Abstract
DNA vaccines have evolved greatly over the last 20 years since their invention, but have yet to become a competitive alternative to conventional protein or carbohydrate based human vaccines. Whilst safety concerns were an initial barrier, the Achilles heel of DNA vaccines remains their poor immunogenicity when compared to protein vaccines. A wide variety of strategies have been developed to optimize DNA vaccine immunogenicity, including codon optimization, genetic adjuvants, electroporation and sophisticated prime-boost regimens, with each of these methods having its advantages and limitations. Whilst each of these methods has contributed to incremental improvements in DNA vaccine efficacy, more is still needed if human DNA vaccines are to succeed commercially. This review foresees a final breakthrough in human DNA vaccines will come from application of the latest cutting-edge technologies, including “epigenetics” and “omics” approaches, alongside traditional techniques to improve immunogenicity such as adjuvants and electroporation, thereby overcoming the current limitations of DNA vaccines in humans
Keywords: DNA vaccine, immunogenicity, adjuvant, epigenetics, RNAi, omics
INTRODUCTION
Unlike conventional protein or polysaccharide based vaccines, DNA vaccines comprise plasmids encoding the vaccine antigen along with a strong eukaryotic promoter used to drive protein expression (Rajcani et al., 2005). Such nucleic acid based vaccines can be delivered intramuscularly, subcutaneously or mucosally with the aim that they will gain access to the cell cytoplasm and thereby induce antigen expression in vivo that, like the protein vaccine they mimic, will then elicit an desired immune response. DNA vaccines have been successfully applied to animal models to variously prevent or treat infectious diseases, cancer, autoimmunity and allergy (Ulmer et al., 1996). On the positive side, the straightforward plasmid structure of DNA vaccines gives them inherent advantages over traditional protein or carbohydrate based vaccines. The one-step cloning of target coding sequence into plasmid vectors offers more convenient development and production when compared to culture and inactivation of whole infectious pathogens or expression and purification of recombinant proteins. Furthermore, by inducing expression of proteins in vivo, antigenic structure is more likely to resemble the native protein structure and include any essential post-translational modifications. From a safety perspective, amplification of the nucleic acids encoding a potential antigen avoids the need to directly handle dangerous pathogens. The convenient manipulation of plasmid DNA in vitro allows easy introduction of beneficial mutations into the antigen coding sequence. In vitro mutation also enables modification of antigen coding sequences to counter rapidly drifting virus strains. Plasmid DNA is stable at room temperature allowing for convenient storage and shipping. In addition to these physical properties, DNA vaccines enable expressed antigens to be presented by both MHC class I and class II complexes, thereby stimulating Th1 and Th2 CD4 and CD8 T cells in addition to B cells (Liu, 2011). To date, veterinary DNA vaccines have been approved for use in fish (infectious haematopoietic necrosis virus), dogs (melanoma), swine (growth hormone releasing hormone) and horses (West Nile virus) (Kutzler and Weiner, 2008). However, success in veterinary approvals has not translated into successful human DNA vaccine applications, with low immunogenicity remaining the Achilles heel of human DNA vaccines. In recent years, many clinical trials have been undertaken on DNA vaccines covering the full range of prophylactic through to therapeutic vaccines vaccines against infections, cancers and a range of other disorders, with details of these studies available through a range of websites including http://www.cancer.gov/clinicaltrials; http://clinicaltrials.gov; http://clinicaltrialsfeeds.org/; http://www.dnavaccine.com/; http://www.niaid.nih.gov/volunteer/vrc/Pages/default.aspx. However, despite more than 100 such clinical trials, more work is still clearly required on design and delivery to lift the immunogenicity of DNA vaccines to the levels required for human regulatory approval and commercial exploitation.
MECHANISM OF ACTION OF DNA VACCINES
In 1990, Wolff et al showed that injection of DNA encoding lactase reporter genes into mouse quadriceps muscle induced sustained protein expression (Wolff et al., 1990). Tang et al. subsequently showed that introducing a plasmid encoding human growth hormone (hGH) into mouse skin induced an antibody response against the expressed protein (Tang et al., 1992), thereby directly mimicking a protein vaccine. Final proof that a DNA encoded antigen could provide effective vaccine protection came from the demonstration that injection of plasmid encoding influenza nuclear protein into mouse muscle generated influenza-specific CD8+ cytotoxic T lymphocytes (CTL)s that then protected the mice from a subsequent influenza challenge (Ulmer et al., 1993).
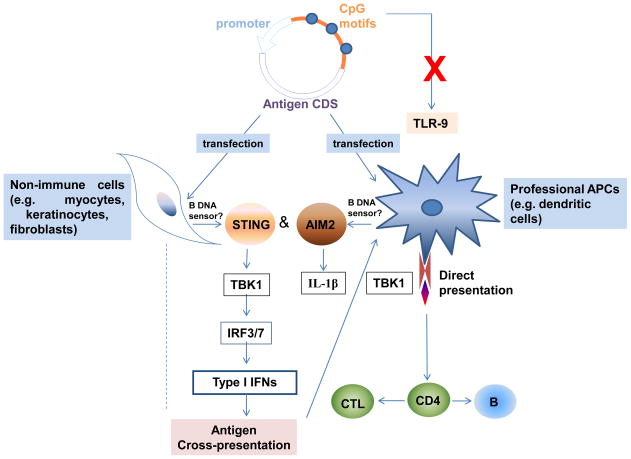
Whilst these studies confirmed the theoretical utility of DNA vaccines, practical considerations remained. For example, DNA inoculation results in antigen expression in the low picogram to nanogram range and most transfected somatic cells are not professional antigen presenting cells (APC). A potential offset is that the sustained low level antigen expression achieved with injected DNA may better prime adaptive immune responses when compared with the short half-life of injected protein antigens. At least three different mechanisms have been proposed to contribute to the immunogenicity of DNA vaccines: 1) DNA-encoded antigens are presented by somatic cells (myocytes or keratinocytes) through their MHC class I pathway to CD8 T cells; 2) DNA immunization results in direct transfection of professional antigen presenting cells (APC) (e.g. dendritic cells); and 3) cross-priming results from transfected somatic cells being phagocytosed by professional APCs which then present the antigens to T cells. Muscle cells are not efficient at presenting antigens via MHC class I, so the latter two mechanisms may be more important to DNA vaccines.
Immunostimulatory elements of plasmid DNA such as unmethylated CpG motifs may also make a contribution to DNA immunogenicity. CpG dinucleotide motifs have a low frequency and are mostly methylated in the mammalian genome. By contrast, bacterial DNA contains many unmethylated CpG motifs enabling this motif to be recognised by mammals as a pathogen associated molecular pattern (PAMP). Unmethylated CpG activates innate immune cells through binding to toll-like receptor (TLR)-9 (Hemmi et al., 2000; Klinman et al., 1997). TLR9 was shown to be important for the effectiveness of a DNA vaccine against lymphocytic choriomeningitis virus (LCMV) in a prime but not in a boost context (Rottembourg et al., 2010). TLR9 on dendritic cells (DCs) was required for efficient priming of CD8+ T cells upon plasmid exposure, in vitro, or single-dose vaccination, in vivo. However, TLR9-deficient mice still respond to DNA, suggesting that CpG motifs are not essential to DNA vaccine action (Babiuk et al., 2004; Tudor et al., 2005). Another study showed that TANK-binding kinase 1 (TBK1), a non-canonical IκB kinase, mediated the adjuvant effect of DNA vaccines in mice (Ishii et al., 2008). It is known that TBK1 can phosphorylate IRF3 and IRF7 to activate the type I interferon genes transcription (Sharma et al., 2003). Furthermore, cytoplasmic DNA can also activate AIM2 (absent in melanoma 2), STING (stimulator of IFN genes) and IRF3-dependent, innate immune pathways (Barber, 2011; Rathinam et al., 2010; Stetson and Medzhitov, 2006) and these may, therefore, also contribute to DNA vaccine action. The current proposed mechanisms of DNA vaccines are summarized in Figure 1.
Figure 1. Mechanisms of DNA vaccines.
Recent studies found that TLR-9 is dispensable for DNA vaccines, while TBK1 and AIM2 pathways were shown to be critical for plasmid DNA induced innate and adaptive immune responses. However, the upstream bona fide B DNA sensor still remains unknown.
TRADITIONAL STRATEGIES FOR DNA VACCINE DESIGN
Over the last twenty years different methods have been developed, as listed in Table 1, to increase the efficacy of DNA vaccines including, as discussed below, codon optimization, alternative promoters, plasmid backbone refinements, inclusion of antigen subcellular targeting systems and genetic adjuvants.
Table 1.
Optimization strategies for DNA vaccines
| Categories | Approaches | Effects | Safety | Practical considerations | |
|---|---|---|---|---|---|
| Pros | Cons | ||||
| Codon optimization | Use of host codons to maximize antigen expression | Most studies have shown increased antigenicity and immune responses (Cheung et al., 2004; Lin et al., 2006; Megati et al., 2008; Ngumbela et al., 2008; Siegismund et al., 2009; Tenbusch et al., 2010; Ternette et al., 2007; Trollet et al., 2009; Zhu et al., 2010). (These studies were carried out in mice) | In some cases no enhancement was observed or codon optimized construct was inferior to native sequence (Dobano et al., 2009; Varaldo et al., 2006). (These studies were carried out in mice) | No safety concerns | When antigen expression level is a problem, codon optimization should be considered |
| Promoter optimization | Use of strong viral promoters (CMV, SV40). | High expression and antigenicity in most cases (Chapman et al., 1991, monkey cells; Cheng et al., 1993, mouse, rabbit and rhesus monkey; Manthorpe et al., 1993, mouse; Wang et al., 2006, mouse). | Some promoters may have suppressive effects on antigenicity or may be inhibited by endogenous cytokines (Cao et al., 2011, mouse; Gribaudo et al., 1993, mouse fibroblast cells; Kerr and Stark, 1992; Vanniasinkam et al., 2006, mouse; Xiang and Ertl, 1995, mouse). | No safety concerns | Strong viral promoters are the first choice to achieve high antigen expression |
| Inducible or endogenous promoters. | Showed greater efficacy for specific DNA vaccines (Cao et al., 2011, mouse; Vanniasinkam et al., 2006, mouse). | Not broadly effective. | |||
| Reduction of bacterial elements | Delete redundant bacterial sequences | e.g. pcDNA3.1 is upgraded to pVAX1 | In some cases, may be time consuming. | Increased safety | More collaboration is needed to standardize new technologies and achieve consistent evaluation. |
| Sucrose induction system | Replace the bacterial selection marker (Luke et al., 2009, mouse; Luke et al., 2011b, rabbit). | ||||
| Mini-circle technology | Maintain the minimum antigen expressing cassette (Jechlinger et al., 2004; Kay et al., 2010, mcDNA preparation technology; Narsinh et al., 2011, stem cells; Osborn et al., 2011, mouse; Zuo et al., 2011, cultured cancer cells and mouse). | ||||
| S/MAR vectors | Generation of episomal vectors | Decrease the integration rate and increase long-term expression (Argyros et al., 2011, cultured cells and mouse; Conese et al., 2004, review). | No commercial standard. Need more evaluation. | Increased safety | This method can be combined with mini-circle technology to achieve maximum safety and efficiency |
| Targeting technologies | Targeting to APCs (ligand fusion and other methods) | Enhanced antigen processing and presentation (Anwar et al., 2005; Argilaguet et al., 2011; Boyle et al., 1998; de Arruda et al., 2004; Faham et al., 2011; Ji et al., 1999; Liu et al., 2011; Lu et al., 2003; Marques et al., 2003; Midha and Bhatnagar, 2009; Niazi et al., 2007; Palumbo et al., 2011; Rigato et al., 2010; Wang et al., 2011b; Yang et al., 2009). (These studies were carried out in mice) | Some studies showed no enhanced effects (Carvalho et al., 2010, cultured cells; Kaur et al., 2009, mouse; Vidalin et al., 1999, mouse). | Potential for interference with the endogenous immune system needs to be evaluated | Targeting effects need to be tested individually. More suitable for anti-cancer DNA vaccines |
| Subcellular targeting (use endogenous trafficking system) | |||||
| Adjuvants | Genetic adjuvants, e.g. cytokines and chemokines | Enhanced immune responses were observed in many studies (Bode et al., 2011, review; Kim et al., 2008, mouse; Kolka et al., 2005, mouse; Liu, 2011, review; Luke et al., 2011a, mouse; Riedl et al., 2006, cultured cells and mouse; Schirmbeck et al., 2002, mouse; Tovey and Lallemand, 2010, review). | Co-expression of inflammatory cytokines or chemokines has the potential for increased side effects | Potential interference with the endogenous immune system needs to be evaluated. | Considering uncertain safety issues may be more suitable for use in therapeutic DNA vaccines, e.g. cancer vaccines |
| Modulation of the pattern recognition receptors, e.g. TLRs, RLRs and NLRs | |||||
| Other adjuvants | |||||
| Epigenetics | Drugs regulating DNA methylation or histone modifications | Some initial promising potential for DNA vaccines (Brooks et al., 2004, rat; Fujisawa et al., 2011, mouse model; Gowda et al., 2011, mouse; Karpf, 2006, review; Pearce and Shen, 2006, review; Riu et al., 2007, mouse; Xu et al., 2011, mouse; Youngblood et al., 2010, review). | Both mechanistic and applied studies are in early stages | Safety issues not known | Need extensive studies, which will provide basis for designing new DNA vaccine strategies. |
| Avoiding epigenetic silencing | |||||
| Epigenetics mechanisms of immune responses | |||||
| RNAi technology | Targeting potential suppressive factors. | Enhanced immune responses were observed in some studies (Geiben-Lynn et al., 2011, mouse; Greenland et al., 2007, mouse; Huang et al., 2008, mouse; Kim et al., 2011, mouse; Kim et al., 2005, mouse; Wang et al., 2011c, mouse). | More studies are needed to identify the appropriate factors to be targeted. | No safety issues have been identified to date but still early days | This technology will serve as a powerful tool when appropriate targets are identified. |
| Systems or “omics” technology | Systems biology | These studies provide valuable guides for DNA vaccine design (Nakaya et al., 2011, human, mouse model; Rappuoli and Aderem, 2011, review; Sette and Rappuoli, 2010, review; Trautmann and Sekaly, 2011, review; Wang et al., 2011d (human and mouse; Xiao et al., 2011, mouse; Yang et al., 2006, in silico and arry study). | Requirement of advanced technology and expensive experiments currently limit use to large companies | No safety issues | These technologies will greatly enhance the rational design of future DNA vaccines. |
| Library technology | |||||
| Proteomics | |||||
| Genomics (reverse vaccinology) | |||||
| Other “omics” and combinations | |||||
CODON OPTIMIZATION
Because codon usage by pathogens is often quite different to mammalian genomes, antigen coding sequences may need optimization for efficient expression in host cells. An early study of a DNA vaccine encoding an H-2Kd-restricted epitope of listeriolysin O (LLO) of L. monocytogenes, showed that in BALB/c mice codon optimization improved CD8 T cell responses against this intracellular bacterium (Uchijima et al., 1998). Codon optimization of DNA encoding bacterial botulinum neurotoxins markedly increased neutralizing antibody titers in mouse models (Trollet et al., 2009). Likewise, codon optimization has been shown to increase immunogenicity and provide better protection against challenge in DNA vaccine against Schistosoma japonicum, influenza virus, HIV, HPV, RSV, and SIV in mouse models (Cheung et al., 2004; Lin et al., 2006; Megati et al., 2008; Ngumbela et al., 2008; Siegismund et al., 2009; Tenbusch et al., 2010; Ternette et al., 2007). Automated algorithms for calculating codon optimization are now available to assist vaccine developers (Harish et al., 2006; Sandhu et al., 2008).
While codon optimization increases antigen expression, this may not, per se, always increase vaccine efficacy. For example, a malaria DNA vaccine study showed more robust CD4+ and CD8+ T cell responses and protection against P. yoelii sporozoite challenge with native, rather than codon-optimized, plasmids in mice (Dobano et al., 2009). Another study in mice using a codon-optimized plasmid encoding the Sm14 antigen of Schistosoma mansoni found that while codon optimization increased Sm14 expression it did not enhance immunity or protection against S. mansoni challenge (Varaldo et al., 2006). The reason for these discrepancies is not known.
PROMOTER DESIGN
The coding sequences in DNA vaccines are driven by polymerase II type promoters. As endogenous mammalian promoters are generally not strong enough for driving high level antigen expression strong virus-derived promoters like cytomegalovirus (CMV) or SV40 promoters (vectors used include pcDNA3.1, pVAX1, pVIVO2, pCI, pCMV and pSV2). Some early studies showed that CMV immediate early enhancer/promoter activity was consistently the strongest explaining its widespread usage in DNA vaccines (Cheng et al., 1993; Manthorpe et al., 1993). The intron A of the CMV immediate-early gene is often included in the complete CMV promoter, as studies showed that secretion of glycoproteins was significantly higher when cells were transfected with intron A-containing plasmids (Chapman et al., 1991). Studies of HIV-1 Env DNA vaccines showed that the strong promoter plays an important role in increasing antigen expression and thereby vaccine immunogenicity in mice (Wang et al., 2006). However, in some cases strong promoter activity can be a problem for DNA vaccine design. For example, the hepatitis C virus (HCV) core protein is a candidate vaccine antigen but has immuno-suppressive properties. To overcome this problem, a hepatitis C virus (HCV) DNA vaccine utilized an inducible in vivo activated Salmonella promoter to drive core protein expression along with a CMV promoter to drive envelope protein 2 (E2) expression. In this way immune responses were generated against both the induced HCV core protein and the E2-protein in BALB/c mice (Cao et al., 2011). However, some studies have shown that the potency of viral promoters does not necessarily correlate with DNA vaccine efficacy. This may be because induction of TNF-α and INF-γ by potent DNA vaccines can paradoxically down-regulate viral promoters and thereby reduce immune responses to the vaccine (Gribaudo et al., 1993; Kerr and Stark, 1992; Vanniasinkam et al., 2006; Xiang and Ertl, 1995). Non-viral promoters such as the MHC class II promoter have also been shown to be effective in DNA vaccines in mice (Vanniasinkam et al., 2006). Thus, although the CMV promoter remains the first choice, in some cases alternative promoters may achieve better immunogenicity for DNA vaccines.
REMOVAL OF BACTERIAL ELEMENTS
Currently used DNA plasmids are composed of bacteria-derived sequences providing replication signals and selection markers necessary for propagation in different E. coli strains. This material may pose safety issues and could have a negative impact on mammalian gene expression by these bacterial elements. For example, the expression vector pcDNA3.1, which has been widely used in DNA vaccines, was modified to generate a new generation of pVAX1 vector, in order to reduce redundant sequences and to change the ampicillin selection marker to kanamycin, given the potential for ampicillin to trigger allergy. The reduced size of the pVAX1 vector permitted cloning of larger DNA fragments and yet retained comparable antigen expression level to the antecedent pcDNA3.1 vector. The advantage of pVAX1 over pcDNA3.1 was demonstrated by the fact that pcDNA3-ANXB1 (pcDNA3-b1) but not pVAX-ANXB1 (pVAX-b1) induced autoimmunity in inoculated mice (Zhou et al., 2011).
The sucrose selection system has recently been developed to remove the need for an antibiotic selection marker. Vectors expressing a 150bp RNA-OUT antisense RNA repressed expression of a chromosomally-integrated, constitutively-expressed, counter-selectable marker (sacB), allowing plasmid selection on sucrose (Luke et al., 2009). Using this antibiotic-free system, a SV40 72 base pair enhancer was further incorporated upstream of the CMV promoter to increase the extra-chromosomal transgene expression or the human T-lymphotropic virus type I (HTLV-I) R region downstream of CMV promoter to increase mRNA translation efficiency. Increased HIV-1 gp120 DNA vaccine-induced neutralizing antibody titers were demonstrated in rabbits using this vector system (Luke et al., 2011b).
Minicircle DNA (mcDNA) technology was developed to completely remove the bacterial backbone by using site-specific recombination based on the ParA resolvase to generate mcDNA (Jechlinger et al., 2004). Another method for mcDNA production used genetically modified E. coli to construct a producer strain that stably expresses a set of inducible minicircle-assembly enzymes, PhiC31 integrase and I-SceI homing endonuclease (Kay et al., 2010). mcDNA technology has been successfully used in gene therapy experiments in mouse models (Osborn et al., 2011; Zuo et al., 2011) and in generation of adult human induced pluripotent stem cells (Narsinh et al., 2011), with potential for this technology to similarly be applied to DNA vaccine design.
SCAFFOLD/MATRIX ATTACHMENT REGION (S/MAR) VECTOR
For some vaccine applications it may be important to have long-term tissue antigen expression. This can be achieved using retrovirus systems that have high transduction efficiency and integration rate. Unfortunately, cases of leukemia (Dave et al., 2004) have resulted from the random integration of therapeutic retrovirus vectors. Episomally-maintained vectors utilizing scaffold/matrix attachment region (S/MAR) have been developed as a potential substitute (Conese et al., 2004). Such vectors include Epstein-Barr virus (EBV) oriP or EBNA1 elements to maintain self-replication. After a single administration, such transgenes have been shown to be expressed in mouse tissues for at least 6 months (Argyros et al., 2011). When minicircle DNA technology was also applied to the episomal vector, higher and more sustained in vitro and in vivo (mouse model) transgene expression was achieved for several months in the absence of selection (Argyros et al., 2011). This suggests that such systems could be applied to vaccine design to prolong antigen expression.
DNA VACCINE TARGETING TECHNOLOGIES
As discussed above, the immune responses induced by DNA vaccines may variously be mediated by antigen presentation by transfected somatic cells, by transfected professional APCs or cross-presentation of antigens by APCs that have ingested apoptotic transfected cells. The ability of non-professional APCs to present antigens on the MHC class II pathway and thereby induce CD4 T helper cells is very limited. Strategies have therefore been developed to target DNA vaccines to professional APCs, such as DCs. Skin, mucosal tissues and lymph nodes contain more DCs than muscle, and thus are useful tissues in which to target DCs. Lymph node targeting vectors were created by designing plasmids encoding human IgG fused to either L-selectin or cytotoxic T-lymphocyte antigen 4 (CTLA4) (Boyle et al., 1998). L-selectin facilitates targeting to lymph nodes by binding to CD34 on endothelial cells, while CTLA4 targets expressed antigen to APCs expressing B7 and both targeting strategies enhanced immune responses. In a recent study, the Hantaan virus (HTNV) nucleocapsid protein was fused with CTLA4 to produce a HTNV DNA vaccine in C57BL/6 mouse model (Liu et al., 2011). In a pig study, increased vaccine immunogenicity was obtained by fusion of ASFV antigen with a single chain antibody variable region against swine HLA-II (Argilaguet et al., 2011). Many more DC targeting methods have been successfully used in mouse models with other targeting molecules, FIRE (F4/80-like receptor) or CIRE (C-type lectin receptor), Cle9A, Flt3, DEC205, or synthetic MHC class II-targeting peptides (Corbett et al., 2005; Daftarian et al., 2011; Kataoka et al., 2011; Lahoud et al., 2011; Njongmeta et al., 2012). These targeting methods differ in regards to their effects. For example, Cle9A and DEC205 but not Cle12A are effective targets for induction of cytotoxic T lymphocyte (CTL) responses with Cle9A shown to enhance antibody production (Lahoud et al., 2011). When DNA vaccines are targeted to Cle9A, FIRE or CIRE, they appear not to need other “danger” signals or adjuvants to elicit a robust immune response (Corbett et al., 2005; Lahoud et al., 2011).
Some specially designed synthetic materials help target plasmids to professional APCs. For example, pDNA-lipoplexes engrafted with flagellin-derived peptides were able to target plasmids to DCs and other APCs in mouse (Faham et al., 2011).
Subcellular targeting is another strategy for enhancing plasmid-encoded antigen processing and/or presentation. This strategy uses host protein trafficking mechanisms to target expressed proteins to particular cellular compartments or mark them for secretion, thereby facilitating antigen processing and presentation. One of the most widely used signal sequence for subcellular targeting is lysosome associated membrane protein 1 (LAMP1). An early study fused the human papillomavirus type 16E7 DNA with LAMP1 to generate a chimeric DNA vaccine targeting the HPV 16E7 antigen to the endosomal compartment, thereby enhancing the immunogenicity of the DNA vaccine (Ji et al., 1999). The LAMP targeting system has subsequently been shown effective in many different model systems (Anwar et al., 2005; de Arruda et al., 2004; Lu et al., 2003; Marques et al., 2003; Rigato et al., 2010; Yang et al., 2009). The LAMP sequence can also be combined with other signal sequences. For example, the protective antigen (PA63) of Bacillus anthracis was fused with tissue plasminogen activator and LAMP1 to generate the pTPA-PA63-LAMP1construct, which was shown to induce high neutralizing antibody titers against anthrax (Midha and Bhatnagar, 2009). Ubiquitin and the human CD1 tail sequence have also been tested as DNA vaccine targeting sequences (Chen et al., 2011; Niazi et al., 2007; Wang et al., 2011b). Such targeting strategies are not infallible, however, as a hepatitis C virus core protein vaccine was not enhanced by fusion with ubiquitin or LAMP1 (Vidalin et al., 1999). A rabies DNA vaccine similarly showed no benefits of MHC class I and II targeting sequences (Kaur et al., 2009). A recent study compared different targeting sequences including a secretion signal, LAMP1 or endoplasmic reticulum (adenovirus e1a) signal which were fused with the green fluorescent protein (GFP)-tagged model genes invariant surface glycoprotein or trans-sialidase from Trypanosoma brucei. For different expressed genes, the effects of cellular targeting varied (Carvalho et al., 2010). A study on intracellular transport and fate of plasmid DNA in mammalian cells showed that plasmid endocytosis can alter the pH value of the late endosome and thereby interfere with antigen processing (Trombone et al., 2007). Thus, when different antigens are fused with lysosome or other targeting sequences, the physical and chemical properties of such non-host proteins may interfere with normal endosomal function. To address this problem, further understanding of the molecular pathways of plasmid-expressed antigens is needed. Recent progress has shown the importance of autophagy pathways in this process (Munz, 2009). Rapamycin-induced autophagy was shown to enhance the presentation of mycobacterial antigen Ag85B, while inhibition of autophagy by 3-methyladenine, or RNAi against beclin-1, attenuated presentation (Jagannath et al., 2009). Similarly, a short polypeptide from the herpes simplex virus type 2 ICP10 gene that can induce antigen aggregation and autophagosomal degradation, enhanced T cell responses when it was co-expressed with chicken ovalbumin (Fu et al., 2010).
Another subcellular targeting method involves inclusion of leader sequence or other localization signals in DNA to allow direct localization of antigens to membranes or secretion outside the cells. (Forns et al., 2000) found that membrane-bound and secreted forms of HCV E2 DNA vaccine induced more E2-specific antibodies compared to the intracellular form (Ma et al., 2002). As DNA vaccines need to get into the nucleus for transcription, strategies of nuclear targeting including liposomes, small peptides and use of a nuclear localization signal (NLS) have also been used to increase antigen expression (Wang et al., 2011a).
DNA VACCINE ADJUVANTS
Vaccine adjuvants function through a range of mechanisms including innate immune activation, antigen depot formation, chemotaxis, antigen uptake and presentation by professional APC and upregulation of co-stimulatory molecules. Conventional vaccine adjuvants including, alum particles (Khosroshahi et al., 2012) and MF59 emulsions (Ott et al., 2002) when mixed with plasmid have also been shown to modesty improve the immunogenicity of DNA vaccines. CpG oligonucleotides that activate TLR9 have also been used as DNA vaccine adjuvants (Bode et al., 2011). Another place in which adjuvants may be relevant is in the DNA prime/protein boost context where the adjuvant is combined with the protein boost to magnify the immune response induced by DNA priming. For example, Advax™, a polysaccharide nanoparticle adjuvant,was successfully combined with a HIV envelope protein boost to enhance the immunogenicity of an env-encoding DNA vaccine (Cristillo et al., 2011; Lobigs et al., 2010; Petrovsky, 2008, 2011; Petrovsky and Cooper, 2011).
Given that DNA vaccines can easily be designed to express cytokines or co-stimulatory molecules, and the nucleic acid sequence per se may serve as an agonist for TLR-9 or other cellular DNA sensors, DNA vaccines can be designed to co-express so called ‘genetic adjuvant’ molecules. Such genetic adjuvants include cytokines, chemokines or immune stimulatory molecules that are expressed from plasmid DNA in cis or trans. Use of genetic adjuvants to enhance DNA vaccine immunogenicity has recently been extensively reviewed and hence won’t be covered in detail here (Liu, 2011; Saade and Petrovsky, 2012; Tovey and Lallemand, 2010).
In addition to these well characterized genetic adjuvant molecules, other strategies for enhancement of DNA vaccines by manipulation at the molecular level could also provide adjuvant effects. For example, the MHC CIITA is a critical regulator of MHC class II expression and co-administration of HPV16 E6 DNA and CIITA DNA resulted in enhanced antigen-specific CD8(+) T cell responses in mouse models (Kim et al., 2008). Retinoic acid-inducible gene I (RIG-I) is an important cellular receptor for dsRNA and RIG-I agonist expressed from the RNA polymerase III promoter enhanced hemagglutinin-specific antibody avidity after intramuscular injection of influenza DNA vaccine in mice (Luke et al., 2011a). A recent study found that upon dsRNA viral infection, the mitochondrial protein MAVS forms prion-like aggregates, which then activate IRF3 (Hou et al., 2011). This suggests that manipulation of MAVS conformation changes during delivery of DNA vaccines or development of new aggregate forming polypeptides may successfully increase DNA vaccine immunogenicity. Other studies have also used a decoy system by fusing antigen to a viral DnaJ-like sequence (J domain) associated with the constitutively expressed host cell stress protein, heat shock protein HSP73. This system supported efficient protein expression including of some unstable and/or toxic antigens (Riedl et al., 2006; Schirmbeck et al., 2002).
DNA VACCINE DELIVERY AND ELECTROPORATION
Another issue in DNA vaccine efficacy is the mode of its delivery. Standard intramuscular injection of naked DNA is very inefficient, with only a tiny fraction of injected DNA being taken up by cells and expressed. An alternative method is to inject DNA vaccines in coated nano- or microparticles, which protect plasmids from degradation and increase phagocytic uptake by professional APCs (Xiang et al., 2010). Plasmid DNA can also be coated on colloidal gold particles and delivered by the “gene gun” method. Although one advantage of “gene gun” is its targeting to Langerhans cells and other professional APCs (Porgador et al., 1998; Stoecklinger et al., 2007), it is limited in dose capacity, which means multiple “shots” at multiple sites are needed for effective immunization. However, the most significant improvement in delivery of DNA vaccines has been electroporation (EP). EP efficiently transfects somatic cells in vivo and its induction of local inflammation also enhances the immune response. Clinical trials have confirmed the efficacy of EP (van Drunen Littel-van den Hurk and Hannaman, 2010). For example, EP of a hepatitis B virus (HBV) DNA, induced potent CTL responses in mice and rabbits (Luxembourg et al., 2006). EP similarly enhanced humoral responses to a DHBV DNA vaccine in ducks (Khawaja et al., 2012). EP was also shown to enhance the immunogenicity in mice of DNA coated microparticles (Barbon et al., 2010). EP therefore remains a promising approach to improve the immunogenicity of DNA vaccines, although the poor tolerability of EP remains a concern in the prophylactic vaccine setting (Sardesai and Weiner, 2011)
USE OF EPIGENETICS IN DNA VACCINE DISIGN
Epigenetics is the study of heritable mechanisms that affect the transcriptional state of a gene, not due to changes in DNA sequence. Epigenetic mechanisms include histone modifications and variants, DNA methylation, chromatin remodeling, RNAi and noncoding RNAs. Epigenetics has been shown to be involved in a wide variety of biological processes including immune system function (Cuddapah et al., 2010; Goldberg et al., 2007). Epigenetic tools may be useful to unravel DNA vaccine mechanisms and to design more potent DNA vaccines. In a study of adenovirus-vector mediated gene delivery, rats were given an intramuscular injection of virus expressing human fibroblast growth factor 4 driven by the CMV promoter, with the ratio of copies of hFGF-4 mRNA per copy of viral DNA decreasing 385-fold between 6 hours and 28 days after the injection, with extensive methylation of the CMV being shown to be responsible for the gene silencing (Brooks et al., 2004). Gene expression using plasmid DNA faces similar silencing effects, suggesting that enhanced antigen expression could be achieved if silencing effects are avoided. Sequences in episomal vectors that originate from bacterial sources play a critical role in transcriptional silencing of transgenes in vivo (Riu et al., 2007). Furthermore, episomal vectors undergo chromatinization in vivo and both persistence and silencing of transgene expression is associated with specific histone modifications. Removal of the bacterial elements by using minicircle DNA technology enabled higher transgene expression, manifested by active histone marks detected by ChIP assays (Riu et al., 2007). This indicates that when plasmids are located within the nucleus, epigenetic regulation of transcription takes place. A study of cancer vaccines confirmed that modulatory agents, including DNA methyltransferase (DNMT) and histone deacetylase (HDAC) inhibitors, enhanced antigen and MHC class I expression (Karpf, 2006). Another study found that active histone modification by HDAC inhibitor enhanced the effectiveness of IL-13 receptor targeted immunotoxin in murine models of human pancreatic cancer (Fujisawa et al., 2011). A study on parasite infection found that a DNA-protein complex was required for the entry of parasite DNA into cells for recognition by TLR9. The nucleosome (histone-DNA complex) was confirmed as the TLR9-binding immunostimulatory component of Plasmodium falciparum that activated DCs (Gowda et al., 2011). Another study found that histones released during tissue injuries can mediate cell death by activating TLR2 and TLR4 (Xu et al., 2011). Thus, plasmid DNA may be reconstituted with histones bearing active modifications to achieve higher antigen expression and increase DNA vaccine immunogenicity. As CD8+ T cells are critical for control of viral infections, epigenetic mechanisms involved in activation of naïve T cells and maintenance of the memory T cell identity (Pearce and Shen, 2006; Youngblood et al., 2010) may also be utilized for DNA vaccine design (Fernandez-Morera et al., 2010).
RNAi TECHNOLOGIES IN DNA VACCINE DESIGN
RNA interference (RNAi) is a post-transcriptional gene silencing process triggered by double-stranded short hairpin RNA (shRNA) structures. Since its discovery, RNAi has mainly been used as a research tool for loss of function studies of target genes but also as a therapeutic method for human diseases (Lares et al., 2010). Due to its ease of production and flexibility in delivery, RNAi technology has potential applications in DNA vaccine design. One way to use RNAi for DNA vaccines would be to use it to block genes that suppress vaccine action. For example, immune responses induced by DNA vaccines are attenuated due to the limited duration of antigen expression in vivo. Due to death of transfected cells, use of shRNA to knock down caspase 12 (Casp12), a cell death mediator that is upregulated after DNA vaccination resulted in increased plasmid luciferase and HIV-gp120 Env antigen expression and higher CD8 T cell and antibody production (Geiben-Lynn et al., 2011). Similarly, RNAi-mediated depletion of the pro-apoptotic proteins Bak and Bax at the time of immunization of HPV16 E7 vaccine prolonged the life of antigen-expressing DCs and increased antitumor effects against E7-expressing tumors (Kim et al., 2005). Fas-mediated apoptosis limits DNA vaccine-induced immune responses (Greenland et al., 2007), and co-delivery of HPV-16 E7 DNA vaccine with DNA expressing shRNA against Fas ligand significantly enhanced CTL responses against E7 (Huang et al., 2008). RNAi may also be used to block immune-suppressive genes that otherwise inhibit vaccine responses. Depletion of Foxo3, a critical suppressive regulator of T cell proliferation, by RNAi increased the efficacy of a HER-2/neu DNA cancer vaccine (Wang et al., 2011c). Similarly knockdown of the IL10 receptor enhanced the potency of a DC vaccine (Kim et al., 2011). Furthermore, blockade of the programmed cell death-1 (PD-1) ligand B7-H1 (PD-L1) by RNAi augmented DC-mediated T cell responses and antiviral immunity in HBV transgenic mice (Jiang, 2012). Thus, use of RNAi against target genes limiting plasmid expression such as apoptosis genes or mediating immune suppressive effects is a powerful new strategy for DNA vaccine enhancement. However, safety issues of use of RNAi to enhance human DNA vaccines still need to be addressed and hence such technologies are most likely to first be applied to therapeutic cancer vaccines rather than more typical prophylactic vaccines.
SYSTEMS OR “OMICS”APPROACHES TO DNA VACCINE DESIGN
Accelerating advances in next-generation sequencing, microarrays, and high throughput proteomics approaches, provide the opportunity to apply these new techniques to DNA vaccine design. One recent proteomics study screened proteins for interaction with plasmid DNA and found that human serum amyloid P (SAP) inhibited plasmid transfection and enhanced plasmid clearance. SAP may contribute to the low efficacy of DNA vaccines in humans, as in other species suppressive effects of SAP are much weaker (Wang et al., 2011d). Hence SAP could, for example, serve as a new siRNA target for enhancing DNA vaccine efficacy, although the feasibility, effectiveness and safety of such an approach would first need to be tested in suitable animal models.
Systems biology approaches have also been used to analyze the molecular signatures that correlate with a positive immunization response. For example, expression levels of CaMKIV kinase at day 3 were negatively correlated with subsequent influenza antibody titers (Nakaya et al., 2011). This provides a successful example of the applications of systems biology to identify biomarkers that predict vaccine effectiveness (Trautmann and Sekaly, 2011). A study of Leptospira interrogans used bioinformatics, comparative genomic hybridization and transcription analysis to screen for candidate antigens from the pathogen’s genome and found 226 candidate genes out of 4727 open reading frames (ORFs) (Yang et al., 2006). This is the concept of reverse vaccinology (Sette and Rappuoli, 2010). A ribosome display of Cryptosporidium parvum cDNA library enabled identification of a new adhesion protein named Cp20, which when included in a pVAX1-Cp20 DNA vaccine, induced antibody and cellular responses and protection (Xiao et al., 2011). Since DNA vaccines are quick and easy to prepare, they are particularly useful for screening potential antigens identified through reverse vaccinology approaches. Thus the development of new DNA vaccines will in future be assisted by next generation sequencing, advanced bioinformatics analysis and other cutting-edge “omics” technologies (Kennedy and Poland, 2011; Poland et al., 2011).
PRIME/BOOST DNA VACCINE STRATEGIES
Whilst DNA vaccines by themselves suffer from low immunogenicity, this is not necessarily true when they are combined with other vaccine modalities in prime-boost type approaches. Regimens like DNA prime/protein boost, DNA prime/viral vector boost (e.g. using adenovirus) have shown major success. An early study in mouse models has shown that a DNA prime followed by a single protein boost of the same modified vaccinia virus Ankara (MVA) antigen induced complete protection in challenges, which was correlated with induction of very high levels of CD8+ T cells (Schneider et al., 1998). A mouse study of Leishmania donovani gp63 vaccine comparing different prime/boost combinations, found that the DNA prime/protein boost regimen was better than DNA/DNA or protein/protein regimens for long-term protection in mouse models (Mazumder et al., 2011). Human studies have also shown superior immune responses of mixed modality prime-boost compared to pure DNA vaccine regimens (Lu et al., 2008). Recent studies have used DNA/protein or DNA/Ad-vector regimens for HIV immunization (Churchyard et al., 2011; De Rosa et al., 2011; Jaoko et al., 2010; Koblin et al., 2011; Ledgerwood et al., 2011). The underlying mechanisms behind the effectiveness of heterogeneous prime-boost regimens are not well understood but DNA priming results in much lower antigen expression compared to protein vaccines, and this may preferentially prime T-helper cell responses with the humoral response subsequently being boosted by the high dose protein or viral vector boost.
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
The immunogenicity of DNA vaccines in humans is limited by low levels of antigen expression, when compared to conventional protein vaccines. DNA vaccines may be able to make up for this limitation by altering plasmid construction to maximize protein expression, targeting of the expressed antigen to professional APC thereby ensuring efficient MHC-I and MHC-II compartment loading, inclusion of a genetic adjuvant, use of electroporation and, where suitable, use of a DNA prime/protein or vector boost approach. As summarized in Figure 2, the development of new technologies provides even greater opportunities to further enhance the efficacy of DNA vaccines. The most likely scenario for the first successful human DNA vaccines is that they will be part of a DNA prime/protein boost vaccine strategy where the initial DNA prime is used to ensure efficient CD8 and CD4 T-cell priming whereas the protein boost is used to maximize antibody production. Notably, more than twenty years from their initial discovery, and after many disappointing human clinical trials of first generation vaccines, DNA vaccines are currently undergoing somewhat of a revival thanks to introduction of more efficient designs and better delivery technologies including electroporation. While many outside the field may still be rightfully skeptical, given the failure to meet early promise, this is not an uncommon phase in new technology introduction and often heralds, final success. This is very reminiscent of the history of monoclonal antibody therapeutics that similarly went through a highly negative phase before all the initial technology problems were solved and they emerged as pharmaceutical blockbusters. DNA vaccine may similarly be just moving past their darkest hour and thereby be soon ready to re-emerge as commercially viable products, most likely initially in the area of therapeutic cancer vaccines.
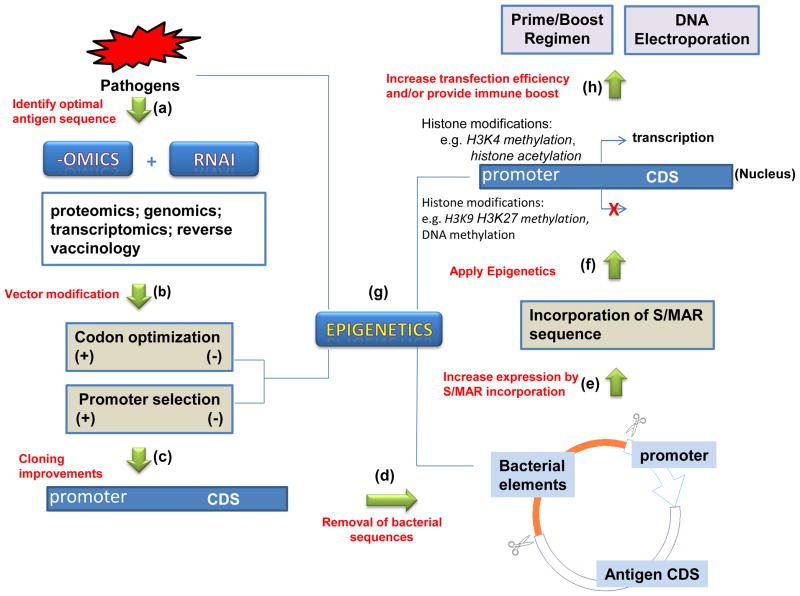
Figure 2. Potential design strategies and technologies for future DNA vaccine development.
(a) With the ease of DNA synthesis and manipulation, a lot of high throughput technologies, e.g. gene expression arrays, proteomics, genomics, transcriptomics, reverse vaccinology and RNAi screening platforms, which were accompanied by comprehensive bioinformatics tools, can efficiently map and find the DNA sequences encoding optimal antigens for vaccine development. (b) After selecting the candidate pathogen DNA sequences, codon optimization and promoter design are the main two steps before cloning into expression vectors. Simple codon conversion and using the strong viral promoter will most likely result in higher gene expression, but controversial effects were also reported. The more precise algorithm is anticipated for this purpose in the field of plasmid DNA based therapy or vaccination. (c) The optimized DNA fragments are then cloned into expression vector to test expression. To further optimize the DNA vaccine, minicircle DNA technology (d) will be used to completely remove the bacterial elements and incorporate S/MAR sequence (e) to enhance expression and safety. (f) and (g) Epigenetics mechanisms are closely related with most of the above steps and will be applied in DNA vaccine design. (h) The immunogenicity of the DNA vaccine construction could be further increased by injection of the optimized construction using new delivery devices (e.g. EP) and/or by using prime/boost regimen. See the article for details.
Literature searching method
The literature referenced in this review was searched by using the PubMed database for literature published prior to 1 March 2012 with keywords (“DNA vaccine” OR “DNA vaccines” OR “DNA vaccination” OR “DNA vaccinations” OR “DNA Immunization” OR “DNA immunizations” OR “gene vaccination” OR “gene vaccinations” OR “genetic vaccine”) alone or along with other related topics (e.g. epigenetics, RNAi. Studies published in the most recent two years were paid more attention, as they represented the most up to date development in the area.
Highlights.
Overview of strengths and weaknesses of previous DNA vaccine optimization strategies
Identifies new technologies (S/MAR vector, mcDNA, RNAi) relevant to DNA vaccine design
“Epigenetics” and “omics” technologies provide exciting new opportunities
Imminent breakthrough in human DNA vaccines incorporating latest technologies
Acknowledgments
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272200800039C and Collaborative Research Contact No. U01AI061142. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Competing financial interests
The authors are employees or directors of Vaxine Pty Ltd, a company developing vaccine technologies including Advax™ adjuvant.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anwar A, Chandrasekaran A, Ng ML, Marques E, August JT. West Nile premembrane-envelope genetic vaccine encoded as a chimera containing the transmembrane and cytoplasmic domains of a lysosome-associated membrane protein: increased cellular concentration of the transgene product, targeting to the MHC II compartment, and enhanced neutralizing antibody response. Virology. 2005;332:66–77. doi: 10.1016/j.virol.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Argilaguet JM, Perez-Martin E, Gallardo C, Salguero FJ, Borrego B, Lacasta A, Accensi F, Diaz I, Nofrarias M, Pujols J, Blanco E, Perez-Filgueira M, Escribano JM, Rodriguez F. Enhancing DNA immunization by targeting ASFV antigens to SLA-II bearing cells. Vaccine. 2011;29:5379–5385. doi: 10.1016/j.vaccine.2011.05.084. [DOI] [PubMed] [Google Scholar]
- Argyros O, Wong SP, Fedonidis C, Tolmachov O, Waddington SN, Howe SJ, Niceta M, Coutelle C, Harbottle RP. Development of S/MAR minicircles for enhanced and persistent transgene expression in the mouse liver. J Mol Med (Berl) 2011;89:515–529. doi: 10.1007/s00109-010-0713-3. [DOI] [PubMed] [Google Scholar]
- Babiuk S, Mookherjee N, Pontarollo R, Griebel P, van Drunen Littel-van den Hurk S, Hecker R, Babiuk L. TLR9−/− and TLR9+/+ mice display similar immune responses to a DNA vaccine. Immunology. 2004;113:114–120. doi: 10.1111/j.1365-2567.2004.01938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN. Cytoplasmic DNA innate immune pathways. Immunol Rev. 2011;243:99–108. doi: 10.1111/j.1600-065X.2011.01051.x. [DOI] [PubMed] [Google Scholar]
- Barbon CM, Baker L, Lajoie C, Ramstedt U, Hedley ML, Luby TM. In vivo electroporation enhances the potency of poly-lactide co-glycolide (PLG) plasmid DNA immunization. Vaccine. 2010;28:7852–7864. doi: 10.1016/j.vaccine.2010.09.078. [DOI] [PubMed] [Google Scholar]
- Bode C, Zhao G, Steinhagen F, Kinjo T, Klinman DM. CpG DNA as a vaccine adjuvant. Expert Rev Vaccines. 2011;10:499–511. doi: 10.1586/erv.10.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle JS, Brady JL, Lew AM. Enhanced responses to a DNA vaccine encoding a fusion antigen that is directed to sites of immune induction. Nature. 1998;392:408–411. doi: 10.1038/32932. [DOI] [PubMed] [Google Scholar]
- Brooks AR, Harkins RN, Wang P, Qian HS, Liu P, Rubanyi GM. Transcriptional silencing is associated with extensive methylation of the CMV promoter following adenoviral gene delivery to muscle. J Gene Med. 2004;6:395–404. doi: 10.1002/jgm.516. [DOI] [PubMed] [Google Scholar]
- Cao J, Chen Z, Ren Y, Luo Y, Cao M, Lu W, Zhao P, Qi Z. Oral immunization with attenuated Salmonella carrying a co-expression plasmid encoding the core and E2 proteins of hepatitis C virus capable of inducing cellular immune responses and neutralizing antibodies in mice. Vaccine. 2011;29:3714–3723. doi: 10.1016/j.vaccine.2011.02.083. [DOI] [PubMed] [Google Scholar]
- Carvalho JA, Azzoni AR, Prazeres DM, Monteiro GA. Comparative analysis of antigen-targeting sequences used in DNA vaccines. Mol Biotechnol. 2010;44:204–212. doi: 10.1007/s12033-009-9229-x. [DOI] [PubMed] [Google Scholar]
- Chapman BS, Thayer RM, Vincent KA, Haigwood NL. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic acids research. 1991;19:3979–3986. doi: 10.1093/nar/19.14.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JH, Yu YS, Liu HH, Chen XH, Xi M, Zang GQ, Tang ZH. Ubiquitin conjugation of hepatitis B virus core antigen DNA vaccine leads to enhanced cell-mediated immune response in BALB/c mice. Hepatitis monthly. 2011;11:620–628. doi: 10.5812/kowsar.1735143X.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Ziegelhoffer PR, Yang NS. In vivo promoter activity and transgene expression in mammalian somatic tissues evaluated by using particle bombardment. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:4455–4459. doi: 10.1073/pnas.90.10.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung YK, Cheng SC, Sin FW, Xie Y. Plasmid encoding papillomavirus Type 16 (HPV16) DNA constructed with codon optimization improved the immunogenicity against HPV infection. Vaccine. 2004;23:629–638. doi: 10.1016/j.vaccine.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Churchyard GJ, Morgan C, Adams E, Hural J, Graham BS, Moodie Z, Grove D, Gray G, Bekker LG, McElrath MJ, Tomaras GD, Goepfert P, Kalams S, Baden LR, Lally M, Dolin R, Blattner W, Kalichman A, Figueroa JP, Pape J, Schechter M, Defawe O, De Rosa SC, Montefiori DC, Nabel GJ, Corey L, Keefer MC. A phase IIA randomized clinical trial of a multiclade HIV-1 DNA prime followed by a multiclade rAd5 HIV-1 vaccine boost in healthy adults (HVTN204) PLoS One. 2011;6:e21225. doi: 10.1371/journal.pone.0021225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conese M, Auriche C, Ascenzioni F. Gene therapy progress and prospects: episomally maintained self-replicating systems. Gene Ther. 2004;11:1735–1741. doi: 10.1038/sj.gt.3302362. [DOI] [PubMed] [Google Scholar]
- Corbett AJ, Caminschi I, McKenzie BS, Brady JL, Wright MD, Mottram PL, Hogarth PM, Hodder AN, Zhan Y, Tarlinton DM, Shortman K, Lew AM. Antigen delivery via two molecules on the CD8- dendritic cell subset induces humoral immunity in the absence of conventional “danger”. European journal of immunology. 2005;35:2815–2825. doi: 10.1002/eji.200526100. [DOI] [PubMed] [Google Scholar]
- Cristillo AD, Ferrari MG, Hudacik L, Lewis B, Galmin L, Bowen B, Thompson D, Petrovsky N, Markham P, Pal R. Induction of mucosal and systemic antibody and T-cell responses following prime-boost immunization with novel adjuvanted human immunodeficiency virus-1-vaccine formulations. The Journal of general virology. 2011;92:128–140. doi: 10.1099/vir.0.023242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Barski A, Zhao K. Epigenomics of T cell activation, differentiation, and memory. Curr Opin Immunol. 2010;22:341–347. doi: 10.1016/j.coi.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daftarian P, Kaifer AE, Li W, Blomberg BB, Frasca D, Roth F, Chowdhury R, Berg EA, Fishman JB, Al Sayegh HA, Blackwelder P, Inverardi L, Perez VL, Lemmon V, Serafini P. Peptide-Conjugated PAMAM Dendrimer as a Universal DNA Vaccine Platform to Target Antigen-Presenting Cells. Cancer research. 2011;71:7452–7462. doi: 10.1158/0008-5472.CAN-11-1766. [DOI] [PubMed] [Google Scholar]
- Dave UP, Jenkins NA, Copeland NG. Gene therapy insertional mutagenesis insights. Science. 2004;303:333. doi: 10.1126/science.1091667. [DOI] [PubMed] [Google Scholar]
- de Arruda LB, Chikhlikar PR, August JT, Marques ET. DNA vaccine encoding human immunodeficiency virus-1 Gag, targeted to the major histocompatibility complex II compartment by lysosomal-associated membrane protein, elicits enhanced long-term memory response. Immunology. 2004;112:126–133. doi: 10.1111/j.1365-2567.2004.01823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa SC, Thomas EP, Bui J, Huang Y, deCamp A, Morgan C, Kalams SA, Tomaras GD, Akondy R, Ahmed R, Lau CY, Graham BS, Nabel GJ, McElrath MJ. HIV-DNA priming alters T cell responses to HIV-adenovirus vaccine even when responses to DNA are undetectable. J Immunol. 2011;187:3391–3401. doi: 10.4049/jimmunol.1101421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobano C, Sedegah M, Rogers WO, Kumar S, Zheng H, Hoffman SL, Doolan DL. Plasmodium: mammalian codon optimization of malaria plasmid DNA vaccines enhances antibody responses but not T cell responses nor protective immunity. Exp Parasitol. 2009;122:112–123. doi: 10.1016/j.exppara.2009.02.010. [DOI] [PubMed] [Google Scholar]
- Faham A, Herringson T, Parish C, Suhrbier A, Khromykh AA, Altin JG. pDNA-lipoplexes engrafted with flagellin-related peptide induce potent immunity and anti-tumour effects. Vaccine. 2011 doi: 10.1016/j.vaccine.2011.07.045. [DOI] [PubMed] [Google Scholar]
- Fernandez-Morera JL, Calvanese V, Rodriguez-Rodero S, Menendez-Torre E, Fraga MF. Epigenetic regulation of the immune system in health and disease. Tissue Antigens. 2010;76:431–439. doi: 10.1111/j.1399-0039.2010.01587.x. [DOI] [PubMed] [Google Scholar]
- Forns X, Payette PJ, Ma X, Satterfield W, Eder G, Mushahwar IK, Govindarajan S, Davis HL, Emerson SU, Purcell RH, Bukh J. Vaccination of chimpanzees with plasmid DNA encoding the hepatitis C virus (HCV) envelope E2 protein modified the infection after challenge with homologous monoclonal HCV. Hepatology (Baltimore, Md) 2000;32:618–625. doi: 10.1053/jhep.2000.9877. [DOI] [PubMed] [Google Scholar]
- Fu X, Tao L, Zhang X. A short polypeptide from the herpes simplex virus type 2 ICP10 gene can induce antigen aggregation and autophagosomal degradation for enhanced immune presentation. Human gene therapy. 2010;21:1687–1696. doi: 10.1089/hum.2010.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa T, Joshi BH, Puri RK. Histone modification enhances the effectiveness of IL-13 receptor targeted immunotoxin in murine models of human pancreatic cancer. Journal of translational medicine. 2011;9:37. doi: 10.1186/1479-5876-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiben-Lynn R, Frimpong-Boateng K, Letvin NL. Modulation of plasmid DNA vaccine antigen clearance by caspase 12 RNA interference potentiates vaccination. Clinical and vaccine immunology : CVI. 2011;18:533–538. doi: 10.1128/CVI.00390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Gowda NM, Wu X, Gowda DC. The nucleosome (histone-DNA complex) is the TLR9-specific immunostimulatory component of Plasmodium falciparum that activates DCs. PLoS One. 2011;6:e20398. doi: 10.1371/journal.pone.0020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenland JR, Geiben R, Ghosh S, Pastor WA, Letvin NL. Plasmid DNA vaccine-elicited cellular immune responses limit in vivo vaccine antigen expression through Fas-mediated apoptosis. J Immunol. 2007;178:5652–5658. doi: 10.4049/jimmunol.178.9.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribaudo G, Ravaglia S, Caliendo A, Cavallo R, Gariglio M, Martinotti MG, Landolfo S. Interferons inhibit onset of murine cytomegalovirus immediate-early gene transcription. Virology. 1993;197:303–311. doi: 10.1006/viro.1993.1591. [DOI] [PubMed] [Google Scholar]
- Harish N, Gupta R, Agarwal P, Scaria V, Pillai B. DyNAVacS: an integrative tool for optimized DNA vaccine design. Nucleic acids research. 2006;34:W264–266. doi: 10.1093/nar/gkl242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS Forms Functional Prion-like Aggregates to Activate and Propagate Antiviral Innate Immune Response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Mao CP, Peng S, Hung CF, Wu TC. RNA interference-mediated in vivo silencing of fas ligand as a strategy for the enhancement of DNA vaccine potency. Human gene therapy. 2008;19:763–773. doi: 10.1089/hum.2007.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, Akira S. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL, Jr, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med. 2009;15:267–276. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- Jaoko W, Karita E, Kayitenkore K, Omosa-Manyonyi G, Allen S, Than S, Adams EM, Graham BS, Koup RA, Bailer RT, Smith C, Dally L, Farah B, Anzala O, Muvunyi CM, Bizimana J, Tarragona-Fiol T, Bergin PJ, Hayes P, Ho M, Loughran K, Komaroff W, Stevens G, Thomson H, Boaz MJ, Cox JH, Schmidt C, Gilmour J, Nabel GJ, Fast P, Bwayo J. Safety and immunogenicity study of Multiclade HIV-1 adenoviral vector vaccine alone or as boost following a multiclade HIV-1 DNA vaccine in Africa. PLoS One. 2010;5:e12873. doi: 10.1371/journal.pone.0012873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jechlinger W, Azimpour Tabrizi C, Lubitz W, Mayrhofer P. Minicircle DNA immobilized in bacterial ghosts: in vivo production of safe non-viral DNA delivery vehicles. J Mol Microbiol Biotechnol. 2004;8:222–231. doi: 10.1159/000086703. [DOI] [PubMed] [Google Scholar]
- Ji H, Wang TL, Chen CH, Pai SI, Hung CF, Lin KY, Kurman RJ, Pardoll DM, Wu TC. Targeting human papillomavirus type 16 E7 to the endosomal/lysosomal compartment enhances the antitumor immunity of DNA vaccines against murine human papillomavirus type 16 E7-expressing tumors. Human gene therapy. 1999;10:2727–2740. doi: 10.1089/10430349950016474. [DOI] [PubMed] [Google Scholar]
- Jiang W. Blockade of B7-H1 enhances dendritic cell-mediated T cell response and antiviral immunity in HBV transgenic mice. Vaccine. 2012;30:758–766. doi: 10.1016/j.vaccine.2011.11.076. [DOI] [PubMed] [Google Scholar]
- Karpf AR. A potential role for epigenetic modulatory drugs in the enhancement of cancer/germ-line antigen vaccine efficacy. Epigenetics. 2006;1:116–120. doi: 10.4161/epi.1.3.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K, Fujihashi K, Oma K, Fukuyama Y, Hollingshead SK, Sekine S, Kawabata S, Ito HO, Briles DE, Oishi K. The nasal dendritic cell-targeting Flt3 ligand as a safe adjuvant elicits effective protection against fatal pneumococcal pneumonia. Infection and immunity. 2011;79:2819–2828. doi: 10.1128/IAI.01360-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur M, Rai A, Bhatnagar R. Rabies DNA vaccine: no impact of MHC class I and class II targeting sequences on immune response and protection against lethal challenge. Vaccine. 2009;27:2128–2137. doi: 10.1016/j.vaccine.2009.01.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay MA, He CY, Chen ZY. A robust system for production of minicircle DNA vectors. Nat Biotechnol. 2010;28:1287–1289. doi: 10.1038/nbt.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy RB, Poland GA. The Top Five “Game Changers” in Vaccinology: Toward Rational and Directed Vaccine Development. OMICS. 2011 doi: 10.1089/omi.2011.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr IM, Stark GR. The antiviral effects of the interferons and their inhibition. Journal of interferon research. 1992;12:237–240. doi: 10.1089/jir.1992.12.237. [DOI] [PubMed] [Google Scholar]
- Khawaja G, Buronfosse T, Jamard C, Guerret S, Zoulim F, Luxembourg A, Hannaman D, Evans C, Hartmann D, Cova L. Enhanced magnitude and breadth of neutralizing humoral response to a DNA vaccine targeting the DHBV envelope protein delivered by in vivo electroporation. Virology. 2012;425:61–69. doi: 10.1016/j.virol.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Khosroshahi KH, Ghaffarifar F, Sharifi Z, D’Souza S, Dalimi A, Hassan ZM, Khoshzaban F. Comparing the effect of IL-12 genetic adjuvant and alum non-genetic adjuvant on the efficiency of the cocktail DNA vaccine containing plasmids encoding SAG-1 and ROP-2 of Toxoplasma gondii. Parasitology research. 2012;111:403–411. doi: 10.1007/s00436-012-2852-7. [DOI] [PubMed] [Google Scholar]
- Kim D, Hoory T, Monie A, Ting JP, Hung CF, Wu TC. Enhancement of DNA vaccine potency through coadministration of CIITA DNA with DNA vaccines via gene gun. J Immunol. 2008;180:7019–7027. doi: 10.4049/jimmunol.180.10.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kang TH, Noh KH, Bae HC, Ahn YH, Lee YH, Choi EY, Chun KH, Lee SJ, Kim TW. Blocking the immunosuppressive axis with small interfering RNA targeting interleukin (IL)-10 receptor enhances dendritic cell-based vaccine potency. Clinical and experimental immunology. 2011;165:180–189. doi: 10.1111/j.1365-2249.2011.04410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Lee JH, He L, Boyd DA, Hardwick JM, Hung CF, Wu TC. Modification of professional antigen-presenting cells with small interfering RNA in vivo to enhance cancer vaccine potency. Cancer research. 2005;65:309–316. [PubMed] [Google Scholar]
- Klinman DM, Yamshchikov G, Ishigatsubo Y. Contribution of CpG motifs to the immunogenicity of DNA vaccines. J Immunol. 1997;158:3635–3639. [PubMed] [Google Scholar]
- Koblin BA, Casapia M, Morgan C, Qin L, Wang ZM, Defawe OD, Baden L, Goepfert P, Tomaras GD, Montefiori DC, McElrath MJ, Saavedra L, Lau CY, Graham BS. Safety and immunogenicity of an HIV adenoviral vector boost after DNA plasmid vaccine prime by route of administration: a randomized clinical trial. PLoS One. 2011;6:e24517. doi: 10.1371/journal.pone.0024517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolka JA, Vreede AP, Roessler BJ. Lipopolysaccharide recognition protein, MD-2, facilitates cellular uptake of E. coli-derived plasmid DNA in synovium. J Gene Med. 2005;7:956–964. doi: 10.1002/jgm.743. [DOI] [PubMed] [Google Scholar]
- Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nat Rev Genet. 2008;9:776–788. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahoud MH, Ahmet F, Kitsoulis S, Wan SS, Vremec D, Lee CN, Phipson B, Shi W, Smyth GK, Lew AM, Kato Y, Mueller SN, Davey GM, Heath WR, Shortman K, Caminschi I. Targeting antigen to mouse dendritic cells via Clec9A induces potent CD4 T cell responses biased toward a follicular helper phenotype. J Immunol. 2011;187:842–850. doi: 10.4049/jimmunol.1101176. [DOI] [PubMed] [Google Scholar]
- Lares MR, Rossi JJ, Ouellet DL. RNAi and small interfering RNAs in human disease therapeutic applications. Trends in biotechnology. 2010;28:570–579. doi: 10.1016/j.tibtech.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledgerwood JE, Wei CJ, Hu Z, Gordon IJ, Enama ME, Hendel CS, McTamney PM, Pearce MB, Yassine HM, Boyington JC, Bailer R, Tumpey TM, Koup RA, Mascola JR, Nabel GJ, Graham BS. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. The Lancet infectious diseases. 2011;11:916–924. doi: 10.1016/S1473-3099(11)70240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CT, Tsai YC, He L, Calizo R, Chou HH, Chang TC, Soong YK, Hung CF, Lai CH. A DNA vaccine encoding a codon-optimized human papillomavirus type 16 E6 gene enhances CTL response and anti-tumor activity. Journal of biomedical science. 2006;13:481–488. doi: 10.1007/s11373-006-9086-6. [DOI] [PubMed] [Google Scholar]
- Liu F, Liang M, Cao S, Liu Q, Zhang Q, Li C, Zhang S, Wang S, Li D. Fusion with extracellular domain of cytotoxic T-lymphocyte-associated-antigen 4 leads to enhancement of immunogenicity of Hantaan virus DNA vaccines in C57BL/6 mice. Virology journal. 2011;8:448. doi: 10.1186/1743-422X-8-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MA. DNA vaccines: an historical perspective and view to the future. Immunol Rev. 2011;239:62–84. doi: 10.1111/j.1600-065X.2010.00980.x. [DOI] [PubMed] [Google Scholar]
- Lobigs M, Pavy M, Hall RA, Lobigs P, Cooper P, Komiya T, Toriniwa H, Petrovsky N. An inactivated Vero cell-grown Japanese encephalitis vaccine formulated with Advax, a novel inulin-based adjuvant, induces protective neutralizing antibody against homologous and heterologous flaviviruses. The Journal of general virology. 2010;91:1407–1417. doi: 10.1099/vir.0.019190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Wang S, Grimes-Serrano JM. Current progress of DNA vaccine studies in humans. Expert Rev Vaccines. 2008;7:175–191. doi: 10.1586/14760584.7.2.175. [DOI] [PubMed] [Google Scholar]
- Lu Y, Raviprakash K, Leao IC, Chikhlikar PR, Ewing D, Anwar A, Chougnet C, Murphy G, Hayes CG, August TJ, Marques ET., Jr Dengue 2 PreM-E/LAMP chimera targeted to the MHC class II compartment elicits long-lasting neutralizing antibodies. Vaccine. 2003;21:2178–2189. doi: 10.1016/s0264-410x(03)00009-4. [DOI] [PubMed] [Google Scholar]
- Luke J, Carnes AE, Hodgson CP, Williams JA. Improved antibiotic-free DNA vaccine vectors utilizing a novel RNA based plasmid selection system. Vaccine. 2009;27:6454–6459. doi: 10.1016/j.vaccine.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke JM, Simon GG, Soderholm J, Errett JS, August JT, Gale M, Jr, Hodgson CP, Williams JA. Coexpressed RIG-I agonist enhances humoral immune response to influenza virus DNA vaccine. J Virol. 2011a;85:1370–1383. doi: 10.1128/JVI.01250-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke JM, Vincent JM, Du SX, Gerdemann U, Leen AM, Whalen RG, Hodgson CP, Williams JA. Improved antibiotic-free plasmid vector design by incorporation of transient expression enhancers. Gene Ther. 2011b;18:334–343. doi: 10.1038/gt.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxembourg A, Hannaman D, Ellefsen B, Nakamura G, Bernard R. Enhancement of immune responses to an HBV DNA vaccine by electroporation. Vaccine. 2006;24:4490–4493. doi: 10.1016/j.vaccine.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Ma X, Forns X, Gutierrez R, Mushahwar IK, Wu T, Payette PJ, Bukh J, Purcell RH, Davis HL. DNA-based vaccination against hepatitis C virus (HCV): effect of expressing different forms of HCV E2 protein and use of CpG-optimized vectors in mice. Vaccine. 2002;20:3263–3271. doi: 10.1016/s0264-410x(02)00304-3. [DOI] [PubMed] [Google Scholar]
- Manthorpe M, Cornefert-Jensen F, Hartikka J, Felgner J, Rundell A, Margalith M, Dwarki V. Gene therapy by intramuscular injection of plasmid DNA: studies on firefly luciferase gene expression in mice. Human gene therapy. 1993;4:419–431. doi: 10.1089/hum.1993.4.4-419. [DOI] [PubMed] [Google Scholar]
- Marques ET, Jr, Chikhlikar P, de Arruda LB, Leao IC, Lu Y, Wong J, Chen JS, Byrne B, August JT. HIV-1 p55Gag encoded in the lysosome-associated membrane protein-1 as a DNA plasmid vaccine chimera is highly expressed, traffics to the major histocompatibility class II compartment, and elicits enhanced immune responses. The Journal of biological chemistry. 2003;278:37926–37936. doi: 10.1074/jbc.M303336200. [DOI] [PubMed] [Google Scholar]
- Mazumder S, Maji M, Das A, Ali N. Potency, efficacy and durability of DNA/DNA, DNA/protein and protein/protein based vaccination using gp63 against Leishmania donovani in BALB/c mice. PLoS One. 2011;6:e14644. doi: 10.1371/journal.pone.0014644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megati S, Garcia-Hand D, Cappello S, Roopchand V, Masood A, Xu R, Luckay A, Chong SY, Rosati M, Sackitey S, Weiner DB, Felber BK, Pavlakis GN, Israel ZR, Smith LR, Eldridge JH, Sidhu MK, Egan MA. Modifying the HIV-1 env gp160 gene to improve pDNA vaccine-elicited cell-mediated immune responses. Vaccine. 2008;26:5083–5094. doi: 10.1016/j.vaccine.2008.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midha S, Bhatnagar R. Anthrax protective antigen administered by DNA vaccination to distinct subcellular locations potentiates humoral and cellular immune responses. European journal of immunology. 2009;39:159–177. doi: 10.1002/eji.200838058. [DOI] [PubMed] [Google Scholar]
- Munz C. Enhancing immunity through autophagy. Annual review of immunology. 2009;27:423–449. doi: 10.1146/annurev.immunol.021908.132537. [DOI] [PubMed] [Google Scholar]
- Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li GM, McCausland M, Kanchan V, Kokko KE, Li S, Elbein R, Mehta AK, Aderem A, Subbarao K, Ahmed R, Pulendran B. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsinh KH, Jia F, Robbins RC, Kay MA, Longaker MT, Wu JC. Generation of adult human induced pluripotent stem cells using nonviral minicircle DNA vectors. Nat Protoc. 2011;6:78–88. doi: 10.1038/nprot.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngumbela KC, Ryan KP, Sivamurthy R, Brockman MA, Gandhi RT, Bhardwaj N, Kavanagh DG. Quantitative effect of suboptimal codon usage on translational efficiency of mRNA encoding HIV-1 gag in intact T cells. PLoS One. 2008;3:e2356. doi: 10.1371/journal.pone.0002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niazi KR, Ochoa MT, Sieling PA, Rooke NE, Peter AK, Mollahan P, Dickey M, Rabizadeh S, Rea TH, Modlin RL. Activation of human CD4+ T cells by targeting MHC class II epitopes to endosomal compartments using human CD1 tail sequences. Immunology. 2007;122:522–531. doi: 10.1111/j.1365-2567.2007.02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njongmeta LM, Bray J, Davies CJ, Davis WC, Howard CJ, Hope JC, Palmer GH, Brown WC, Mwangi W. CD205 antigen targeting combined with dendritic cell recruitment factors and antigen-linked CD40L activation primes and expands significant antigen-specific antibody and CD4(+) T cell responses following DNA vaccination of outbred animals. Vaccine. 2012 doi: 10.1016/j.vaccine.2011.12.110. [DOI] [PubMed] [Google Scholar]
- Osborn MJ, McElmurry RT, Lees CJ, DeFeo AP, Chen ZY, Kay MA, Naldini L, Freeman G, Tolar J, Blazar BR. Minicircle DNA-based gene therapy coupled with immune modulation permits long-term expression of alpha-L-iduronidase in mice with mucopolysaccharidosis type I. Mol Ther. 2011;19:450–460. doi: 10.1038/mt.2010.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott G, Singh M, Kazzaz J, Briones M, Soenawan E, Ugozzoli M, O’Hagan DT. A cationic sub-micron emulsion (MF59/DOTAP) is an effective delivery system for DNA vaccines. Journal of controlled release : official journal of the Controlled Release Society. 2002;79:1–5. doi: 10.1016/s0168-3659(01)00545-4. [DOI] [PubMed] [Google Scholar]
- Palumbo RN, Zhong X, Wang C. Polymer-mediated DNA vaccine delivery via bystander cells requires a proper balance between transfection efficiency and cytotoxicity. Journal of controlled release : official journal of the Controlled Release Society. 2011 doi: 10.1016/j.jconrel.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Shen H. Making sense of inflammation, epigenetics, and memory CD8+ T-cell differentiation in the context of infection. Immunol Rev. 2006;211:197–202. doi: 10.1111/j.0105-2896.2006.00399.x. [DOI] [PubMed] [Google Scholar]
- Petrovsky N. Freeing vaccine adjuvants from dangerous immunological dogma. Expert Rev Vaccines. 2008;7:7–10. doi: 10.1586/14760584.7.1.7. [DOI] [PubMed] [Google Scholar]
- Petrovsky N. The vaccine renaissance. Hum Vaccin. 2011;7:149–152. doi: 10.4161/hv.7.2.14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovsky N, Cooper PD. Carbohydrate-based immune adjuvants. Expert Rev Vaccines. 2011;10:523–537. doi: 10.1586/erv.11.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland GA, Ovsyannikova IG, Kennedy RB, Haralambieva IH, Jacobson RM. Vaccinomics and a New Paradigm for the Development of Preventive Vaccines Against Viral Infections. OMICS. 2011 doi: 10.1089/omi.2011.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porgador A, Irvine KR, Iwasaki A, Barber BH, Restifo NP, Germain RN. Predominant role for directly transfected dendritic cells in antigen presentation to CD8+ T cells after gene gun immunization. The Journal of experimental medicine. 1998;188:1075–1082. doi: 10.1084/jem.188.6.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajcani J, Mosko T, Rezuchova I. Current developments in viral DNA vaccines: shall they solve the unsolved? Rev Med Virol. 2005;15:303–325. doi: 10.1002/rmv.467. [DOI] [PubMed] [Google Scholar]
- Rappuoli R, Aderem A. A 2020 vision for vaccines against HIV, tuberculosis and malaria. Nature. 2011;473:463–469. doi: 10.1038/nature10124. [DOI] [PubMed] [Google Scholar]
- Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl P, Fissolo N, Reimann J, Schirmbeck R. A stress protein-facilitated antigen expression system for plasmid DNA vaccines. Methods in molecular medicine. 2006;127:41–53. doi: 10.1385/1-59745-168-1:41. [DOI] [PubMed] [Google Scholar]
- Rigato PO, Maciel M, Jr, Goldoni AL, Piubelli O, de Brito CA, Fusaro AE, Eurico de Alencar LX, August T, Azevedo Marques ET, Jr, da Silva Duarte AJ, Sato MN. Immunization of neonatal mice with LAMP/p55 HIV gag DNA elicits robust immune responses that last to adulthood. Virology. 2010;406:37–47. doi: 10.1016/j.virol.2010.06.050. [DOI] [PubMed] [Google Scholar]
- Riu E, Chen ZY, Xu H, He CY, Kay MA. Histone modifications are associated with the persistence or silencing of vector-mediated transgene expression in vivo. Mol Ther. 2007;15:1348–1355. doi: 10.1038/sj.mt.6300177. [DOI] [PubMed] [Google Scholar]
- Rottembourg D, Filippi CM, Bresson D, Ehrhardt K, Estes EA, Oldham JE, von Herrath MG. Essential role for TLR9 in prime but not prime-boost plasmid DNA vaccination to activate dendritic cells and protect from lethal viral infection. J Immunol. 2010;184:7100–7107. doi: 10.4049/jimmunol.0803935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saade F, Petrovsky N. Technologies for enhanced efficacy of DNA vaccines. Expert Rev Vaccines. 2012;11:189–209. doi: 10.1586/erv.11.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu KS, Pandey S, Maiti S, Pillai B. GASCO: genetic algorithm simulation for codon optimization. In silico biology. 2008;8:187–192. [PubMed] [Google Scholar]
- Sardesai NY, Weiner DB. Electroporation delivery of DNA vaccines: prospects for success. Curr Opin Immunol. 2011;23:421–429. doi: 10.1016/j.coi.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmbeck R, Kwissa M, Fissolo N, Elkholy S, Riedl P, Reimann J. Priming polyvalent immunity by DNA vaccines expressing chimeric antigens with a stress protein-capturing, viral J-domain. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2002;16:1108–1110. doi: 10.1096/fj.01-0993fje. [DOI] [PubMed] [Google Scholar]
- Schneider J, Gilbert SC, Blanchard TJ, Hanke T, Robson KJ, Hannan CM, Becker M, Sinden R, Smith GL, Hill AV. Enhanced immunogenicity for CD8+ T cell induction and complete protective efficacy of malaria DNA vaccination by boosting with modified vaccinia virus Ankara. Nat Med. 1998;4:397–402. doi: 10.1038/nm0498-397. [DOI] [PubMed] [Google Scholar]
- Sette A, Rappuoli R. Reverse vaccinology: developing vaccines in the era of genomics. Immunity. 2010;33:530–541. doi: 10.1016/j.immuni.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- Siegismund CS, Hohn O, Kurth R, Norley S. Enhanced T- and B-cell responses to simian immunodeficiency virus (SIV)agm, SIVmac and human immunodeficiency virus type 1 Gag DNA immunization and identification of novel T-cell epitopes in mice via codon optimization. The Journal of general virology. 2009;90:2513–2518. doi: 10.1099/vir.0.013730-0. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Stoecklinger A, Grieshuber I, Scheiblhofer S, Weiss R, Ritter U, Kissenpfennig A, Malissen B, Romani N, Koch F, Ferreira F, Thalhamer J, Hammerl P. Epidermal langerhans cells are dispensable for humoral and cell-mediated immunity elicited by gene gun immunization. J Immunol. 2007;179:886–893. doi: 10.4049/jimmunol.179.2.886. [DOI] [PubMed] [Google Scholar]
- Tang DC, DeVit M, Johnston SA. Genetic immunization is a simple method for eliciting an immune response. Nature. 1992;356:152–154. doi: 10.1038/356152a0. [DOI] [PubMed] [Google Scholar]
- Tenbusch M, Grunwald T, Niezold T, Storcksdieck Genannt Bonsmann M, Hannaman D, Norley S, Uberla K. Codon-optimization of the hemagglutinin gene from the novel swine origin H1N1 influenza virus has differential effects on CD4(+) T-cell responses and immune effector mechanisms following DNA electroporation in mice. Vaccine. 2010;28:3273–3277. doi: 10.1016/j.vaccine.2010.02.090. [DOI] [PubMed] [Google Scholar]
- Ternette N, Tippler B, Uberla K, Grunwald T. Immunogenicity and efficacy of codon optimized DNA vaccines encoding the F-protein of respiratory syncytial virus. Vaccine. 2007;25:7271–7279. doi: 10.1016/j.vaccine.2007.07.025. [DOI] [PubMed] [Google Scholar]
- Tovey MG, Lallemand C. Adjuvant activity of cytokines. Methods in molecular biology (Clifton, NJ) 2010;626:287–309. doi: 10.1007/978-1-60761-585-9_19. [DOI] [PubMed] [Google Scholar]
- Trautmann L, Sekaly RP. Solving vaccine mysteries: a systems biology perspective. Nat Immunol. 2011;12:729–731. doi: 10.1038/ni.2078. [DOI] [PubMed] [Google Scholar]
- Trollet C, Pereira Y, Burgain A, Litzler E, Mezrahi M, Seguin J, Manich M, Popoff MR, Scherman D, Bigey P. Generation of high-titer neutralizing antibodies against botulinum toxins A, B, and E by DNA electrotransfer. Infection and immunity. 2009;77:2221–2229. doi: 10.1128/IAI.01269-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombone AP, Silva CL, Lima KM, Oliver C, Jamur MC, Prescott AR, Coelho-Castelo AA. Endocytosis of DNA-Hsp65 alters the pH of the late endosome/lysosome and interferes with antigen presentation. PLoS One. 2007;2:e923. doi: 10.1371/journal.pone.0000923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudor D, Dubuquoy C, Gaboriau V, Lefevre F, Charley B, Riffault S. TLR9 pathway is involved in adjuvant effects of plasmid DNA-based vaccines. Vaccine. 2005;23:1258–1264. doi: 10.1016/j.vaccine.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Uchijima M, Yoshida A, Nagata T, Koide Y. Optimization of codon usage of plasmid DNA vaccine is required for the effective MHC class I-restricted T cell responses against an intracellular bacterium. J Immunol. 1998;161:5594–5599. [PubMed] [Google Scholar]
- Ulmer JB, Donnelly JJ, Parker SE, Rhodes GH, Felgner PL, Dwarki VJ, Gromkowski SH, Deck RR, DeWitt CM, Friedman A, et al. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science. 1993;259:1745–1749. doi: 10.1126/science.8456302. [DOI] [PubMed] [Google Scholar]
- Ulmer JB, Sadoff JC, Liu MA. DNA vaccines. Curr Opin Immunol. 1996;8:531–536. doi: 10.1016/s0952-7915(96)80042-2. [DOI] [PubMed] [Google Scholar]
- van Drunen Littel-van den Hurk S, Hannaman D. Electroporation for DNA immunization: clinical application. Expert Rev Vaccines. 2010;9:503–517. doi: 10.1586/erv.10.42. [DOI] [PubMed] [Google Scholar]
- Vanniasinkam T, Reddy ST, Ertl HC. DNA immunization using a non-viral promoter. Virology. 2006;344:412–420. doi: 10.1016/j.virol.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Varaldo PB, Miyaji EN, Vilar MM, Campos AS, Dias WO, Armoa GR, Tendler M, Leite LC, McIntosh D. Mycobacterial codon optimization of the gene encoding the Sm14 antigen of Schistosoma mansoni in recombinant Mycobacterium bovis Bacille Calmette-Guerin enhances protein expression but not protection against cercarial challenge in mice. FEMS immunology and medical microbiology. 2006;48:132–139. doi: 10.1111/j.1574-695X.2006.00133.x. [DOI] [PubMed] [Google Scholar]
- Vidalin O, Tanaka E, Spengler U, Trepo C, Inchauspe G. Targeting of hepatitis C virus core protein for MHC I or MHC II presentation does not enhance induction of immune responses to DNA vaccination. DNA and cell biology. 1999;18:611–621. doi: 10.1089/104454999315024. [DOI] [PubMed] [Google Scholar]
- Wang G, Pan L, Zhang Y. Approaches to improved targeting of DNA vaccines. Hum Vaccin. 2011a;7:1271–1281. doi: 10.4161/hv.7.12.17983. [DOI] [PubMed] [Google Scholar]
- Wang Q, Lei C, Wan H, Liu Q. Improved Cellular Immune Response Elicited by a Ubiquitin-Fused DNA Vaccine Against Mycobacterium tuberculosis. DNA and cell biology. 2011b doi: 10.1089/dna.2011.1309. [DOI] [PubMed] [Google Scholar]
- Wang S, Farfan-Arribas DJ, Shen S, Chou TH, Hirsch A, He F, Lu S. Relative contributions of codon usage, promoter efficiency and leader sequence to the antigen expression and immunogenicity of HIV-1 Env DNA vaccine. Vaccine. 2006;24:4531–4540. doi: 10.1016/j.vaccine.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Wang ST, Chang CC, Yen MC, Tu CF, Chu CL, Peng YT, Chen DY, Lan JL, Lin CC. RNA interference-mediated silencing of Foxo3 in antigen-presenting cells as a strategy for the enhancement of DNA vaccine potency. Gene Ther. 2011c;18:372–383. doi: 10.1038/gt.2010.146. [DOI] [PubMed] [Google Scholar]
- Wang Y, Guo Y, Wang X, Huang J, Shang J, Sun S. Serum amyloid P component facilitates DNA clearance and inhibits plasmid transfection: implications for human DNA vaccine. Gene Ther. 2011d doi: 10.1038/gt.2011.67. [DOI] [PubMed] [Google Scholar]
- Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- Xiang SD, Selomulya C, Ho J, Apostolopoulos V, Plebanski M. Delivery of DNA vaccines: an overview on the use of biodegradable polymeric and magnetic nanoparticles. Wiley interdisciplinary reviews. Nanomedicine and nanobiotechnology. 2010;2:205–218. doi: 10.1002/wnan.88. [DOI] [PubMed] [Google Scholar]
- Xiang Z, Ertl HC. Manipulation of the immune response to a plasmid-encoded viral antigen by coinoculation with plasmids expressing cytokines. Immunity. 1995;2:129–135. doi: 10.1016/s1074-7613(95)80001-8. [DOI] [PubMed] [Google Scholar]
- Xiao D, Yin C, Zhang Q, Li JH, Gong PT, Li SH, Zhang GC, Gao YJ, Zhang XC. Selection and identification of a new adhesion protein of Cryptosporidium parvum from a cDNA library by ribosome display. Exp Parasitol. 2011 doi: 10.1016/j.exppara.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Xu J, Zhang X, Monestier M, Esmon NL, Esmon CT. Extracellular histones are mediators of death through TLR2 and TLR4 in mouse fatal liver injury. J Immunol. 2011;187:2626–2631. doi: 10.4049/jimmunol.1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HL, Zhu YZ, Qin JH, He P, Jiang XC, Zhao GP, Guo XK. In silico and microarray-based genomic approaches to identifying potential vaccine candidates against Leptospira interrogans. BMC genomics. 2006;7:293. doi: 10.1186/1471-2164-7-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Sun K, Srinivasan KN, Salmon J, Marques ET, Xu J, August JT. Immune responses to T-cell epitopes of SARS CoV-N protein are enhanced by N immunization with a chimera of lysosome-associated membrane protein. Gene Ther. 2009;16:1353–1362. doi: 10.1038/gt.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood B, Davis CW, Ahmed R. Making memories that last a lifetime: heritable functions of self-renewing memory CD8 T cells. Int Immunol. 2010;22:797–803. doi: 10.1093/intimm/dxq437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Wang F, Zhang Y, Yang F, Wang Y, Sun S. Down-regulation of Prdx6 contributes to DNA vaccine induced vitiligo in mice. Mol Biosyst. 2011;7:809–816. doi: 10.1039/c0mb00181c. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Lu F, Dai Y, Wang X, Tang J, Zhao S, Zhang C, Zhang H, Lu S, Wang S. Synergistic enhancement of immunogenicity and protection in mice against Schistosoma japonicum with codon optimization and electroporation delivery of SjTPI DNA vaccines. Vaccine. 2010;28:5347–5355. doi: 10.1016/j.vaccine.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Zuo Y, Wu J, Xu Z, Yang S, Yan H, Tan L, Meng X, Ying X, Liu R, Kang T, Huang W. Minicircle-oriP-IFNgamma: A Novel Targeted Gene Therapeutic System for EBV Positive Human Nasopharyngeal Carcinoma. PLoS One. 2011;6:e19407. doi: 10.1371/journal.pone.0019407. [DOI] [PMC free article] [PubMed] [Google Scholar]