Abstract
In this study, we investigated lentiviral vector development and transduction efficiencies in well-differentiated primary cultures of pig airway epithelia (PAE) and wild-type pigs in vivo. We noted gene transfer efficiencies similar to that observed for human airway epithelia (HAE). Interestingly, feline immunodeficiency virus (FIV)-based vectors transduced immortalized pig cells as well as pig primary cells more efficiently than HIV-1–based vectors. PAE express TRIM5α, a well-characterized species-specific lentiviral restriction factor. We contrasted the restrictive properties of porcine TRIM5α against FIV- and HIV-based vectors using gain and loss of function approaches. We observed no effect on HIV-1 or FIV conferred transgene expression in response to porcine TRIM5α overexpression or knockdown. To evaluate the ability of GP64-FIV to transduce porcine airways in vivo, we delivered vector expressing mCherry to the tracheal lobe of the lung and the ethmoid sinus of 4-week-old pigs. One week later, epithelial cells expressing mCherry were readily detected. Our findings indicate that pseudotyped FIV vectors confer similar tropisms in porcine epithelia as observed in human HAE and provide further support for the selection of GP64 as an appropriate envelope pseudotype for future preclinical gene therapy studies in the porcine model of cystic fibrosis (CF).
Keywords: airway epithelia, cystic fibrosis, gene therapy, GP64, large animal model, lung, pig, sinus
Introduction
Mice with null CFTR mutations1,2,3,4 or specific disease-associated CFTR mutations5,6,7 have provided many insights into cystic fibrosis (CF) disease; however, they do not recapitulate several aspects of the lung disease pathogenesis. The recent development of alternative CF animal models8,9,10 provides a new resource for preclinical therapeutic studies. Importantly, the CF pig exhibits many similarities to CF in humans including multiorgan system involvement (sinus, lung, pancreas, intestine, liver) and the presence of an airway mucosal host defense defect at birth.9,11
The cell types comprising the conducting airway epithelium in pigs and humans are similar, and notably lack the ~50% bronchiolar exocrine cells typical of mice. The pig bronchial epithelium is pseudostratified and contains ciliated, basal, and goblet cells, and abundant submucosal glands (reviewed in ref. 12). Importantly, the distribution of submucosal glands in the conducting airways and the CFTR-dependent and -independent secretion of liquid and macromolecules is similar to human submucosal glands.13,14,15,16 Thus, pig airway cell composition is much more akin to human airways than are those of mice.
In addition to anatomy and physiology, the cell biology of humans is likely to be more similar to pigs than mice (reviewed in ref. 12). Indeed, some cellular restriction factors of lentiviral vectors may not be present in mice, but can limit gene transfer to pigs or humans. For example, murine cells do not express a restricting TRIM5α and are permissive to both HIV and feline immunodeficiency virus (FIV).17 Similar to primate variants of TRIM5α, ungulate variants can restrict retroviral transduction.18,19 The predicted protein sequence of porcine TRIM5α is phylogenetically close to the broadly restrictive bovine TRIM5α protein,18 suggesting that porcine TRIM5α may also be broadly restrictive. For gene therapy to be successful, there is a need to match the optimal choice of viral vector to the appropriate preclinical model.
Optimizing vector design for gene transfer to the airways is an important current focus for CF therapeutic development (reviewed in refs. 20,21). Here, we report engineering a lentiviral vector system for improved efficacy in the wild-type porcine airways. These results have implications for pulmonary therapeutic applications and will help shape the design for viral vector correction studies in the recently developed pig CF model.9
Results
Comparison of primate and non-primate lentiviral vectors in porcine cells
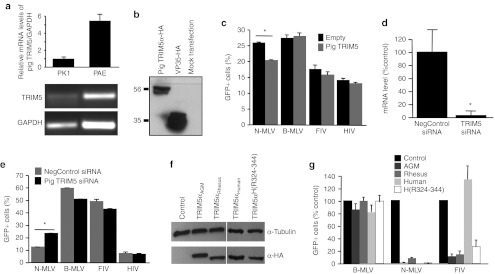
Species-specific host cell restriction factors are known barriers for viral vector transduction. Porcine cells are not widely used for lentiviral gene transfer studies and little effort has been put forth optimizing or comparing viral vector transduction efficacy in this animal model. We generated matched vesicular stomatitis virus G protein (VSVG)-pseudotyped FIV and HIV-based vectors expressing enhanced green fluorescent protein (GFP) driven by the cytomegalovirus promoter (Figure 1a). In addition, these self-inactivating vectors contain the wPRE and cPPT elements.22 Murine leukemia viruses (MLVs) are subtyped as N-tropic (N-MLV) or B-tropic (B-MLV) according to their infectivity in certain strains of mice. Importantly, N-MLV infection is consistently restricted by Trim5α from mammalian species whereas B-MLV is resistant to the Trim5α restriction.18,23 As such, N-MLV and B-MLV vectors expressing GFP serve as controls for Trim5α restriction. We transduced human cervical (HeLa), bovine kidney (MDBK), and porcine kidney epithelia cells (PK1) with FIV, HIV, N-MLV, and B-MLV at multiplicity of infection (MOIs) ranging from 0.5 to 5 (Figure 1b). FIV- and HIV-based vectors transduced HeLa cells with similar efficacy and, consistent with previous observations,18 both had restricted expression in MDBK cells. Interestingly, FIV transduced PK1 cells approximately tenfold better than HIV across all MOIs tested. No restriction of FIV was apparent in PK1 cells compared with B-MLV, which typically displayed higher levels of transduction in most cell types tested, including airway cell lines (data not shown) and HeLa cells (Figure 1b). We next contrasted the lentiviral transduction patterns in well-differentiated primary cultures of porcine airway epithelia (PAE). PAEs derived from wild-type pig trachea were transduced with N-MLV, B-MLV, FIV, or HIV-1 at MOIs of 5 or 20 (Figure 1b). Consistent with PK1 cells, we observed greater transduction efficacy of FIV as compared with HIV. We next compared the transduction efficiency of FIV and HIV in four different immortalized cell lines derived from human lung epithelia. While higher level of transduction was seen with FIV compared to HIV in H441 cells, similar transduction efficiencies with FIV or HIV were observed in the rest of the cell lines (Figure 1c). Transduction pattern differences of FIV and HIV-1 in porcine cells, but not in human-derived cells, suggests the existence of species-specific restriction factors against HIV-1 in porcine cells.
Figure 1.
Transduction patterns of HIV- and FIV-based vectors. (a) Schematics of FIV and HIV transgene vectors expressing GFP are shown. ΨPackaging signal. (b) VSVG-pseudotyped HIV, FIV, N-MLV, and B-MLV expressing GFP were applied to human (HeLa), cow (MDBK), or pig (PK1) cell lines at an MOI = 0.5, 1, and 5 to examine transduction efficiency. Pig primary airway epithelia (PAE) were transduced with MOI = 5 and 20. GFP-positive cells were counted 4 days post-transduction by FACS. n = 4. (c) HIV-1 or FIV lentiviral vectors were applied at the same MOI to four airway-derived cell lines and the HeLa cervical adenocarcinoma cell line. Transduction efficiencies were measured by FACS analysis of GFP-positive cells and data normalized to HIV-1 efficiency. Data are presented as the mean ± SE, n = 6. B-MLV, B-tropic murine leukemia virus; CMV, cytomegalovirus; eGFP, enhanced green fluorescent protein; FACS, fluorescence-activated cell sorting; FIV, feline immunodeficiency virus; MOI, multiplicity of infection; N-MLV, N-tropic murine leukemia virus; VSVG, vesicular stomatitis virus G protein.
A porcine TRIM5 ortholog was identified from sequence homology and predicted protein domain similarity to known TRIM proteins.18 A previous study reported that HIV-1 infection is restricted in cell lines originated from pigs,24 but there has been little focus on porcine TRIM5. Bovine TRIM5 was previously demonstrated to have broad restrictive properties against many lentiviruses including HIV-1 and FIV.18,19,24 We hypothesized that porcine TRIM5 could restrict both FIV and HIV vectors because of its close phylogenetic similarity to bovine TRIM5. Using porcine-specific primers, porcine TRIM5α mRNA was successfully amplified from PK1 and PAE cells (Figure 2a). A hemaglutinin (HA)-tagged porcine TRIM5α was transiently transfected into HT1080 cell lines and expression was confirmed by western blot (Figure 2b). Cells transfected with empty plasmid or porcine TRIM5α were transduced with N-MLV, B-MLV, FIV, or HIV-1 at MOIs of 5. B-MLV transduction was not restricted in the presence of porcine TRIM5, as expected (Figure 2c). While restriction of N-MLV was observed in porcine TRIM5-expressing cells, no restriction was observed for FIV or HIV transduction.
Figure 2.
Species-specific restriction. (a) Total RNA was isolated from PK1 and PAE and mRNA levels of pig TRIM5α were determined using reverse transcription-PCR. The levels were normalized to pig GAPDH. (b) Protein expression of HA-tagged TRIM5α, or an HA-tagged VP35 positive control, was determined by western blot using an anti-HA antibody. (c) VSVG-pseudotyped HIV, FIV, N-MLV, or B-MLV expressing GFP (MOI = 5) were applied to HT1080 cells transiently transfected with empty plasmid or pig TRIM5α. Transduction efficiencies were measured as percentage of GFP-positive cell 48 hours after the transduction using flow cytometry. *P < 0.005, n = 3. (d) Silencing of porcine TRIM5 was confirmed by transfecting PK1 cells with 10 nmol/l of siRNA against porcine TRIM5 and compared with negative control siRNA (10 nmol/l). *P < 0.001. (e) Twenty-four hours after siRNA transfection, PK1 cells were transduced with VSVG-pseudotyped HIV, FIV, N-MLV, and B-MLV expressing GFP (MOI = 5); 48 hours after the transduction, transduction efficiencies were measured as percentage of GFP-positive cell using flow cytometry. *P < 0.01, n = 3. (f) MDTF cell lysates probed with an anti-HA antibody or α-tubulin demonstrate that all stably transduced cell lines express equivalent levels of TRIM5α. (g) The indicated stably transfected MDTF cells were transduced with B-MLV, N-MLV, or FIV at an MOI yielding 30–40% GFP-positive cells on parental MDTF cells. Percent transduction is normalized to parental control MDTF cells. Bars indicate means ± SE (n = 12). α-HA, α-hemaglutinin; B-MLV, B-tropic murine leukemia virus; FIV, feline immunodeficiency virus; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GFP, green fluorescent protein; MDTF, Mus dunni tail fibroblast; MOI, multiplicity of infection; N-MLV, N-tropic murine leukemia virus; PAE, primary airway epithelium; siRNA, small-interfering RNA; VSVG, vesicular stomatitis virus G protein.
A small-interfering RNA was designed to knockdown porcine TRIM5α mRNA, and efficacy was confirmed by real-time PCR in PK1 cells (Figure 2d). Knockdown of porcine TRIM5α successfully relieved restriction of N-MLV in PK1 cells; however, knockdown of porcine TRIM5α had no effect on the transduction efficiency of FIV- or HIV-based vectors (Figure 2e). Taken together these data suggest that FIV is more suitable than HIV as a vector platform for preclinical studies in pigs. Yet, the difference in transduction efficiency cannot be attributed to a TRIM5α-mediated restriction mechanism.
Restriction of FIV by primate TRIM5α
To determine whether human or nonhuman primate TRIM5α restricts FIV, we used Mus dunni tail fibroblast (MDTF) cells stably expressing TRIM5α from multiple primate species including: human (TRIM5αhu), rhesus macaque (TRIM5αrh), and African green monkey (TRIM5αAGM).25,26 In addition, MDTF cells expressing a chimeric human TRIM5α protein that contains the rhesus B30.2/SPRY domain hypervariable region comprising residues 325-344 (TRIM5αhu(rh325-344)) were also tested. This chimera contains the residues responsible for capsid specificity.26 MDTF cells expressing no TRIM5α serve as the baseline. All TRIM5α proteins were HA-tagged and cell lines confirmed by immunoblotting to express equivalent protein levels (Figure 2f). Cells were transduced with GFP expressing N-MLV, B-MLV, or FIV pseudotyped with VSVG. As expected, no significant restriction of B-MLV was observed; however, N-MLV virus was restricted by TRIM5α expressed from each of the tested MDTF cell lines (Figure 2g). Following transduction with FIV, restriction was seen in cells expressing TRIM5α from both nonhuman primate species tested (Figure 2g). Human TRIM5α did not restrict FIV; whereas, TRIM5αhu(rh325-344) restricted expression to levels similar to TRIM5αrh. These data suggest that based on TRIM5α restriction patterns, pigs may be better suited to FIV-based preclinical studies than nonhuman primates.
Lentiviral transduction of primary pig airway cells in vitro
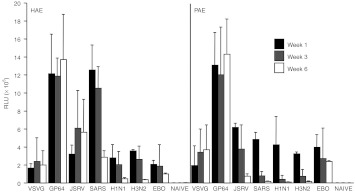
Pseudotyping lentiviral vectors with viral envelope glycoproteins is a strategy to alter vector tropism. We compared the efficacy of pseudotyped FIV to transduce the apical surface of well-differentiated primary cultures of airway epithelia derived from human lung donors (human airway epithelia (HAE)) or porcine donors (PAE). FIV expressing Gaussia (secreted) luciferase driven by the Rous sarcoma virus promoter and pseudotyped with the indicated envelope glycoprotein was applied to the apical surface of HAE or PAE (MOI = 30) for 16 hours. At 1-, 3-, and 6-week timepoints post-transduction, cell washings were collected and the levels of luciferase expression were quantified (Figure 3).
Figure 3.
Envelope glycoprotein screen. FIV expressing secreted luciferase was pseudotyped with the indicated envelope glycoproteins and applied to the apical surface of HAE or PAE at an MOI of 30. Washings were collected at 1-, 3-, and 6-week timepoints and expression was quantified by luciferase assay. Each bar represents one culture each from four independent human or pig specimens. FIV, feline immunodeficiency virus; HAE, human airway epithelium; MOI, multiplicity of infection; PAE, primary airway epithelium; RLU, relative light units; VSVG, vesicular stomatitis virus G protein.
VSVG is the most commonly used envelope for pseudotyping and serves as the standard for comparison. FIV pseudotyped with VSVG transduced HAE and PAE with approximately equal efficacy and expression persisted 6 weeks in culture. We previously reported that the envelope glycoprotein from Jaaksietke sheep retrovirus (JSRV) efficiently pseudotypes FIV with concentrated titers typically exceeding 1 × 109 TU/ml.27,28 Here, we observed that at the early timepoints, the levels of transgene expression from JSRV-FIV were similar between HAE and PAE, but by 6 weeks post-transduction, expression was waning in PAE. When FIV was pseudotyped with the S-protein from severe acute respiratory syndrome (SARS) coronavirus, the expression steadily declined in both HAE and PAE. The SARS glycoprotein was the only candidate that displayed a clear species preference. We evaluated the HA and neuraminidase from either the 1918 H1N1 influenza strain or a common H3N2 strain of swine influenza (A/SwMN593/99). Waning expression was observed for FIV pseudotyped with both sets of influenza virus glycoproteins.
We previously reported that pseudotyping FIV with either the Ebola envelope glycoprotein29 or GP64 from Autographa californica baculovirus results in apical entry into polarized primary cultures of HAE.30,31 In this study, we observed that GP64-pseudotyped FIV transduced the apical surface of both HAE and PAE with the greatest efficacy of the envelopes tested. Furthermore, expression persisted 6 weeks in culture. Based on these results, we elected to carry forward GP64 as our pseudotyping candidate for in vivo studies in the pig airways.
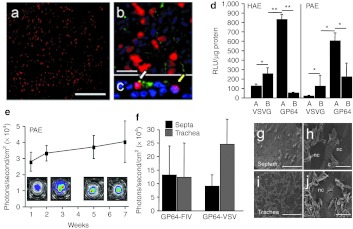
To determine the percent of PAE cells transduced in vitro, we delivered GP64-FIV expressing the mCherry reporter gene driven by the Rous sarcoma virus promoter (MOI = 150). One week post-transduction, the efficacy of gene transfer was examined visually by fluorescence microscopy (Figure 4a–c) or quantified by fluorescence-activated cell sorting (FACS). FACS analysis demonstrated that 10 ± 6.5% were mCherry positive (n = 3 donors, one culture/donor). By counterstaining for β-tubulin, a marker for cilia, we determined that at 1 week post-transduction, both ciliated and nonciliated surface epithelial cells were transduced (Figure 4c). This pattern of transduction is consistent with previous observations in mouse nasal epithelia (P.L. Sinn, M. Oakland, unpublished data).
Figure 4.
In vitro transduction of primary cultures. GP64-FIV expressing mCherry was applied to the apical surface of (a) porcine airway epithelia (PAE) at an MOI of 150. Bar = 100 µm. Cells were counterstained with β-tubulin (cilia marker, green) and To-Pro 3 (nuclei, blue). Bar = 20 µm. Using fluorescence microscopy expression was confirmed in both ciliated, white arrow, and nonciliated cells, yellow arrow ((b) en face view; (c) vertical section). (d) Primary cultures were transduced with GP64-FIV or VSVG vector applied to the apical (Ap) or basolateral (BL) surface. Four days after initial vector incubation, cells were harvested and the β-galactosidase activity was quantified and normalized to total protein. Each bar represents one culture each from four independent human or pig specimens. *P < 0.01; **P < 0.00001. PAE cells were transduced with GP64-FIV expressing firefly luciferase. (e) Persistence of expression was followed using bioluminescent imaging. Representative cultures are shown, inset. n = 4. (f) GP64-FIV or GP64-VSV expressing firefly luciferase were applied to the apical surface of PAE derived from the trachea or septa. n = 4. Well-differentiated PAE isolated from nasal (g,h) septa and (i,j) trachea of pigs were imaged using scanning electron microscopy. Representative ciliated (c) or nonciliated cells (nc) are indicated. Bars = 100 µm (in g,i) or 20 µm (in h,j). HAE, human airway epithelium; MOI, multiplicity of infection; RLU, relative light units; VSVG, vesicular stomatitis virus G protein.
To confirm that FIV pseudotyped with GP64 or VSVG has similar apical or basolateral transduction preference in PAE as previously observed in HAE, vector expressing β-galactosidase was applied to the apical or basolateral surface of the epithelial sheet for 4 hours (MOI = 30). Four days following vector delivery, gene transfer was quantified by measuring β-galactosidase activity. As shown (Figure 4d), we replicate previous observations that GP64-FIV preferentially transduces HAE at the apical surface and VSVG-FIV preferentially transduces HAE at the basolateral surface.30 Furthermore, at the same MOI, β-galactosidase activity following apical application of GP64-FIV was greater than that achieved with basolateral VSVG-FIV vector application. Using GP64-FIV expressing a firefly luciferase reporter gene and longitudinal bioluminescent imaging, persistent gene expression was observed for 7 weeks in PAE with no significant decline (Figure 4e).
We next measured GP64-FIV–mediated transduction efficiency in porcine nasal and tracheal epithelia. We cultured primary cells derived from nasal septa and trachea of wild-type newborn pigs. GP64-FIV expressing firefly luciferase was applied apically for 24 hours (MOI = 5) and luciferase activity assays were performed a week later. GP64-FIV transduced porcine nasal epithelia with equal efficacy as tracheal epithelia (Figure 4f). Interestingly, in mouse-derived PAE cells, GP64-FIV transduces septal-derived cells with greater efficacy than tracheal-derived cells (P.L. Sinn, M. Oakland, P.B. McCray, unpublished observations). A VSV-based vector pseudotyped with GP64 was also used to transduce both porcine nasal and tracheal epithelial cells. In PAE cells, we observed a trend of improved GP64-VSV transduction in tracheal-derived epithelial cells (Figure 4f). Both septa- and tracheal-derived cultures were imaged using scanning electron microscopy. Electron microscopy confirmed that cultures were well differentiated, and similar apical surface morphology was observed for cultures from both anatomical regions (Figure 4g–j).
Lentiviral transduction of pig airways in vivo
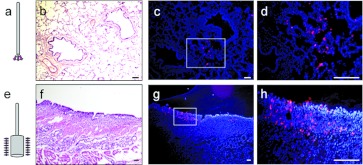
To evaluate the ability of GP64-FIV to transduce porcine pulmonary airways in vivo, we delivered GP64-FIV expressing mCherry vector to the tracheal lobe of 4-week-old pigs. The animals were sedated and a Pentax FB-10X bronchoscope (outer diameter 3.5 mm) was introduced orotracheally. The bronchoscope was passed into the tracheal lobe via the tracheal bronchus and, for the initial lung delivery strategy, a PE20 catheter (Figure 5a) was passed distally under direct visualization until it wedged. Two milliliter of GP64-FIV mCherry vector (5 × 108 TU) was delivered in a solution of 1% methylcellulose. One week later, animals were euthanized and the tracheal lobe was fixed, frozen, and sectioned. Alternate sections were stained with hematoxylin and eosin to visualize cellular morphology (Figure 5b,f). As shown, cells expressing mCherry were readily detected (Figure 5c,d). No mCherry expression was observed in the left lower lobe of the same animals (data not shown). We observed that a bolus delivery of vector through a polyethylene catheter resulted in mCherry expression that was mainly restricted to the distal airways and alveoli.
Figure 5.
In vivo gene transfer to the porcine lung. (a) Using a bronchoscope, GP64-FIV was delivered to the tracheal lobe of 4-week-old pigs. A bolus volume was delivered to the pig airways through a wedged catheter. Lungs were examined 1 week later by fluorescence microscopy ((c) low power; (d) high power). (b) Serial sections were H&E stained. (e) A separate cohort of pigs received a bolus volume of GP64-FIV delivered through a drug infusion balloon. As before, lungs were examined 1 week later by fluorescence microscopy ((g) low power; (h) high power) and (f) serial sections were H&E stained. Bars = 50 µm. H&E, hematoxylin and eosin.
We next contrasted polyethylene catheter-mediated vector delivery to vector delivery using a drug infusion balloon catheter designed for treating pulmonary thrombi (Figure 5e). As before, a bronchoscope guided the catheter to the tracheal lobe and 1 ml of GP64-FIV mCherry vector (5 × 108 TU) was delivered. The balloon remained inflated for ~3 minutes while vector was applied directly to the conducting airway. As shown, isolated regions of abundant mCherry expression were observed in conducting airway epithelial cells of the tracheal lobe (Figure 5g,h). As before, no mCherry expression was observed in the left lower lobe of the same animals (data not shown).
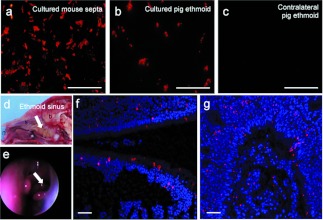
Gene transfer to sinuses is relevant because CF pigs develop spontaneous sinusitis similar to humans and sinuses may be a site of initial bacterial infection and seeding of the lungs.32 We determined whether the FIV-mediated mCherry expression in the transduced epithelial cells in vivo persisted after the cells were isolated and allowed to differentiate at an air-liquid interface in vitro. For initial pilot experiments, we delivered vector to the nasal epithelia of mice. After septa from mice were collected and the epithelial cells were cultured, mCherry expression was readily observed (Figure 6a). The epithelial cells were allowed to fully differentiate at an air-liquid interface for 4 weeks before lysates were collected for FACS analysis. Based on FACS analysis, 5.5 ± 0.3% of culture mouse septa were mCherry positive. Encouraged by these results, we repeated the process in the pig ethmoid sinus. As before, the epithelial cells were allowed to fully differentiate at an air-liquid interface for 4 weeks. Again, mCherry expression was readily observed in cultured left ethmoid sinus epithelia (Figure 6b) but not the contralateral untransduced ethmoid sinus (Figure 6c). By visual count, we determined that 4.5 ± 0.9% of surface cells derived from the left ethmoid sinus were mCherry positive. This technique may provide a strategy to confirm in vivo CFTR gene transfer efficacy in future experiments.
Figure 6.
Ethmoid sinus in vivo gene transfer. (a) GP64-FIV expressing mCherry was delivered to mice via nasal instillation (7 doses over 7 days, 1 dose/day). One week later, the septa were collected and the epithelia were cultured. mCherry was visualized by fluorescence microscopy. One dose of GP64-FIV expressing mCherry was delivered to the ethmoid sinus of pigs. One week later, (b) the transduced sinus and the (c) contralateral sinus were collected, cultured, and visualized by fluorescence microscopy. Bars = 100 µm. (d) GP64-FIV expressing mCherry (2 ml total volume in 1% methylcellulose) was delivered to the ethmoid sinus. n, nasal opening; s, septal bone; b, brain. (e) The ethmoid sinus was visualized with a rigid fiber optic endoscope. One week following vector delivery, the ethmoid sinus was removed, fixed, frozen, and sectioned. (f,g) mCherry expression (red) was visualized by fluorescence microscopy. Tissues were co-stained with DAPI nuclear stain (blue). Bars = 50 µm. DAPI, 4',6-diamidino-2-phenylindole.
Lastly, we investigated whether FIV vector could efficiently transduce sinus epithelia in a non-CF pig. In the same cohorts of animals that received vector in the tracheal lobe, 1 ml of GP64-FIV vector (titer 5 × 108 TU/ml) was delivered in a solution of 1% methylcellulose to the left ethmoid sinus (Figure 6d) via direct nasal instillation guided by a rigid fiber-optic endoscope (Figure 6e). Seven days later, animals were killed and the ethmoid sinus was fixed, frozen, sectioned, and transgene expression was examined by fluorescence imaging. The ethmoid sinus was dissected and sectioned in such a way that both transverse (Figure 6f) and longitudinal (Figure 6g) planes were observed. Widespread mCherry expression was observed in both surface and basal epithelial cells. No expression was observed in the contralateral sinus (data not shown).
Discussion
In this study, we engineered and validated a lentiviral vector for preclinical gene transfer experiments in porcine airways. A goal of these studies was to examine the porcine respiratory tract tropism of lentiviral vector pseudotypes with known preference for transducing the apical surface of airway epithelia. We and others reported successful lentiviral pseudotyping with glycoproteins from diverse viral families such as filovirus,29 coronavirus,33 orthomyxovirus,30 retrovirus,27,28 and baculovirus.30,31 However, the most effective envelope glycoprotein candidate for lentiviral-mediated pulmonary gene transfer we have identified to date is GP64. In mice, GP64-FIV conferred persistent gene transfer in vivo, consistent with progenitor cell transduction.31,34
We compared FIV- and HIV-1–based lentiviral vectors for their ability to transduce pig airway epithelia cells in vitro. We reproducibly observed better transduction for FIV than HIV-1 in both immortalized PK1 cells and primary cultures of PAE cells. This unexpected finding led us to investigate the transduction efficiency of FIV and HIV-1 vectors in multiple human airway cell lines. We conclude that the two lentiviruses have similar transduction patterns in human cells, displaying no significant differences in efficiency. However, both lentiviral vectors were less efficient than control B-MLV in porcine cells, suggesting that some restriction is present. Whether the observed restriction is viral vector or host cell-specific has not yet been determined. A possible explanation is that another host cell restriction factor identifies incoming lentiviral but not oncoretroviral capsid proteins.
VSV has been well documented to require an acidic cellular compartment for infection.35 Likewise, wild-type baculovirus,36 JSRV,37 Ebolavirus,38 and Influenza A,39 utilize the acidic interior pH of the endosome to induce a conformational change in the glycoprotein and insert the fusion peptide into the host membrane. There is some debate as to where fusion occurs with wild-type SARS-coronavirus. Initially, it was thought to enter cells through direct fusion with the plasma membrane;40 however, more recent evidence suggests that virus entry may also involve pH-dependent endocytosis.41 Of the envelope glycoproteins listed, only FIV pseudotyped with VSVG and GP64 have been demonstrated to transduce cells via a low-pH endosome route.30 The observation that not all envelope glycoproteins resulted in persistent expression in primary cells was unexpected. The mechanism for this result is currently unknown but may be dependent upon the cell types transduced and the entry or endocytosis pathway directed by interactions between envelope glycoproteins and their receptors.
TRIM5α is perhaps the best studied species-specific lentiviral barrier to transduction. Our data suggest that porcine TRIM5α is inefficient at preventing transduction of FIV or HIV-1. This was a surprising observation due to the sequence conservation between porcine TRIM5α and bovine TRIM5α. We note that sequence divergence does exist in a key region of the SPRY domain thought to be important for capsid binding.42 We are currently investigating this in more detail.
Our observation that FIV is not restricted by human TRIM5α varies with observations by Saenz et al.17 in which FIV was restricted by TRIM5αhu when overexpressed in Crandell feline kidney cells. However, our findings are similar to the lack of restriction observed when TRIM5αhu was expressed in a canine cell line.17 Endogenous TRIM proteins and/or unnamed proteins with sequence homology to TRIM5α could be responsible for the discrepant findings in these cell lines. Hetero-multimers of TRIM family members can occur.43,44 It is possible that interaction between endogenous proteins alters the function of expressed exogenous TRIM5α either positively or negatively. For example, a BLAST of the human TRIM5α coiled-coil amino acid sequence (residues 130-231, ref. 19) against the dog, cat, and mouse genomes reveals 44–51% identity and 64–72% similarity. There may be enough conservation in the human TRIM5α coiled-coil region to allow multimerization with other species variants resulting in dominant negative activity, which could render the protein inactive. This could explain the lack of restriction we observed in MDTF cells and Saenz et al.17 observed in canine cells.
Both AGM and rhesus TRIM5α restricted FIV-mediated gene transfer; therefore, other non-primate animal models may be better choices for preclinical studies. Interestingly, the FIV transduction patterns in HAE equaled or exceeded those of HIV. The enhanced efficiency of gene transfer of FIV vectors over HIV vectors in porcine cells suggests that this vector may be better suited for lentiviral vector development in a pig CF model. This result also emphasizes the importance of assessing potential host-specific lentiviral restriction factors when selecting animal models for preclinical studies.
We performed the mCherry marker virus experiment in pigs that were 4 weeks old. We suspect that the smaller surface area and rapid growth of immature pigs may help the transduction efficiency of lentiviral vectors in vivo. Indeed, for a gene therapy strategy to be effective in humans, early intervention may also likely be necessary. Importantly, newborn screening for CF is now implemented nationwide and allows earlier diagnosis, within days after birth. This provides the unique opportunity to introduce therapy before the chronic disease is established.
Estimating in vivo gene transfer efficiency is challenging as the transduction is heterogeneous, with some areas showing high expression and others with lower expression. We have seen a similar pattern with previous marker vector studies in rabbits and mice.31,45 We estimate the range of transduction efficiencies in the pig airways to be from <1 to 7%. Based on previous studies, expression of CFTR in as few as 6–10% of airway epithelia is sufficient to restore the function of chloride ion transport.46 However, another study suggested higher levels of correction (25%) may be required to normalize sodium ion transport and mucociliary clearance.47 Further studies are required to determine the preferred gene transfer targets and the level of CFTR correction required to prevent or slow disease progression.
We believe that higher transduction efficiencies might be obtained with repeated administration of the vector or with vectors of higher titer.31 Minimal humoral immune responses were observed after repeated topical delivery of a GP64-FIV to murine nasal epithelia.31 We showed additive increases in transgene expression with repeat dosing. Innate and adaptive immune responses to viral vectors will vary depending on the purity of the vector preparation, the envelope pseudotype, the dose delivered, the target tissue, the animal model, the route of delivery, and the number of administrations. Future studies will investigate these approaches in the large, genetically diverse porcine animal model.
This study addresses the optimal design of a lentiviral-based therapeutic reagent for preclinical studies in a pig model of CF. Neither humans with CF nor CF pigs have respiratory disease at birth, but disease does spontaneously develop in the sinus48 and lungs during the initial postnatal months.11 Further, a bacterial eradication defect and reduced airway surface liquid pH was recently identified to be present at birth in the CF pig model.11,49 These phenotypes offer the opportunity to alter disease course in measurable ways through gene transfer.
Materials and Methods
Vector production. The FIV vector system utilized in this study22,50 expressed either secreted (Gaussia) luciferase, mCherry, or firefly luciferase. Pseudotyped FIV vector particles were generated by transient transfection, concentrated 250-fold by centrifugation, and titered using real-time PCR as previously described.22 The following envelope glycoproteins from the following viruses were used in this study: Autographa californica multicapsid nucleopolyhedrovirus (GP64),30 Indiana strain of VSVG, JSRV,28 SARS,33 1918 strain of human influenza (H1N1, a gift from Dr Gary Kobinger, Public Health Agency of Canada, Winnipeg, Manitoba), swine Influenza A/Sw/MN/593/99 (H3N2, a gift from Dr Chris Olsen, The University of Wisconsin, Madison, WI), and Ebola Zaire.29 Viral envelope gene sequences were PCR amplified and TA-cloned into the mammalian expression plasmids pcDNA3.1-TOPO (K4800-01; Invitrogen, Carlsbad, CA) using the manufacturer's protocol.
MLV vectors were generated by a three plasmid transient transfection procedure described previously.51 HIV vectors (gifts from Dr John J Rossi, Beckman Research Institute of the City of Hope, Duarte, CA) were generated by a four plasmid transient transfection as previously described.52 N-MLV, B-MLV, HIV, and FIV expressed GFP under control of a cytomegalovirus promoter. Viral titers were determined by visually counting GFP-positive HT1080 cells (CCL-121; ATCC, Manassas, VA) following limiting dilutions of 250-fold centrifuge-concentrated supernatants.
Primary cultures of airway epithelia. Airway epithelial cells were isolated from humans or pigs and grown at the air-liquid interface as described previously.53 Briefly, the culturing process is as follows: tissues undergo enzymatic dispersion; epithelial cells are dissociated; and seeded onto collagen-coated semipermeable membranes (0.6 cm2, Millicell-PCF; Millipore, Billerica, MA).53 Twenty-four hours after seeding, the cells are allowed to grow and differentiate at the air-liquid interface. All preparations used were well differentiated (>3–4 weeks old; resistance >1,000 ohm × cm2). This study was approved by the Institutional Review Board at the University of Iowa (Iowa City, Iowa).
In vitro viral vector administration. Primary cultures of HAE or PAE were transduced with pseudotyped FIV by diluting vector preparations in serum-free media to achieve the desired MOI and applying 100 µl of the solution to the apical cell surface. After incubation for 4 hours at 37 °C, the vector was removed and cells were further incubated at 37 °C, for 4 days. To transduce epithelia with pseudotyped FIV from the basolateral side, the Millicell culture insert containing the epithelium was turned over and vector applied to the basolateral surface for 4 hours in 100 µl of media. Following the 4-hour transduction, the vector was removed and the culture insert was turned upright and allowed to incubate at 37 °C, 5% CO2, for 4 days.
The A549 (no. CCL-185; ATCC) and H441 (no. HTB-174; ATCC) epithelial cell lines are derived from human lung carcinomas. IB3,54 and HBE55 cell lines are transformed HAE cells from CF and non-CF subjects, respectively. MDTF, IB3, HT1080, MDBK (CCL-22; ATCC), and 293T (CRL-11268; ATCC) cells were maintained in DMEM (Gibco, Grand Island, NY), and 10% fetal bovine serum (FBS). MDTF cells stably expressing TRIM5α were maintained under neomycin selection with 1 mg/ml Geneticin (Gibco). A549 cells were maintained in 50:50 DMEM/Ham's F-12 (Gibco), 10% FBS, 1% L-glutamine. H441 cells were maintained in RPMI (Gibco), 10% FBS. HBE cells were maintained in MEM (Gibco), 10% FBS. Pig kidney cells, PK1 (CL-101; ATCC), were maintained in Medium 199 (11150059; Invitrogen) supplemented with 3% FBS. In addition, each medium was supplemented with penicillin (100 U/ml) and streptomycin (100 µg/ml).
In vivo viral vector administration. To analyze mCherry expression in vivo, FIV3.2 mCherry virus (5 × 108 TU) was delivered to the ethmoid sinus and tracheal lobe of 4-week-old pigs. The pigs were anesthetized using ketamine/xylazine (20 mg/kg; 2 mg/kg). Nasal passageways were opened using Afrin-coated gauze and 2 ml of viral vector resuspended in 1% methylcellulose56 was delivered to right ethmoid sinus via syringe and tube. To deliver 2 ml of viral vector to the tracheal lobe, a pediatric bronchoscope lined with a PE20 catheter or drug infusion balloon (ClearWay; Atrium, Hudson, NH) was used to ensure accurate delivery. Seven days following vector delivery, the pigs were euthanized and the ethmoid sinuses and lungs were collected for further analysis.
Fluorescence analysis of in vivo mCherry expression. Ethmoid sinuses and lung lobes were collected and fixed overnight in 4% paraformaldehyde. The tissues were then placed in 15% sucrose for 8 hours and transferred to 30% sucrose overnight, all steps carried out at 4 °C. Tissues were frozen into OCT blocks using 2-methylbutane and liquid nitrogen. Frozen tissues were stored at −80 °C and sectioned using a microtome at −35 °C. Fluorescence microscopy was then used to analyze sectioned tissues for mCherry expression in the ethmoid sinus and tracheal lobe. To quantify the percentage of FIV-transduced cells in cultured ethmoid sinus tissue, 6–7 representative fields were examined under ×20 magnification. The number of mCherry-positive cells was divided by the number of DAPI-stained nuclei to determine the percent positive transduction. Approximately 600–900 cells were counted from each section.
Immunohistochemistry and confocal microscopy. Well-differentiated primary porcine epithelial cells transduced with GP64-FIV expressing mCherry were rinsed with phosphate-buffered saline (PBS), fixed in 2% paraformaldehyde for 10 minutes, and rinsed with PBS. Cells were permeabilized with 0.2% Triton-X100 followed by incubation in 5% fetal calf serum overnight at 4 °C. After rinsing with PBS, cells were incubated with 1:200 of FITC-conjugated β-tubulin (Sigma, St Louis, MO) for 1 hour. Cells were rinsed and 1:1,000 of To-Pro-3 nuclear stain was applied for 10 minutes. Sample images were captured using Zeiss 710 confocal microscope (Carl Zeiss, Jena, Germany).
Electron microscopy. Scanning electron microscopy was used to examine cell morphology after fixation in 2.5% glutaraldehyde (0.1 mol/l sodium cacodylate buffer, pH 7.4) for 30 minutes. After serial alcohol dehydration, samples were treated with hexamethyldisilazane (Polysiences, Warrington PA) and mounted on stubs. Samples were examined under a Hitachi S-4800 electron microscope (Hitachi, Dallas, TX).
Trim5α detection. For the reverse transcription-PCR assays, RNA was extracted from cells using the RNeasy Mini Kit (74104; Qiagen, Valencia, CA) following the manufacturer's protocol. cDNA was generated using SuperScript II Reverse Transcriptase (18064-022; Invitrogen). Reverse transcription-PCR for the pig species variant of TRIM5 was performed using specific primers (pigTRIM5-F 5′ TGTCCAGGCAGAGTTTATG CGACT 3′ and pigTRIM5-R 5′ TCCAAATTTGCGCGCGTATCT CAC 3′) and pig GAPDH (pigGAPDH-F 5′ GGCAAAGTGG ACATTGTCG 3′ and pigGAPDH-R 5′ GGGATGATGTTCTGGG CAG 3′). Thirty cycles of PCR were performed (95 °C for 30 seconds, 54 °C for 30 seconds, 72 °C for 60 seconds). For western blots, cells were lysed with Galacto-Light Plus lysis solution (Tropix, Grand Island, NY) and loading volumes were determined based on BCA assay using Pierce BCA protein assay kit (23227; Thermo scientific, Rockford, IL). Proteins were separated on a 10% Tris-HCl polyacrylamide gel (161-1155; Bio-Rad, Hercules, CA) and transferred to a nitrocellulose membrane (162-0145; Bio-Rad). The blot was blocked overnight in blocking buffer (5% milk-0.05% Tween-20 in PBS), followed by a wash in 1× PBS. The membrane was probed with a 1:500 dilution of rabbit monoclonal anti-HA primary antibody (H6708; Sigma) for 2 hours followed by three rinses in wash buffer. HRP-conjugated goat anti-rabbit IgG (1:1,000, 31460; Thermo scientific) was used as the secondary antibody for 1 hour. Blots were developed using SuperSignal West Pico Chemiluminescent Substrate kit (34080; Pierce, Rockford, IL) and exposed to Kodak BioMax XAR film (165 1454; Kodak, Rochester, NY).
Pig TRIM5α cloning and knockdown. PCR primers to amplify pig TRIM5α from a cDNA clone (Genbank accession no. AY970971, kindly provided by Dr Timothy Smith, USDA/ARS US Meat Animal Research Center, Clay Center, NE) were designed (TRIM5ClaIFwd 5′ AGAatcgatGAAGCTGGGACGAGCAGGA 3′ and TRIM5HASphIRev 5′ CGAgcatgcTCAAGC GTAGTCTGGGACGTCGTATGGGTAAGAGCCCGGCGAGC ACAGA 3′) which introduced a ClaI restriction site at the 5′ end and SphI restriction site at the 3′ end. The PCR product was ligated into pCR2.1-TOPO (45-0641; Invitrogen). Following ClaI and SphI enzymatic digestion, pig TRIM5 was ligated into pCAGGs vector plasmid57 creating pCAGGs-pigTRIM5. Dicer substrate RNAi duplexes against pig TRIM5 were designed using Integrated DNA Technologies (IDT, Coralville, IA) online RNAi design software. Three duplexes were tested individually (100 and 10 nmol/l) by reverse transfecting 4 × 104 cells per well of PK1 cells in 48-well dishes using Lipofectamine RNAiMAX transfection reagent (13778; Invitrogen). Same amount of negative control small-interfering RNA (5′ AUACGCGUAUUAUACGCGAUUAACGAC 3′, and 5′ CGUUAAUCGCGUAUAAUACGCGUAT 3′) was used as a comparison. Two days after transfection, mRNA levels of porcine TRIM5 were measured by real-time PCR using the TaqMan Universal PCR Master kit (4304437; ABI, Grand Island, NY). The primers and a probe used to detect porcine TRIM5 are as follows: The upstream primer TRIM5F445 (5′ TAGGCAT GTGGCCAACATAGTGGA 3′), the downstream primer TRIM5 R581 (5′ AGCAAATGACCTTCCCATCCTCCT 3′), and the probe TRIM5P501 (5′ 56FAM-TGGCGCAGA-ZEN-AGGGAAATCTCT GTGTA-3IABkFQ 3′). The levels were normalized to pig GAPDH. Primers and a probe used to detect porcine GAPDH are as follows: The upstream primer (5′ GGCATGGCCTTCCGTGT 3′), the downstream primer (5′ GCCCAGGATGCCCTTGAG 3′), and the probe (5′ 56FAM-CCTGCTTCA-ZEN-CCACCTTCTTGATG TCATCAT-3IABkFQ 3′). The duplex that yielded the greatest knockdown (TRIM5po S 5′ CCACUAAUUUCGUCAGGGAAACA TT 3′, and TRIM5po AS 5′ AAUGUUUCCCmUGmACGAAAUU mAGmUGmGmGmU 3′) was used for subsequent knockdown experiments.
Porcine TRIM5 was silenced by reverse transfecting PK1 cells with siTRIM5po. Briefly, 5 × 104 cells were seeded in 24-well plates. Cells were transfected with 10 nmol/l of siTRIM5po or 10 nmol/l of negative control small-interfering RNA using Lipofectamine RNAiMAX transfection reagent. Twenty-four hours after the incubation, cells were transduced with viral vectors and TRIM5 restriction assays were performed.
TRIM5 restriction assay. Stably transduced MDTF cells expressing either TRIM5α from human (TRIM5αhu), African green monkey (TRIM5αAGM), rhesus (TRIM5αrh), a human/rhesus chimera (TRIM5αhu(rh325-344)), or a TRIM5α null MDTF control cell line (kindly provided by Theodora Hatziioannou and Paul D Bieniasz)25,26 were seeded in 24-well dishes (105 cells/well) yielding ~50% confluency 24 hours later. HT1080 cells were seeded in 24-well dishes (2 × 104 cells/well). After 18 hours of incubation, cells were transiently transfected with empty plasmid or pCAGGs-pig TRIM5 using lipofectamine 2000 reagent (11668; Invitrogen). Twenty-four hours after the seeding of MDTF cells or 48 hours after the seeding of HT1080 cells, cells were transduced with appropriate MOI or MOI predetermined for achieving ~30–40% GFP-positive cells by fluorescence cell sorting (FACScan; Becton-Dickinson, San Diego, CA) on TRIM5α null control cells. Viruses tested included N-MLV, B-MLV, and FIV, all expressing GFP. All transductions were carried out for 1 hour at 37 °C with 5 µg/ml polybrene (107689; Sigma). Two days post-transduction, the percentage of GFP-positive cells was determined by flow cytometry.
Statistics. Unless otherwise noted, all numerical data are presented as the mean ± SE. Statistical analyses were performed using Student's t-test.
Acknowledgments
We thank Christine Wohlford-Lenane, Paula Ludwig, and Peter Taft for their valuable technical assistance. We also thank Gary Kobinger and Christopher Olsen for providing candidate envelope glycoproteins as well as Theodora Hatziioannou and Paul D Bieniasz for providing TRIM5α-expressing cell lines. Abhay Divekar and Susan Haynes provided valuable consultations concerning the catheter selection. This work was supported by National Institutes of Health grants: P01 HL-51670, P01 HL-091842, R01 HL-105821, and the Roy J. Carver Charitable Trust. We also acknowledge the support of the In Vitro Models and Cell Culture Core, Gene Transfer Vector Core, and Cell Morphology Core, partially supported by the Center for Gene Therapy for Cystic Fibrosis (NIH P30 DK-54759) and the Cystic Fibrosis Foundation. We thank Ken Cornetta and Daniela Bishof of the NIH National Heart, Lung, and Blood Institute Gene Therapy Resource Program for their support and consultation. The authors declared no conflict of interest.
References
- Clarke LL., and, Boucher RC. Chloride secretory response to extracellular ATP in human normal and cystic fibrosis nasal epithelia. Am J Physiol. 1992;263 2 Pt 1:C348–C356. doi: 10.1152/ajpcell.1992.263.2.C348. [DOI] [PubMed] [Google Scholar]
- O'Neal WK, Hasty P, McCray PB, Jr, Casey B, Rivera-Pérez J, Welsh MJ.et al. (1993A severe phenotype in mice with a duplication of exon 3 in the cystic fibrosis locus Hum Mol Genet 21561–1569. [DOI] [PubMed] [Google Scholar]
- Dorin JR, Dickinson P, Alton EW, Smith SN, Geddes DM, Stevenson BJ.et al. (1992Cystic fibrosis in the mouse by targeted insertional mutagenesis Nature 359211–215. [DOI] [PubMed] [Google Scholar]
- Ratcliff R, Evans MJ, Cuthbert AW, MacVinish LJ, Foster D, Anderson JR.et al. (1993Production of a severe cystic fibrosis mutation in mice by gene targeting Nat Genet 435–41. [DOI] [PubMed] [Google Scholar]
- Zeiher BG, Eichwald E, Zabner J, Smith JJ, Puga AP, McCray PB., Jret al. (1995A mouse model for the delta F508 allele of cystic fibrosis J Clin Invest 962051–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney SJ, Alton EW, Smith SN, Lunn DP, Farley R, Lovelock PK.et al. (1996Cystic fibrosis mice carrying the missense mutation G551D replicate human genotype-phenotype correlations EMBO J 15955–963. [PMC free article] [PubMed] [Google Scholar]
- Colledge WH, Abella BS, Southern KW, Ratcliff R, Jiang C, Cheng SH.et al. (1995Generation and characterization of a delta F508 cystic fibrosis mouse model Nat Genet 10445–452. [DOI] [PubMed] [Google Scholar]
- Rogers CS, Hao Y, Rokhlina T, Samuel M, Stoltz DA, Li Y.et al. (2008Production of CFTR-null and CFTR-DeltaF508 heterozygous pigs by adeno-associated virus-mediated gene targeting and somatic cell nuclear transfer J Clin Invest 1181571–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CS, Stoltz DA, Meyerholz DK, Ostedgaard LS, Rokhlina T, Taft PJ.et al. (2008Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs Science 3211837–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Sui H, Fisher JT, Yan Z, Liu X, Cho HJ.et al. (2010Disease phenotype of a ferret CFTR-knockout model of cystic fibrosis J Clin Invest 1203149–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltz DA, Meyerholz DK, Pezzulo AA, Ramachandran S, Rogan MP, Davis GJ.et al. (2010Cystic fibrosis pigs develop lung disease and exhibit defective bacterial eradication at birth Sci Transl Med 229ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CS, Abraham WM, Brogden KA, Engelhardt JF, Fisher JT, McCray PB., Jret al. (2008The porcine lung as a potential model for cystic fibrosis Am J Physiol Lung Cell Mol Physiol 295L240–L263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis SK, Corboz MR, Taylor AE., and, Ballard ST. Effect of anion transport inhibition on mucus secretion by airway submucosal glands. Am J Physiol. 1997;272 2 Pt 1:L372–L377. doi: 10.1152/ajplung.1997.272.2.L372. [DOI] [PubMed] [Google Scholar]
- Inglis SK, Corboz MR., and, Ballard ST. Effect of anion secretion inhibitors on mucin content of airway submucosal gland ducts. Am J Physiol. 1998;274 5 Pt 1:L762–L766. doi: 10.1152/ajplung.1998.274.5.L762. [DOI] [PubMed] [Google Scholar]
- Joo NS, Saenz Y, Krouse ME., and, Wine JJ. Mucus secretion from single submucosal glands of pig. Stimulation by carbachol and vasoactive intestinal peptide. J Biol Chem. 2002;277:28167–28175. doi: 10.1074/jbc.M202712200. [DOI] [PubMed] [Google Scholar]
- Choi HK, Finkbeiner WE., and, Widdicombe JH. A comparative study of mammalian tracheal mucous glands. J Anat. 2000;197 Pt 3:361–372. doi: 10.1046/j.1469-7580.2000.19730361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz DT, Teo W, Olsen JC., and, Poeschla EM. Restriction of feline immunodeficiency virus by Ref1, Lv1, and primate TRIM5alpha proteins. J Virol. 2005;79:15175–15188. doi: 10.1128/JVI.79.24.15175-15188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Z, Vandegraaff N, O'huigin C, Song B, Yuan W, Xu C.et al. (2006Evolution of a cytoplasmic tripartite motif (TRIM) protein in cows that restricts retroviral infection Proc Natl Acad Sci USA 1037454–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylinen LM, Keckesova Z, Webb BL, Gifford RJ, Smith TP., and, Towers GJ. Isolation of an active Lv1 gene from cattle indicates that tripartite motif protein-mediated innate immunity to retroviral infection is widespread among mammals. J Virol. 2006;80:7332–7338. doi: 10.1128/JVI.00516-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakland M, Sinn PL., and, McCray PB., Jr Advances in cell and gene-based therapies for cystic fibrosis lung disease. Mol Ther. 2012;20:1108–1115. doi: 10.1038/mt.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesenbach U., and, Alton EW. Progress in gene and cell therapy for cystic fibrosis lung disease. Curr Pharm Des. 2012;18:642–662. doi: 10.2174/138161212799315993. [DOI] [PubMed] [Google Scholar]
- Sinn PL, Goreham-Voss JD, Arias AC, Hickey MA, Maury W, Chikkanna-Gowda CP.et al. (2007Enhanced gene expression conferred by stepwise modification of a nonprimate lentiviral vector Hum Gene Ther 181244–1252. [DOI] [PubMed] [Google Scholar]
- Song B, Javanbakht H, Perron M, Park DH, Stremlau M., and, Sodroski J. Retrovirus restriction by TRIM5alpha variants from Old World and New World primates. J Virol. 2005;79:3930–3937. doi: 10.1128/JVI.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J.et al. (1999Species-specific, postentry barriers to primate immunodeficiency virus infection J Virol 7310020–10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Perez-Caballero D, Yang A, Cowan S., and, Bieniasz PD. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc Natl Acad Sci USA. 2004;101:10774–10779. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D, Hatziioannou T, Yang A, Cowan S., and, Bieniasz PD. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J Virol. 2005;79:8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinn PL, Burnight ER, Shen H, Fan H., and, McCray PB., Jr Inclusion of Jaagsiekte sheep retrovirus proviral elements markedly increases lentivirus vector pseudotyping efficiency. Mol Ther. 2005;11:460–469. doi: 10.1016/j.ymthe.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Sinn PL, Penisten AK, Burnight ER, Hickey MA, Williams G, McCoy DM.et al. (2005Gene transfer to respiratory epithelia with lentivirus pseudotyped with Jaagsiekte sheep retrovirus envelope glycoprotein Hum Gene Ther 16479–488. [DOI] [PubMed] [Google Scholar]
- Sinn PL, Hickey MA, Staber PD, Dylla DE, Jeffers SA, Davidson BL.et al. (2003Lentivirus vectors pseudotyped with filoviral envelope glycoproteins transduce airway epithelia from the apical surface independently of folate receptor alpha J Virol 775902–5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinn PL, Burnight ER, Hickey MA, Blissard GW., and, McCray PB., Jr Persistent gene expression in mouse nasal epithelia following feline immunodeficiency virus-based vector gene transfer. J Virol. 2005;79:12818–12827. doi: 10.1128/JVI.79.20.12818-12827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinn PL, Arias AC, Brogden KA., and, McCray PB., Jr Lentivirus vector can be readministered to nasal epithelia without blocking immune responses. J Virol. 2008;82:10684–10692. doi: 10.1128/JVI.00227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainz JG, Michl R, Pfister W., and, Beck JF. Cystic fibrosis upper airways primary colonization with Pseudomonas aeruginosa: eradicated by sinonasal antibiotic inhalation. Am J Respir Crit Care Med. 2011;184:1089–1090. doi: 10.1164/ajrccm.184.9.1089. [DOI] [PubMed] [Google Scholar]
- Jia HP, Look DC, Hickey M, Shi L, Pewe L, Netland J.et al. (2006Infection of human airway epithelia by SARS coronavirus is associated with ACE2 expression and localization Adv Exp Med Biol 581479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinn PL, Anthony RM., and, McCray PB., Jr Genetic therapies for cystic fibrosis lung disease. Hum Mol Genet. 2011;20 R1:R79–R86. doi: 10.1093/hmg/ddr104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannsdottir HK, Mancini R, Kartenbeck J, Amato L., and, Helenius A. Host cell factors and functions involved in vesicular stomatitis virus entry. J Virol. 2009;83:440–453. doi: 10.1128/JVI.01864-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SX, Han Y., and, Blissard GW. Palmitoylation of the Autographa californica multicapsid nucleopolyhedrovirus envelope glycoprotein GP64: mapping, functional studies, and lipid rafts. J Virol. 2003;77:6265–6273. doi: 10.1128/JVI.77.11.6265-6273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand P, Côté M, Zheng YM, Albritton LM., and, Liu SL. Jaagsiekte sheep retrovirus utilizes a pH-dependent endocytosis pathway for entry. J Virol. 2008;82:2555–2559. doi: 10.1128/JVI.01853-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt CL, Lennemann NJ., and, Maury W. Filovirus entry: a novelty in the viral fusion world. Viruses. 2012;4:258–275. doi: 10.3390/v4020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M. Influenza virus entry. Adv Exp Med Biol. 2012;726:201–221. doi: 10.1007/978-1-4614-0980-9_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qinfen Z, Jinming C, Xiaojun H, Huanying Z, Jicheng H, Ling F.et al. (2004The life cycle of SARS coronavirus in Vero E6 cells J Med Virol 73332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang P, Liu K, Guo F, Zhang Y, Zhang G.et al. (2008SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway Cell Res 18290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li Y, Stremlau M, Yuan W, Song B, Perron M.et al. (2006Functional replacement of the RING, B-box 2, and coiled-coil domains of tripartite motif 5alpha (TRIM5alpha) by heterologous TRIM domains J Virol 806198–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mische CC, Javanbakht H, Song B, Diaz-Griffero F, Stremlau M, Strack B.et al. (2005Retroviral restriction factor TRIM5alpha is a trimer J Virol 7914446–14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L.et al. (2001The tripartite motif family identifies cell compartments EMBO J 202140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Slepushkin V, Zabner J, Keshavjee S, Johnston JC, Sauter SL.et al. (1999Feline immunodeficiency virus vectors persistently transduce nondividing airway epithelia and correct the cystic fibrosis defect J Clin Invest 104R55–R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LG, Olsen JC, Sarkadi B, Moore KL, Swanstrom R., and, Boucher RC. Efficiency of gene transfer for restoration of normal airway epithelial function in cystic fibrosis. Nat Genet. 1992;2:21–25. doi: 10.1038/ng0992-21. [DOI] [PubMed] [Google Scholar]
- Zhang L, Button B, Gabriel SE, Burkett S, Yan Y, Skiadopoulos MH.et al. (2009CFTR delivery to 25% of surface epithelial cells restores normal rates of mucus transport to human cystic fibrosis airway epithelium PLoS Biol 7e1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EH, Pezzulo AA, Meyerholz DK, Potash AE, Wallen TJ, Reznikov LR.et al. (2012Sinus hypoplasia precedes sinus infection in a porcine model of cystic fibrosis Laryngoscope 1221898–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzulo AA, Tang XX, Hoegger MJ, Alaiwa MH, Ramachandran S, Moninger TO.et al. (2012Reduced airway surface pH impairs bacterial killing in the porcine cystic fibrosis lung Nature 487109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JC, Gasmi M, Lim LE, Elder JH, Yee JK, Jolly DJ.et al. (1999Minimum requirements for efficient transduction of dividing and nondividing cells by feline immunodeficiency virus vectors J Virol 734991–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Cowan S, Goff SP, Bieniasz PD., and, Towers GJ. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 2003;22:385–394. doi: 10.1093/emboj/cdg042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, Bauer G, Michienzi A, Yee JK, Lee NS, Kim J.et al. (2003Inhibition of HIV-1 infection by lentiviral vectors expressing Pol III-promoted anti-HIV RNAs Mol Ther 8196–206. [DOI] [PubMed] [Google Scholar]
- Karp PH, Moninger TO, Weber SP, Nesselhauf TS, Launspach JL, Zabner J.et al. (2002An in vitro model of differentiated human airway epithelia. Methods for establishing primary cultures Methods Mol Biol 188115–137. [DOI] [PubMed] [Google Scholar]
- Zeitlin PL, Lu L, Rhim J, Cutting G, Stetten G, Kieffer KA.et al. (1991A cystic fibrosis bronchial epithelial cell line: immortalization by adeno-12-SV40 infection Am J Respir Cell Mol Biol 4313–319. [DOI] [PubMed] [Google Scholar]
- Cozens AL, Yezzi MJ, Kunzelmann K, Ohrui T, Chin L, Eng K.et al. (1994CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells Am J Respir Cell Mol Biol 1038–47. [DOI] [PubMed] [Google Scholar]
- Sinn PL, Shah AJ, Donovan MD., and, McCray PB., Jr Viscoelastic gel formulations enhance airway epithelial gene transfer with viral vectors. Am J Respir Cell Mol Biol. 2005;32:404–410. doi: 10.1165/rcmb.2004-0410OC. [DOI] [PubMed] [Google Scholar]
- Niwa H, Yamamura K., and, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]