIa indicated that the catalytic activity of FIXa-desEGF1 toward the substrate has been dramatically impaired in the presence of the protein cofactor (Fig. 4). Thus, in contrast to apparent Km and kcat values of 67 nM and 75 nM/min/nM, respectively, observed for the wild-type FIXa activation of FX in the presence of FVIIIa, the corresponding values for the FIXa-desEGF1 activation of FX were 86.5 nM and 0.88 nM/min/nM (Table 1), suggesting ~100-fold impairment in the rate of substrate activation by the mutant protease.
Figure 1.
Time course analysis of wild-type and mutant FIX activation by the TF-FVIIa complex. A, the time course of activation of FIX (◯) and FIX-desEGF1 (●) (1 μM each) by FVIIa (1 nM) in complex with relipidated dcTF (5 nM) was monitored in TBS/Ca2+ at room temperature. At each time point, aliquots of the activation reactions were transferred to wells of a 96-well plate containing 20 mM EDTA and the rate of FIXa generation was measured by an amidolytic activity assay as described under “Materials and methods”. B, the same as A except that the activation time course analysis was carried out by FVIIa (50 nM) in complex with soluble TF (200 nM) in solution.
Figure 2.
Time course and concentration-dependence of FX activation by recombinant FIXa derivatives on PCPS vesicles. A, the time course of activation of FX (300 nM) by FIXa (◯) and FIXa-desEGF1 (●) (20 nM each) was monitored on PCPS (50 μM) vesicles in TBS/Ca2+ at room temperature. At each time point, aliquots of the activation reactions were transferred to wells of a 96-well plate containing 20 mM EDTA and the rate of FXa generation was measured by an amidolytic activity assay as described under “Materials and methods”. B, the same as A except that the FX concentration-dependence (x-axis) of activation by FIXa (◯) and FIXa-desEGF1 (●) (20 nM each) was monitored on PCPS (50 μM) vesicles. Solid lines in panel B are nonlinear regression fits of kinetic data (three independent measurements) according to the Michaelis-Menten equation. The Km and kcat constants are presented in Table 1.
Table 1.
Kinetic constants for the activation of FX by FIXa derivatives in the intrinsic Tenase complex and the dissociation constants (Kd(app) and KD) for their interaction with FVIIIa
| Km(app) (nM) | kcat (nM/min/nM) | kcat/Km(app) (nM/min) | Kd(app) (nM) | KD (nM) | |
|---|---|---|---|---|---|
| FIXa | |||||
| FX, PCPS, Ca2+ | 108.7±11.4 | 0.009±0.0003 | 8.3 × 10−5 | - | - |
| FX, PCPS, FVIIIa, Ca2+ | 67.2±2.9 | 74.9±1.6 | 1.1±0.07 | 17.6±1.4 | - |
| PCPS, FVIIIa, Ca2+ | - | - | - | - | 1.5±1.0 |
| FIXa-desEGF1 | |||||
| FX, PCPS, Ca2+ | 136.1±15.9 | 0.011±0.0004 | 8.1 × 10−5 | - | - |
| FX, PCPS, FVIIIa, Ca2+ | 86.5±4.2 | 0.88±0.01 | 0.01±0.0006 | 189.5 ±34.3 | - |
| PCPS, FVIIIa, Ca2+ | - | - | - | - | 135±14 |
| GD-FIXa | |||||
| PCPS, FVIIIa, Ca2+ | - | - | - | - | 310±22 |
The kinetic parameters Km(app) and kcat were determined from the concentration dependence of FX activation by FIXa derivatives on PCPS vesicles in TBS/Ca2+ in the absence and presence of a saturating concentration of FVIIIa as described under “Materials and methods”. The Kd(app) values for the interactions with FVIIIa were determined from the saturable cofactor concentration dependent FXa generation by each FIXa derivative at room temperature as described under “Materials and methods”. All values are the average of at least 3 measurements ± standard deviations. The Kd(app) values for FVIIIa are derived from Fig. 3; the Km(app), kcat values in the presence of FVIIIa are derived from Fig. 4; and KD values at equilibrium for FVIIIa are derived from Fig. 5.
Figure 3.
Factor VIIIa-concentration-dependence of FX activation by recombinant FIXa derivatives on PCPS vesicles. A, FIXa (100 pM) was incubated with FX (300 nM) in the presence of increasing concentrations of FVIIIa (1.5–100 nM) on PCPS vesicles (50 μM) in TBS/Ca2+. Following 1–2 min of activation at room temperature the reaction was terminated by addition of 20 mM EDTA and the rate of FXa generation was measured by an amidolytic activity assay as described under “Materials and methods”. B, the same as A except that FIXa-desEGF1 (5 nM) was incubated with FX in the presence of increasing concentrations of FVIIIa as indicated on x-axis. Solid lines in both panels are nonlinear regression fits of kinetic data (three independent measurements) to a hyperbolic equation, yielding Kd(app) values of 17.6 ± 1.4 nM for FIXa (panel A) and 189.5 ± 34.3 nM for FIXa-desEGF1 interaction with FVIIIa. These values are presented in Table 1.
Figure 4.
Concentration dependence of FX activation by recombinant FIXa derivatives in the intrinsic Tenase complex. A, the concentration dependence of FX (4–500 nM) activation by FIXa (100 pM) was measured in the presence of a saturating concentration of FVIIIa (50 nM) on PCPS vesicles (50 μM) in TBS/Ca2+. Following 1–2 min of activation at room temperature the reaction was terminated by addition of 20 mM EDTA and the rate of FXa generation was measured by an amidolytic activity assay using SpFXa as described under “Materials and methods”. B, the same as A except that the activation of FX was monitored by FIXa-desEGF1 (5 nM) in complex with 500 nM FVIIIa in TBS/Ca2+. The solid lines are nonlinear regression analysis of kinetic data (three independent measurements) according to the Michaelis-Menten equation. The Km and kcat constants are presented in Table 1.
Analysis of the FIXa-FVIIIa interaction by equilibrium binding measurements
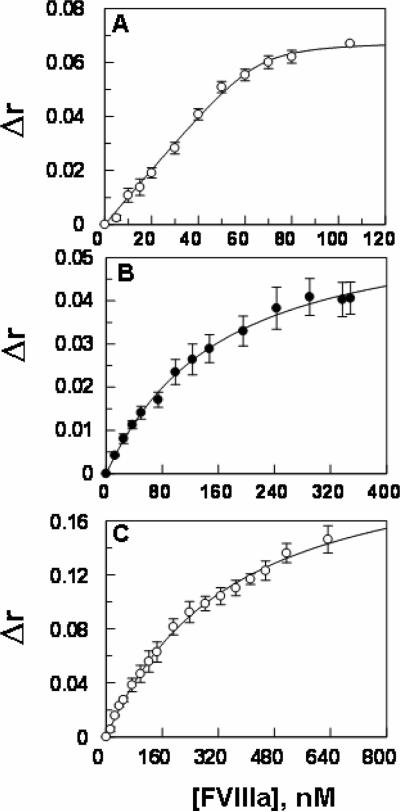
Previous results have indicated the fluorescence anisotropy of the fluorescein (Fl-FPR)-labeled porcine FIXa is enhanced upon interaction with porcine FVIIIa, yielding a KD of ~2 nM for the protease-cofactor interaction (25). However, our titration of human Fl-FPR-FIXa with human FVIIIa did not result in any change in the fluorescence anisotropy of the labeled protein (data not shown). To determine whether a OG488-based assay, similar to that used for the analysis of FXa-FVa interaction (26,27), can be set up for measuring the affinity of human FIXa for FVIIIa at equilibrium, we labeled the active sites of FIXa derivatives with OG488-EGR and monitored their fluorescence anisotropy in the presence of FVIIIa on PCPS vesicles. The results presented in Fig. 5 indicated that the binding of OG488-EGR labeled FIXa derivatives with FVIIIa is associated with different extents of changes in the anisotropy of the labeled proteins. Thus, the OG488-labeled wild-type FIXa interacted with FVIIIa on PCPS vesicles with a KD of ~1.5 nM (Fig. 5A). By contrast, OG488-EGR-FIXa-desEGF exhibited a dramatically weaker KD value of ~135 nM for FVIIIa under the same experimental conditions (Fig. 5B). Interestingly, the OG488-EGR labeled GD-FIXa exhibited a much larger increase in anisotropy and yielded a KD value of above 300 nM for FVIIIa under the same experimental conditions (Fig. 5C). These direct binding data clearly suggest that the NH2-termianl EGF domain of FIXa has an interactive-site for FVIIIa in the intrinsic Tenase complex.
Figure 5.
Enhancement in the anisotropy of the OG488-EGR labeled FIXa derivatives upon interaction with FVIIIa on PCPS vesicles. A, a fixed concentration of the OG488-EGR labeled wild-type FIXa (50 nM) was titrated with increasing concentrations of FVIIIa (0.25–120 nM) on PCPS vesicles (50 μM) in TBS containing 0.1% PEG 8000 and 5 mM Ca2+ at 25 °C. KD (1.5 ± 1.1 nM, n =3) of FIXa for interaction with FVIIIa was calculated from the saturable changes in the anisotropy (Δr) of the labeled protein according to a quadratic binding equation as described under “Materials and methods”. B, the same as panel A except that OG488-EGR labeled FIXa-desEGF1 (50 nM) was used in the FVIIIa titration yielding a KD of (135.4 ± 13.7 nM, n =3) for the interaction of the mutant protease with the cofactor. C, the same as above except that OG488-EGR labeled GD-FIXa (50 nM) was used in the FVIIIa titration yielding a KD of (308.6 ± 22.4 nM, n =3) for the interaction of the mutant protease with the cofactor.
Spectral characterization of fluorescein labeled FIX/IXa derivatives
The active-sites of FIXa and FIXa-desEGF1 were also labeled with the fluorescein dye attached to the tripeptidyl inhibitor Fl-FPR which covalently binds to His57 of the catalytic triad. In the case of the Cys195 mutant of FIX, the fluorescein dye was directly incorporated to the Cys195 mutant residue in both in the zymogenic (FIX) and activated forms (FIXa) of the molecule. The maximum excitation and emission wavelengths for all fluorescein labeled proteins were determined to be identical values of 493 nm and 521 nm, respectively (data not shown). The anisotropy of Fl-FPR labeled proteins in the absence and presence of PCPS vesicles, respectively, determined to be 0.10 and 0.11 for FIXa; 0.10 and 0.12 for FIXa-desEGF1; 0.14 and 0.15 for Cys195 mutant of zymogen FIX and protease FIXa (Table 2). FVIIIa did not significantly change the anisotropies of membrane-bound proteins. The anisotropy of an OR containing PCPS preparation with a density of 0.9 × 10−4 OR/Å2 was determined to be 0.10.
Table 2.
The distance of closet approach between the donor fluorescein dyes in the active-sites of FIX/FIXa derivatives and the acceptor rhodamine dyes incorporated into PCPS vesicles and anisotropy (r) values for the labeled proteins.
| Donor Enzyme | Ro | L | r | r(+PCPS) |
|---|---|---|---|---|
| Fl-FPR-FIXa | 49 ±1 | 75 ±2 | 0.10 | 0.11 |
| Fl-FPR-FIXa-desEGF1 | 46 ±1 | 65 ±2 | 0.10 | 0.12 |
| Fl-FIXa-Cys195 | 49 ±1 | 81 ±3 | 0.14 | 0.15 |
| Fl-FIX-Cys195 | 49 ±1 | 84 ±3 | 0.14 | 0.15 |
The Ro and L values (in Å) were determined based on the efficiency of energy transfer due to decreases in the emission intensity of the donor fluorescein dyes in the active-sites of proteins (20–50 nM) upon their titration with PCPS vesicles containing the acceptor OR dyes with densities of 0.9–2.5 × 10−4 OR/Å2 in 0.1 M NaCl, 0.05 M Hepes, pH 7.4 containing 5 mM Ca2+, assuming a random orientation of transition dipoles for donor and acceptor dyes (κ2 = 2/3) as described in “Materials and methods”. The anisotropy values (r) for fluorescein in the active-sites of the proteins were determined at excitation and emission wavelengths of 493 nm and 521 nm, respectively.
Membrane-dependent fluorescence resonance energy transfer (FRET)
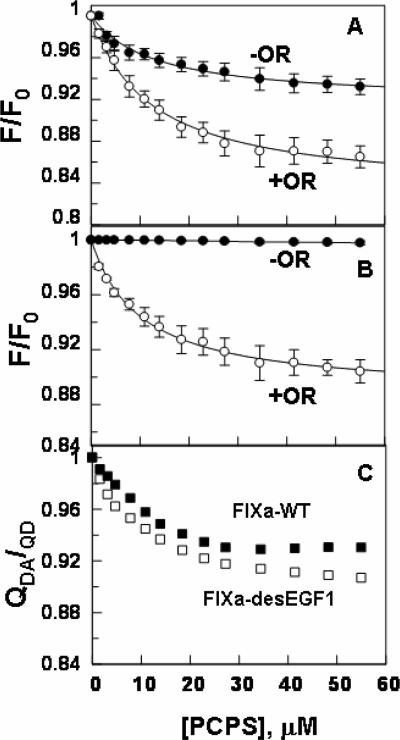
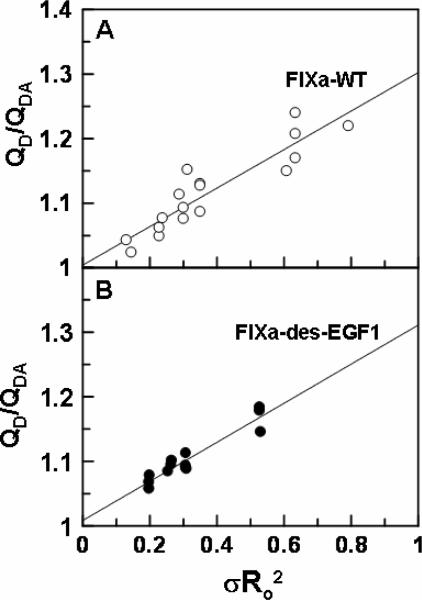
The PCPS concentration dependent changes in the emission intensity of the donor fluorescein dye in the active-site grooves of both Fl-FPR-FIXa and Fl-FIXa-desEGF1, in the absence and presence of the acceptor dye OR, are presented in Fig. 6. While the fluorescein emission intensity was decreased for FIXa on PCPS vesicles both lacking and containing OR (Fig. 6A), the emission intensity of the donor dye in the active-site pocket of FIXa-desEGF1 increased only in the presence of OR (Fig. 6B). Nevertheless, the decrease in the emission intensity of fluorescein in the active-site of FIXa in the presence of OR was significantly greater than that in its absence. These results suggest that the decreases in the emission intensities in both cases are primarily due to singlet-singlet energy transfer between fluorescein in the active-sites and OR at the surface of PCPS vesicles. The OR-independent change in the emission intensity of the donor dye in the active-site of FIXa likely represents a conformational change in the active-site groove of the protease which has also been observed in a similar previous study (21). The energy transfer in all cases was Ca2+-dependent since it was abolished when EDTA was added to the cuvettes at the end of PCPS titrations. The EDTA reversible ratio of the donor quantum yields in the presence and absence of the acceptor (QDA/QD), which was calculated according to equation 1, are presented in Fig. 6C. The results demonstrate ~2–3% larger OR-dependent decrease in the emission intensity of fluorescein in the active-site of Fl-FPR-FIXa-desEGF1 (at the same acceptor density), possibly suggesting that the fluorescent dye is closer to the membrane surface in the active-site of des-EGF1, thus leading to more efficient singlet-singlet energy transfer in the mutant protease. Assuming a random orientation of transition dipoles for donor and acceptor dyes (κ2 = 2/3) (21), the singlet-singlet energy transfer equation 2 was used to calculate the average distance of the closest approach (L) between fluorescein in the active-site pocket of FIXa and FIXa-desEGF1 and OR on PCPS vesicles at several acceptor densities of 0.9 × 10−4 – 2.5 × 10−4 OR/Å2. The average L values for Fl-FPR-FIXa and FIXa-desEGF1 were determined to be 75 ±2 Å (±SD, n = 16) and 65 ± 2 Å (±SD, n = 11), respectively (Table 2). These results suggest that, relative to wild-type FIXa, the fluorescein dye in the active-site pocket of FIXa-desEGF1 is 10 Å closer to the membrane surface assuming a κ2 value of 2/3 for both proteins. The linear dependence of the plot of QD/QDA on the product between Ro2 and the acceptor density (σRo2) for both FIXa and FIXa-desEGF1 (Fig. 7) suggested that the magnitude of the energy transfer correlates with the density of the acceptor OR at the membrane surface as previously demonstrated (20–22).
Figure 6.
Titration of Fl-FIXa and Fl-FIXa-desEGF1 with PCPS vesicles. A, changes in the emission intensity of fluorescein in the active-site of Fl-FPR inhibited FIXa (50 nM) were monitored upon titration with PCPS vesicles containing (◯) or lacking OR (●) as described under “Materials and methods”. B, the same as panel A except that Fl-FIXa-desEGF1 was used in the titration. C, data of panels A and B are re-plotted for both Fl-FIXa (■) and Fl-FIXadesEGF1 (□) according to equation 1 as described under “Materials and methods”. Acceptor densities were 1.4 × 10−4 OR/Å2 for both FRET measurements.
Figure 7.
Linear dependence of FRET upon acceptor density. The Ro values for Fl-FPR-FIXa (panel A) and Fl-FIXa-desEGF1 (panel B) were determined by titrations with OR containing PCPS vesicles at acceptor densities of 0.9–2.5 χ 10−4 OR/Å2 as described under “Materials and methods”.
FRET measurements for FIX/FIXa-Cys195 proteins
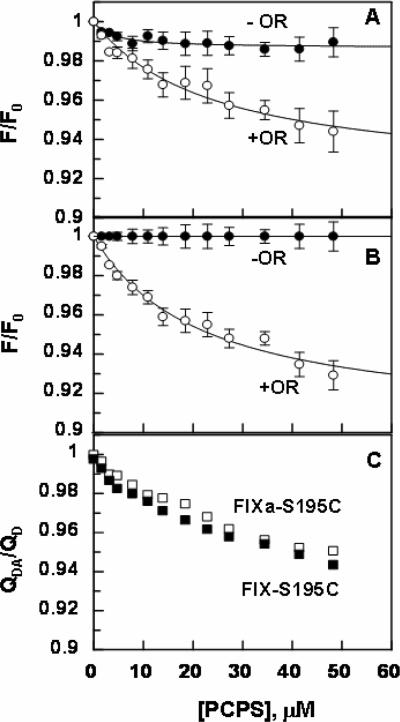
Since fluorescein is not actually tethered to the catalytic pocket but to a site near the P4/P3 binding site of the protease in the Fl-EGR labeled FIXa, we decided to determine the average L value for the FIXa-Cys195 mutant in which the fluorescein dye is directly attached to residue 195. Furthermore, we labeled the zymogenic form of the FIX-Cys195 mutant and conducted the same FRET measurements with the labeled mutant proteins. The titration of either active or zymogen forms of Cys195 mutant with PCPS vesicles lacking OR did not result in significant changes in the emission intensity of the donor fluorescein dye attached to the catalytic residue of the mutant proteins (Fig. 8). However, there was ~5–7% OR-dependent decrease in the emission intensity of fluorescein attached to Cys195 in both proteins (Fig. 8). The L values were determined from the EDTA reversible ratio of QDA/QD (Fig. 8C) according to equations 1 and 2 assuming κ2 = 2/3 as described above. Similar values of 84 ±2 Å (±SD, n = 7) for the zymogenic and 81 ±2Å (±SD, n = 5) for the active form of Cys195 labeled proteins were determined (Table 2), suggesting that residue 195 in both forms is located far above the membrane surface and that the actual distance of the active-site of FIXa relative to the membrane surface, assuming a κ2 value of 2/3 for both proteins (21), may be slightly higher (~6 Å) than the distance obtained using Fl-FPR labeled protease. Similarly, there was no effect on the emission intensity of residue 195 by FVIIIa, possibly suggesting that the cofactor has no conformational effect on the catalytic residue of FIXa in the intrinsic Tenase complex. The comparable L values obtained for both the zymogen and activated forms of Cys195 proteins suggest that the activation peptide of FIX loops out of the molecule and does not contribute to the height of the active-site from the membrane surface. Similar to results presented above (Fig. 7), the plots of QD/QDA exhibited a linear dependence on the product between Ro2 and the acceptor density (σRo2) for all FRET measurements with Cys195 proteins (data not shown).
Figure 8.
Titration of Fl-FIXa-Cys195 and Fl-FIX-Cys195 with PCPS vesicles. A, changes in the emission intensity of fluorescein, covalently attached to Cys195 of the FIXa mutant (50 nM), were monitored upon titration with PCPS vesicles containing (◯) or lacking OR (●) as described under “Materials and methods”. B, the same as panel A except that fluorescein-labeled FIX-Cys195 zymogen was used for the titration. C, data of panels A and B are re-plotted for both fluorescein-labeled FIXa-Cys195 (□) and FIX-Cys195 (■) according to equation 1 as described under “Materials and methods”. Acceptor densities were 1.4 × 10−4 OR/Å2 for both FRET measurements.
Discussion
The results of this study demonstrate that the EGF1 domain of FIXa contributes to the catalytic specificity of intrinsic Tenase by at least two different mechanisms. First, this motif provides a functionally critical interactive-site for FVIIIa in the activation complex. This hypothesis is derived from the direct binding studies, yielding ~100-fold weaker affinity for the interaction of FIXa-desEGF1 with the cofactor on PCPS vesicles. Further support for this hypothesis may come from the observation that the catalytic efficiency of FIXa-desEGF1 toward FX was decreased by 100-fold in the presence of FVIIIa. The mutant exhibited normal activity for the substrate in the absence of the cofactor, ruling out the possibility that the mutant has not folded properly. The direct binding data supports the kinetic data that the binding affinity of FIXa-desEGF1 for FVIIIa has been impaired. The binding data with the GD-FIXa appears to suggest that the Gla-domain of FIXa also interacts with the cofactor, however, the possibility that EGF1 of FIXa is not able to interact with FVIIIa in the absence of the Gla-domain cannot be ruled out from this observation. Nevertheless, it has been previously demonstrated that synthetic peptides derived from the Gla-domain of FIXa inhibit the catalytic activity of the protease toward FX in a FVIIIa-dependent manner in the absence of phospholipid vesicles (12). Thus, the weaker affinity of GD-FIXa for FVIIIa in the intrinsic Tenase complex is likely due to the phospholipid-independent loss of an interaction between the cofactor and the Gla-domain of the protease that is missing in the mutant protease.
Previous FRET measurements have indicated that the active-site of FIXa is located ~89 Å above the membrane surface and that the interaction of FIXa with PCPS and/or FVIIIa is associated with a conformational change in the active-site of the protease (21). In this study, the distance of closest approach (L) between the donor fluorescein dye in the active-site of FIXa and OR dyes on the PCPS vesicles was determined to be ~75Å for the Fl-FPR labeled FIXa and ~81Å for the fluorescein labeled FIXa-Cys195. By contrast, the corresponding L value for the Fl-FPR labeled FIXa-desEGF1 was determined to be ~65 Å, suggesting that the distance of the active-site of the mutant is ~10 Å closer to the membrane surface. Thus, a second function for EGF1 domain is to maintain the active-site of FIXa at an appropriate topography and height above the membrane surface in order for the protease to optimally interact and cleave the scissile bond on the activation peptide of the substrate. In the case of FXa activation of prothrombin in the prothrombinase complex, it has been hypothesized that a function for FVa in the activation complex is to maintain both the active-site of the protease and the activation peptide of the substrate at a similar height above the membrane surface (20,28), thereby effectively mediating the cleavage of the scissile bond of the substrate. In this context, it appears that a spacer function for EGF1 of FIXa also contributes to its effective activation of FX in the intrinsic Tenase complex. This is derived from the observation that the catalytic activity of FIXa-desEGF1 toward FX was markedly impaired in the presence of FVIIIa, though the mutant exhibited normal catalytic activity toward the substrate in the absence of the cofactor. These results suggest that the free protease has sufficient flexibility to access the activation peptide of the substrate even though its active-site is closer to the membrane surface by ~10 Å. The defect in the catalytic function of FIXa-desEGF1 in intrinsic Tenase may be due to the loss of the EGF1-FVIIIa interactive site in the mutant protease. It should be emphasized that these data cannot directly provide evidence for this hypothesis since the possibility that complex formation with FVIIIa on PCPS vesicles traps the mutant protease in a stable unproductive complex with its active-site residue spatially being located ~10 Å below the scissile bond of the substrate cannot be ruled out. However, a dramatically weaker binding of FIXa-desEGF1 to FVIIIa, as measured by a direct binding assay, clearly suggests that EGF1 of FIXa contains an interactive-site for the cofactor.
Previous FRET results have indicated that FVa alters the topography of the membrane-bound FXa in the prothrombinase complex (20,28). This hypothesis is based on the findings that the distance of the of the active-site pocket of FXa relative to the membrane surface in the prothrombinase complex is ~3–7 Å higher than the same distance in the free form of the protease (20,28). Nevertheless, our spectral studies failed to provide any support for a similar effect for FVIIIa on the active-site pocket of FIXa since the emission intensities of the fluorescent dyes in the active-sites of all FIXa derivatives (including FIXa-Cys195) remained unaltered in the presence of the protein cofactor. These observations are in keeping up with the hypothesis that the cofactor function of FVIIIa is primarily mediated through the cofactor altering the conformation of the secondary exosites/surface loops that are located remote from the catalytic pocket of the protease (6). The observation that the interaction of the fluorescein labeled FIXa-Cys195 mutant (label is directly attached to the catalytic residue) with PCPS did not alter the spectral properties of the label, suggests that the conformational effect observed for the PCPS interaction with Fl-FPR-FIXa may involve sites in the proximity of the P4/P3 binding site, where the fluorescent dye is expected to be located in the catalytic groove of the FPR inhibited FIXa.
A spacer function for EGF1 also appears to be critical for the recognition and activation of the substrate FIX by FVIIa-TF since the activation of the FIX-desEGF1 zymogen was dramatically impaired by the extrinsic Tenase complex. By contrast, FXIa, which does not require the Gla-dependent interaction of the substrate with PCPS, exhibited an essentially normal activity toward the EGF1 deletion mutant of the FIX zymogen. In the context of this model of zymogen activation on the membrane surface by extrinsic Tenase, the observation that the L value obtained for Fl-FIXa-Cys195 was similar to the same value for the zymogen form of the mutant suggests that the activation peptide of FIX, containing 35 residues, is not in an elongated configuration, but rather the sequence is horizontally looping out of the body of the molecule without contributing to the height of the membrane-bound zymogen. Given that the active-site of FVIIa in extrinsic Tenase is also located far above the membrane surface (~75 Å) (29), it is not surprising that FVIIa does not recognize the FIX-desEGF1 zymogen as a substrate since the scissile bond on the activation peptide of the mutant is expected to be spatially aligned at a much lower height than the active-site of the protease. It should be noted that, in addition to a spacer function, EGF1 of FIX may also possess an interactive-site for TF in extrinsic Tenase. In support of this hypothesis, an interactive-site for EGF1 of FX in TF has been reported (30). Further support for this hypothesis is provided by the observation that, similar to the membrane-bound complex, FVIIa in complex with the soluble TF exhibited dramatically lower activity toward the EGF1 deletion mutant of FIX in solution. In summary, these results suggest that EGF1 of FIX/FIXa plays at least two essential roles: It provides recognition sites for the cofactors (TF and FVIIIa) and functions as a spacer to spatially align and maintain the activation peptide and active-site of FIX/FIXa at an appropriate height above the membrane surface for optimal catalysis.
Acknowledgment
We thank Audrey Rezaie for proofreading the manuscript.
The research discussed herein was supported by grants awarded by the National Heart, Lung, and Blood Institute of the National Institute of Health (HL 101917 and HL 62565 to ARR).
Footnotes
1These authors contributed equally to this study.
Conflict-of-interest disclosure The authors declare no competing financial interest.
References
- 1.Jackson CM, Nemerson Y. Blood coagulation. Ann Rev Biochem. 1980;49:765–811. doi: 10.1146/annurev.bi.49.070180.004001. [DOI] [PubMed] [Google Scholar]
- 2.Furie B, Furie BC. The molecular basis of blood coagulation. Cell. 1988;53:505–18. doi: 10.1016/0092-8674(88)90567-3. [DOI] [PubMed] [Google Scholar]
- 3.Davie EW, Fujikawa K, Kisiel W. The coagulation cascade: Initiation, maintenance and regulation. Biochemistry. 1991;30:10363–70. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 4.Mann KG. Biochemistry and physiology of blood coagulation. Thromb Haemost. 1999;82:165–74. [PubMed] [Google Scholar]
- 5.Thompson AR. Structure, function, and molecular defects of factor IX. Blood. 1986;67:565–72. [PubMed] [Google Scholar]
- 6.Hopfner K-P, Brandstetter H, Karcher A, et al. Converting blood coagulation factor IXa into factor Xa: dramatic increase in amidolytic activity identifies important active site determinants. EMBO J. 1997;16:6626–35. doi: 10.1093/emboj/16.22.6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mertens K, Celie PHN, Kolkman JA, et al. Factor VIII-factor IX interactions: molecular sites involved in zymogen-cofactor complex assembly. Thromb Haemost. 1999;82:209–17. [PubMed] [Google Scholar]
- 8.Chang J, Jin J, Lollar P, et al. Changing residue 338 in human factor IX from arginine to alanine causes an increase in catalytic activity. J Biol Chem. 1998;273:12089–94. doi: 10.1074/jbc.273.20.12089. [DOI] [PubMed] [Google Scholar]
- 9.Stenflo J. Structure-function relationships of epidermal growth factor modules in vitamin K-dependent clotting factors. Blood. 1991;78:1637–51. [PubMed] [Google Scholar]
- 10.Mathur A, Bajaj SP. Protease and EGF1 domains of factor IXa play distinct role in binding to factor VIIIa. Importance of helix 330 (helix 162 in chymotrypsin of protease domain of factor IXa in its interaction with factor VIIIa. J Biol Chem. 1999;274:18477–86. doi: 10.1074/jbc.274.26.18477. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt AE, Bajaj SP. Structure-function relationships in factor IX and factor IXa. Trends Cardiovasc Med. 2003;13:39–45. doi: 10.1016/s1050-1738(02)00210-4. [DOI] [PubMed] [Google Scholar]
- 12.Blostein MD, Furie BC, Rajotte I, et al. The Gla domain of factor IXa binds to factor VIIIa in the tenase complex. J Biol Chem. 2003;278:31297–302. doi: 10.1074/jbc.M302840200. [DOI] [PubMed] [Google Scholar]
- 13.Chang JY, Monroe DM, Stafford DW, et al. Replacing the first epidermal growth factor-like domain of factor IX with that of factor VII enhances activity in vitro and in canine hemophilia B. J Clin Invest. 1997;100:886–92. doi: 10.1172/JCI119604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin SW, Smith KJ, Welsch D, et al. Expression and characterization of human factor IX and factor IX-factor X chimeras in mouse C127 cells. J Biol Chem. 1990;265:144–50. [PubMed] [Google Scholar]
- 15.Bode W, Mayr I, Baumann U, et al. The refined 1.9 A crystal structure of human alpha-thrombin: interaction with D-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment. EMBO J. 1989;8:3467–75. doi: 10.1002/j.1460-2075.1989.tb08511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang L, Gopalakrishna K, Manithody C, et al. Expression, purification and characterization of factor IX derivatives using a novel vector system. Protein Expr Purif. 2006;50:196–202. doi: 10.1016/j.pep.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 17.Yang L, Manithody C, Olson ST, et al. Contribution of basic residues of the autolysis loop to the substrate and inhibitor specificity of factor IXa. J Biol Chem. 2003;278:25032–8. doi: 10.1074/jbc.M302174200. [DOI] [PubMed] [Google Scholar]
- 18.Rezaie AR, Fiore MM, Neuenschwander PF, et al. Expression and purification of a soluble tissue factor fusion protein with an epitope for an unusual calcium-dependent antibody. Prot Exp Purif. 1992;3:453–60. doi: 10.1016/1046-5928(92)90062-2. [DOI] [PubMed] [Google Scholar]
- 19.Neuenschwander PF, Bianco-Fisher E, Rezaie AR, et al. Phosphatidylethanolamine augments factor VIIa-tissue factor activity: Enhancement of sensitivity to phosphatidylserine. Biochemistry. 1995;34:13988–93. doi: 10.1021/bi00043a004. [DOI] [PubMed] [Google Scholar]
- 20.Qureshi SH, Yang L, Yegneswaran S, et al. FRET studies with factor X mutants provide insight into the topography of the membrane-bound factor X/Xa. Biochem J. 2007;407:427–33. doi: 10.1042/BJ20070735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mutucumarana VP, Duffy EJ, Lollar P, et al. The active site of factor IXa is located far above the membrane surface and its conformation is altered upon association with factor VIIIa: A fluorescence study. J Biol Chem. 1992;267:17012–21. [PubMed] [Google Scholar]
- 22.Yegneswaran S, Smirnov MD, Safa O, et al. Relocating the active site of activated protein C eliminates the need for its protein S cofactor. J Biol Chem. 1999;274:5462–8. doi: 10.1074/jbc.274.9.5462. [DOI] [PubMed] [Google Scholar]
- 23.Huang C, Mason JT. Geometric packing constraints in egg phosphatidylcholine vesicles. Proc Natl Acad Sci (USA) 1978;75:308–10. doi: 10.1073/pnas.75.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bock PE. Active site selective labeling of serine proteases with spectroscopic probes using thioester peptide chloromethyl ketones: Demonstration of thrombin labeling using Nα-[(acetylthio)acetyl]-D-Phe-Pro-Arg-CH2Cl. Biochemistry. 1988;27:6633–9. doi: 10.1021/bi00417a063. [DOI] [PubMed] [Google Scholar]
- 25.Duffy EJ, Parker ET, Mutucumarana VP, et al. Binding of factor VIIIa and factor VIII to factor IXa on phospholipid vesicles. J Biol Chem. 1992;267:17006–11. [PubMed] [Google Scholar]
- 26.Qureshi SH, Yang L, Manithody C, et al. Membrane-dependent interaction of factor Xa and prothrombin with factor Va in the prothrombinase complex. Biochemistry. 2009;48:5034–41. doi: 10.1021/bi900240g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betz A, Krishnaswamy S. Regions remote from the site of cleavage determine macromolecular substrate recognition by the prothrombinase complex. J Biol Chem. 1998;273:10709–18. doi: 10.1074/jbc.273.17.10709. [DOI] [PubMed] [Google Scholar]
- 28.Husten EJ, Esmon CT, Johnson AE. The active site of blood coagulation factor Xa: its distance from the phospholipid surface and its conformational sensitivity to components of the prothrombinase complex. J Biol Chem. 1987;262:12953–62. [PubMed] [Google Scholar]
- 29.McCallum CD, Hapak RC, Neuenschwander PF, et al. The location of the active site of blood coagulation factor VIIa above the membrane surface and its reorientation upon association with tissue factor. A fluorescence energy transfer study. J Biol Chem. 1996;271:28168–75. doi: 10.1074/jbc.271.45.28168. [DOI] [PubMed] [Google Scholar]
- 30.Manithody C, Yang L, Rezaie AR. Identification of a basic region on tissue factor that interacts with the first epidermal growth factor-like domain of factor X. Biochemistry. 2007;46:3193–9. doi: 10.1021/bi6025193. [DOI] [PMC free article] [PubMed] [Google Scholar]