Abstract
Field cancerization effects as well as isolated tumor cell foci extending well beyond the invasive tumor margin have been described previously to account for local recurrence rates following breast conserving surgery despite adequate surgical margins and breast radiotherapy. To look for evidence of possible tumor cell contamination or field cancerization by genetic effects, a pilot study (Study 1: 12 sample pairs) followed by a verification study (Study 2: 20 sample pairs) were performed on DNA extracted from HER2-positive breast tumors and matching normal adjacent mammary tissue samples excised 1–3 cm beyond the invasive tumor margin. High-resolution molecular inversion probe (MIP) arrays were used to compare genomic copy number variations, including increased HER2 gene copies, between the paired samples; as well, a detailed histologic and immunohistochemical (IHC) re-evaluation of all Study 2 samples was performed blinded to the genomic results to characterize the adjacent normal tissue composition bracketing the DNA-extracted samples. Overall, 14/32 (44 %) sample pairs from both studies produced genome-wide evidence of genetic aberrations including HER2 copy number gains within the adjacent normal tissue samples. The observed single-parental origin of monoallelic HER2 amplicon haplotypes shared by informative tumor–normal pairs, as well as commonly gained loci elsewhere on 17q, suggested the presence of contaminating tumor cells in the genomically aberrant normal samples. Histologic and IHC analyses identified occult 25–200 μm tumor cell clusters overexpressing HER2 scattered in more than half, but not all, of the genomically aberrant normal samples re-evaluated, but in none of the genomically normal samples. These genomic and microscopic findings support the conclusion that tumor cell contamination rather than genetic field cancerization represents the likeliest cause of local clinical recurrence rates following breast conserving surgery, and mandate caution in assuming the genomic normalcy of histologically benign appearing peritumor breast tissue.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-012-2290-3) contains supplementary material, which is available to authorized users.
Keywords: HER2/ERBB2-amplified breast tumors, Normal adjacent tissue
Introduction
Breast conserving surgery (BCS) is a well-accepted standard of care for the treatment of both early stage breast cancer and preinvasive ductal carcinoma in situ (DCIS) [1]. Following initial randomized studies of BCS, efforts were made by pathologists studying mastectomy specimens to estimate the amount of occult breast cancer multifocality and thereby predict the likelihood of recurrence after BCS, as well as guide the extent of surgical resection margin needed to remove all invasive tumor cells. Despite initial reports of multifocality rates as high as 40–60 % [2, 3], definitions of multifocality restricted to residual tumor foci outside simulated quadrantectomies led to lower estimates of 28–38 % [4, 5]. Later mastectomy studies, eliminating more diffusely spreading invasive phenotypes from consideration (e.g., invasive lobular cancers) and further restricting primary tumors to those most eligible for BCS (stages Tis, T1–2), demonstrated that a 2-cm clear surgical margin would still leave behind occult tumor foci in up to 42 % of cases, a third of which being invasive tumor cell clusters and the remainder being occult noninvasive foci of DCIS [6].
Over the >20 years that have elapsed since the completion of six prospectively randomized trials showing that BCS followed by whole breast irradiation yielded overall survival rates comparable to mastectomy, studies of occult residual tumor burden following BCS have declined because of the impact of whole breast irradiation in significantly reducing but not eliminating potentially life-threatening loco-regional recurrences, the majority of which occur near the primary tumor site [7, 8]. However, technical, cosmetic, and economic issues still beg the question of how much normal breast tissue should be removed during BCS to achieve clean surgical margins and further minimize loco-regional recurrences [7, 8]. Moreover, with BCS, the current standard of care and an ever-growing need to extract normal host tissue DNA and RNA from the same small BCS specimen as control samples for the many high-resolution genetic and epigenetic assays now being performed on breast tumors [9]; there is renewed concern about the molecular normalcy of morphologically normal peritumor mammary tissue. Adding fuel to these clinical and technical questions are several newly recognized issues: (i) a negative margin of any width does not indicate the absence of residual occult breast tumor cells outside that margin [7]; (ii) among BCS eligible patients, the likelihood of multifocality and residual tumor volume elsewhere in the breast does not correlate with tumor size [6, 7], but is determined by tumor phenotype with triple-negative (ER-negative, PR-negative, HER2-negative) and HER2-positive breast cancers likeliest to recur [7, 10]; and (iii) “field cancerization,” characterized by genetic and epigenetic alterations in histologically normal-appearing tissue is common within 5 cm of a breast tumor margin and thought to provide a microenvironment conducive to new tumor formation [11, 12].
The term “field cancerization” was coined over 50 years ago [13], but more recently has been redefined to apply to a morphologically normal-appearing microenvironment surrounding various tumor types (e.g., breast, prostate, lung, head and neck, gastrointestinal, and skin cancers) that, when assayed by high-resolution molecular assays, exhibits a gradient of genetic and/or epigenetic aberrancies relative to more remote non-tumor-bearing areas in that organ [11, 12]. In 1996, Smith and coworkers [14] first demonstrated genetic loss of heterozygosity (LOH) in morphologically normal lobules adjacent to most breast cancers bearing that same LOH; and they postulated that lost tumor suppressor gene(s) at those loci characterized a breast tumor initiating genetic field defect. Later genetic studies subsequently showed that shortened telomeric DNA and four to five times more prevalent LOH loci characterized the field cancerization of normal-appearing tissues within 1 cm of microscopically defined tumor margins [12]. More recently, gene expression profiling of the peritumor breast microenvironment by different groups has revealed the presence of two distinct RNA expression subtypes [15, 16]: one “normal” microenvironment, independent of distance from tumor, and another “active” microenvironment, its presence dependent on distance from the tumor edge and independent of tumor phenotype yet associated with poorer patient prognosis, and its signature consistent with extracellular remodeling, wound healing, fibrosis, and epithelial-to-mesenchymal transition (EMT) [16]. Whether or not this epigenetic EMT-like phenotype within histologically normal cancer-adjacent mammary tissue also reflects any underlying genetic field cancerization effect remains unclear, as neither of those two gene expression studies evaluated genomic aberrations in the microdissected peritumor tissue, and the investigators concluded that this EMT-like phenotype was a host reaction unrelated to any specific breast cancer phenotype [15, 16].
In order to address the questions of residual breast tumor burden within the peritumor microenvironment and the genomic status of morphologically normal breast tissue surrounding aggressive breast cancers of the same phenotype, we performed a pilot study (Study 1, n = 12 cases) followed by a verification study (Study 2, n = 20 cases) on DNA samples isolated from HER2-positive breast tumors (T) and their normal-appearing mammary tissue (N), excised from 1 to 3 cm beyond the invasive tumor margin. High-resolution molecular inversion probe (MIP) arrays (20K array for Study 1, 300K array for Study 2) were used to examine differences in genome copy number gains and losses between the T/N pairs; such MIP arrays are capable of sensitive quantification of allele-specific copy number changes (ranging from 1 to 60 altered copies per locus) across the entire genome as well as accurate determination of allele-specific composition within the HER2 amplicon [17–20]. We also reasoned that increased HER2 gene copies would be easier to detect than loss of normal gene copy numbers arising from occult tumor cells situated within a field of genomically normal tissue. For correlation with the 300K MIP array findings in Study 2, a more detailed microscopic re-evaluation of all N samples was performed by independent study pathologists blinded to the genomic data and employing histologic and immunohistochemical (IHC) assays to characterize the adjacent normal tissue composition bracketing the N sample cores extracted for DNA. While these retrospective studies produced microscopic evidence of extratumoral cancer cell foci in 20 % (4/20) of the N samples from the HER2-positive cases (Study 2), the 300K MIP arrays detected excess HER2 gene copy numbers along with other genomic aberrations consistent with those found in the adjacent HER2-positive tumors in 35 % (7/20) of the N samples, along with unrelated genomic aberrations in another 10 % (2/20) of samples.
Methods
Study samples
Under multi-institutional review board approval, all cryobanked (−80 °C without fixatives) tissue samples studied (32 sample pairs, average sample wet weight >100 mg) were received in two different dry ice batches (Study 1, archived before 2006; Study 2, archived before 2009) from the Stiftung Tumorbank Basel with each sample pair coded and delinked from all personal identifiers. All tumors were HER2-positive based on a well-validated quantitative immunoassay for HER2 protein expression performed in Basel [21], with only the Study 2 samples accompanied by limited annotation (age-at-diagnosis, ER and PR status, tumor size, nodal status), as summarized in Table 2, not including treatment or follow-up data. Swiss pathologists had microscopically confirmed that all the tumor samples contained >50 % invasive breast cancer cells, and that the normal-appearing adjacent mammary tissues had been excised minimally 1 cm away but no more than 3 cm away from the invasive tumor margin.
Table 2.
Summary of Study 2 adjacent normal (N) breast tissue constituents including fat, muscle, and either benign (normal or usual ductal hyperplasia, UDH) or malignant epithelium, as well as MIP array determined copy number normalcy or abnormality (CNA) and HER2 copy number gain and HER2 IHC assay (1–3+) of cancer (CA) cells identified in the N samples
| Sample pair # | N # | N stroma (yes = 1) | N fat (yes = 1) | N muscle (yes = 1) | N epi (yes = 1) | N epi-UDH (yes = 1) | N CA-epi (yes = 1) | N CA-epi HER2 (IHC) | N HER2 Gain (MIP, yes = 1) | N Other CNA (MIP, yes = 1) | N normal CN (MIP, yes = 1) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 21N | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| 2 | 22N | 1 | 1 | 0 | 0 | 0 | 1 | 3 | 1 | 1 | 0 |
| 3 | 23N | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| 4 | 24N | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| 5 | 25N | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| 6 | 26N | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | |
| 7 | 27N | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | |
| 8 | 28N | 1 | 1 | 0 | 0 | 0 | 1 | 3 | 1 | 1 | 0 |
| 9 | 29N | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | |
| 10 | 30N | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 11 | 31N | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | |
| 12 | 32N | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | |
| 13 | 33N | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | |
| 14 | 34N | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | |
| 15 | 35N | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 16 | 36N | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | |
| 17 | 37N | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |
| 18 | 38N | 0 | 1 | 0 | 0 | 0 | 1 | 3 | 1 | 1 | 0 |
| 19 | 39N | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | |
| 20 | 40N | 1 | 1 | 0 | 1 | 0 | 1 | 3 | 1 | 1 | 0 |
Sample processing for microscopy, DNA extraction, and MIP microarrays
Cryosectioning was performed on the central core of each sample (10 sections, 50 μm each) for DNA extraction, with flanking tissue used to generate at least five 10-μm frozen section slides for microscopic analysis and correlation with DNA studies. Frozen sections from the paired tumor and normal adjacent tissue samples were independently evaluated (blinded to DNA results) by pathologists (AL, FW) within the Clinical Pathology Laboratory of the University of California at San Francisco (UCSF) using hematoxylin and eosin staining (H&E, recording malignant and benign components including epithelial, fat, stromal, and muscle cell content) and IHC assay of epithelial HER2 expression by Hercep Test (DAKO, 0–3 scale), as previously described [22]. Core sections were DNA extracted as previously described for HER2 gene analysis by comparative genomic hybridization [23], using a standardized UCSF protocol involving proteinase K digestion, phenol/chloroform/isoamyl alcohol (PCI) extraction, and ammonium acetate/ethanol precipitation followed by TE resuspension and RNase A digestion (http://waldman.ucsf.edu/protocols/dna.frozen.htm), and then repeated extraction and precipitation. TE redissolved DNA frozen in 96-well plates (~200 ng DNA in 50 μl/well) was shipped to Affymetrix, Inc., Santa Clara, CA (Yuker Wang), where Study 1 sample pairs were analyzed on a 20K MIP array platform and Study 2 samples were later analyzed on a 300K MIP array platform, and all pre-processed data returned for further determination of allele-specific copy number aberrations (CNA) [17–20].
Allelic copy number determination, genomic aberrations, and visualization tools
Pre-processed MIP array data for each sample were analyzed as previously described [19, 20] after first being segmented by a modified version of a circular binary segmentation (CBS) algorithm appropriate for single-nucleotide polymorphism (SNP) data [24, 25]. All programming scripts were performed using R (http://www.rproject.org/). Inputs into the modified algorithm were the A and B allele values on the copy number scale for every SNP (after removing those SNPs with call rate below 90 %). In the first round of segmentation, square root of the total copy number (sum of A and B alleles) was used to stabilize the variance; otherwise, the original copy number scale was used. In the second round, the absolute differences between the A and B alleles of all heterozygotes were segmented. The first segmentation was performed with a type I error of 0.009 and the second was performed with a type I error of 0.001, which maintained a typical CBS type I error of 0.01. Allele-specific means were estimated from the means of the total copy numbers and means of the differences between the A and B alleles of the heterozygotes, with the lower allele-specific mean referred to as the minor copy number and the upper allele-specific mean referred to as the major copy number. The minor copy numbers were combined across adjacent segments if t tests corresponding to the minimum SNP values were not significant by a cutoff p value of 0.05. The same was true of the major copy number and maximum SNP values. Heterozygotes were identified using an iterative binning method based on the minimum allele frequency across all SNPs. The bin between values 0 and 0.25 of size at least 0.05 that contained the lowest ratio of minimum allele values to bin size was first identified, then any SNP with a minimum allele value below the bin was estimated to be homozygous, while any above the bin was estimated to be heterozygous; SNPs in the bin were estimated to be homozygous or heterozygous in the same proportion as those outside the bin, and those with lower values estimated to be heterozygous. Allelic and total copy number differences and frequencies were analyzed using the Bioconductor R packages aCGH and DNAcopy [26–28]. For some graphic purposes, segmentation files were normalized by log2 median centering each sample separately resulting in an estimated normal copy number at zero, with positive values indicating an increase in copy number and negative numbers indicating a decrease in copy number. Visualization was also performed with a custom drawing script in python using the matplotlib visualization library, with data represented in the box-plot summary view available on the UCSC Cancer Genomics Browser [29]. Visualization of the ERBB2/HER2 amplicon on chromosome 17 was performed using the Integrative Genomics Viewer (IGV) [30].
Results
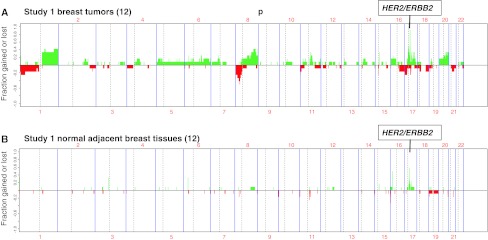
In the pilot study (Study 1), genome-wide allele-specific copy number analysis was performed using 20K MIP arrays with 12 paired DNA samples extracted from cryobanked HER2-positive breast tumor samples and their adjacent normal tissue, as described in “Methods” section. The genome-wide total CNA determined from these lower resolution MIP arrays are summarized in Fig. 1, showing the CNA fraction gained or lost for each probe across all 12 tumors (a) and their adjacent normal tissue samples (b). This study was initially intended to document monoallelic amplification in breast tumors that had been scored as HER2-positive prior to archiving by quantitative immunoassay [21], and to look for evidence of genomically abnormal peritumor field defects based on earlier evidence of monoallelic HER2 transcription present in normal-appearing breast epithelium adjacent to HER2-positive tumors [23]. Individual genomic profiles confirmed the presence of a 17q12–21 HER2 amplicon in 10/12 (85 %) Study 1 tumors, only half of which appeared to show a monoallelic haplotype (i.e., single-parental origin). The minimal common region of this HER2 amplicon haplotype within these five tumors was ~400 kbp in size and consisted of the following known genes (centromere to telomere direction): CRKRS, NEURODL, PPP1R1B, STARD3, TCAP, PNMT, PERLD1, ERBB2/HER2, C17orf37, GRB7, and IKZF3. As well, all of these HER2-positive tumors appeared genomically consistent with those described in a larger high-resolution genomic study [31] of HER2-positive breast cancers by the following criteria: (i) near 40 % frequency of other chromosome 17q amplification peaks within 17q11–12, 17q21, and 17q23; (ii) >20 % frequency of other chromosome copy number gains/amplifications at 1q, 8q, 11q, and 20q loci; and (iii) near 20 % frequency of copy number losses in other chromosomes at 1p, 8p, 16q, 17p, and 18q loci. Most strikingly, however, was the detection of HER2 amplification in nearly 60 % of the matched adjacent normal samples in Study 1 (Fig. 1B), and the finding in 5/12 normals of the same monoallelic amplicon haplotype as found in their paired tumor samples. Uncertain whether these Study 1 observations signified the presence in these adjacent tissues of cancerization with normal-appearing epithelial cells displaying amplified HER2 but few other genomic aberrations, or the presence of a few contaminating malignant cells genomically similar to the nearby tumor, we launched Study 2 to verify these findings and better define the histologic nature of the normal-appearing tissue adjacent to HER2-positive breast cancers.
Fig. 1.
Frequency of significant allelic gain (green) or loss (red) based on 20K MIP array platform analysis of 12 paired Study 1 samples, plotted as a function of genomic location, with HER2-positive tumor frequencies shown above (A) and their adjacent normal breast tissue frequencies shown below (B). The 17q chromosome location of the HER2/ERBB2 amplicon is indicated above each panel. Vertical axis labels indicate fraction of samples with gained (0 to +1.0) or lost (0 to −1.0) allelic probes
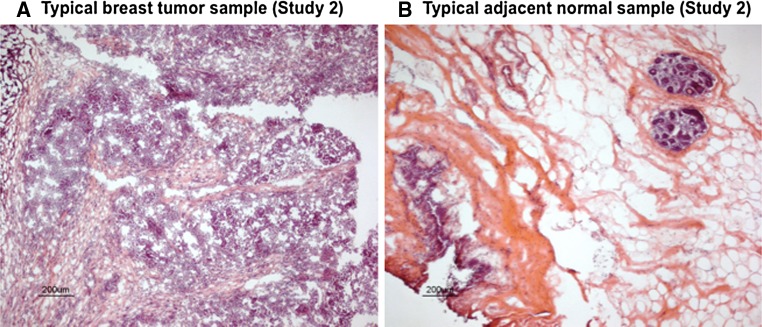
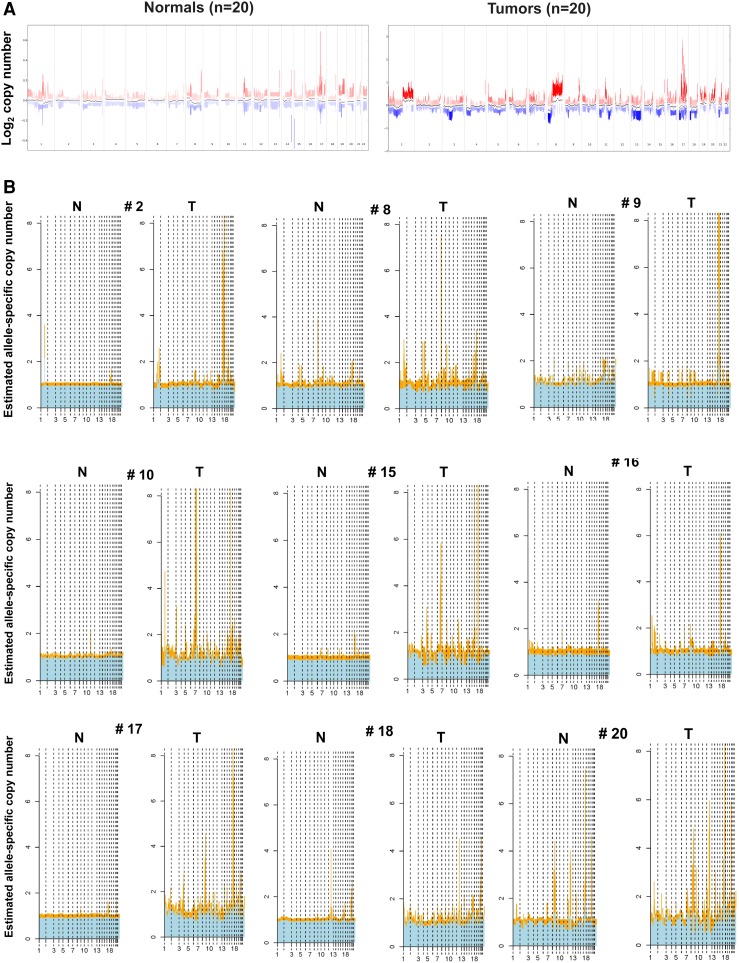
Representative histologic (H&E) appearances of a HER2-positive tumor and a normal-appearing mammary tissue samples from Study 2 are shown in Fig. 2. Fully detailed characteristics for each of the 20 Study 2 tumors (age-at-diagnosis, tumor size and nodal status, ER/PR status, HER2 receptor expression, and HER2 gene copy number) are given in Table 1; as well, the corresponding adjacent normal tissue sample characteristics (histologic, IHC, MIP) are summarized in Table 2. Importantly, these HER2-positive tumors appear to be representative of those found clinically as 75 % were diagnosed at >50 years of age, half of them lacked hormone receptor expression (ER-neg, PR-neg), 60 % were node-negative, and 80 % presented as T1–2 lesions (Table 1). Also shown in Table 1, all Study 2 tumors contained HER2 amplicons (up to 918 kbp in size, from chromosome 17 positions 34179009–35097207) and MIP array estimated HER2 copy number gains averaging 13.9 allelic copies/tumor and ranging from as few as 3 to as many as 56.7 copies/tumor, well within the quantitative dynamic range of the 300K MIP array platform [17–20]. Despite significant resolution (probe density) differences in the MIP arrays used in the two studies, a similar profile of genomic gains and losses across all chromosomes was apparent between the HER2-positive tumors in Studies 1 and 2. Likewise, when comparing the genome-wide log2 total copy number gains and losses between the 20 normal and tumor samples (summary box-plots in Fig. 3A), as seen with the Study 1 sample pairs, the Study 2 normals showed a striking genome-wide profile resemblance to the summary tumor profile despite the difference in their copy number scales (lower copy number values for normals than tumors), and consistent with the fact that 11 of the 20 normal samples (# 1, 3, 4, 5, 6, 7, 11, 12, 13, 14, 19) possessed perfectly normal allele-specific copy numbers across their entire genomes as would be expected of all genomically normal tissues (Supplementary Fig. 1). Thus, the summary profile of 20 normals showing allelic copy numbers that vary from 1 (log2 = 0, Fig. 3A) reflects the genomic gain and loss contributions from the aberrant genomes of only 9 of the 20 Study 2 normal samples (# 2, 8, 9, 10, 15, 16, 17, 18, 20), whose individual sample profiles are shown next to their tumor sample profiles in Fig. 3B. Of note, there was no statistical differences in age-at-diagnosis, tumor size, nodal involvement, hormone receptor (ER, PR) status, tumor HER2 expression level or estimated HER2 copy numbers between sample pairs with or without detectable genomic aberrations in their adjacent normal tissues.
Fig. 2.
Histologic (H&E) images from representative Study 2 cryosections of a HER2-positive cancer (A) and a normal-appearing cancer-adjacent sample (B). All cancer-adjacent samples were characterized for constituent histologic elements including fat, stroma, and epithelium as illustrated above and summarized in Table 2
Table 1.
Summary of Study 2 tumor (T) characteristics including age-at-diagnosis, MIP array estimate of HER2 gene copy number, HER2 protein overexpression level by ELISA, estrogen and progesterone receptor (ER, PR) status, tumor size, stage (T1–4), and nodal (N0–1) status
| Sample pair # | T # | Age | T HER2 CN (MIP) | T HER2 U/mg (ELISA) | T ER | T PR | T cm | Tumor stage | Nodal stage |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21T | 50 | 12.2 | 659 | neg | neg | 3 | 2 | 0 |
| 2 | 22T | 57 | 7 | 372 | pos | pos | 1.7 | 1 | 0 |
| 3 | 23T | 39 | 7.8 | 362 | neg | neg | 2.5 | 2 | 1 |
| 4 | 24T | 38 | 3.8 | 359 | pos | neg | 2.7 | 2 | 0 |
| 5 | 25T | 47 | 5.5 | 429 | pos | neg | 3 | 4 | 1 |
| 6 | 26T | 53 | 17.5 | 420 | neg | neg | 2.4 | 2 | 0 |
| 7 | 27T | 78 | 18 | 282 | pos | pos | 3 | 2 | 1 |
| 8 | 28T | 79 | 4.5 | 838 | neg | neg | 2.5 | 2 | 0 |
| 9 | 29T | 46 | 25.8 | 430 | neg | neg | 3.5 | 2 | 1 |
| 10 | 30T | 39 | 29.9 | 326 | neg | neg | 2.5 | 2 | 1 |
| 11 | 31T | 54 | 5 | 608 | pos | neg | 4 | 2 | 1 |
| 12 | 32T | 54 | 14.1 | 493 | pos | pos | 1.8 | 1 | 0 |
| 13 | 33T | 55 | 18.1 | 481 | neg | neg | 2.5 | 2 | 0 |
| 14 | 34T | 54 | 56.7 | 814 | pos | pos | 5.5 | 3 | 1 |
| 15 | 35T | 77 | 12 | 446 | neg | neg | 6.2 | 4 | 1 |
| 16 | 36T | 68 | 9.8 | 627 | neg | neg | 2.4 | 2 | 0 |
| 17 | 37T | 52 | 18.1 | 697 | neg | neg | 3 | 2 | 0 |
| 18 | 38T | 52 | 3.6 | 401 | pos | neg | 3 | 2 | 0 |
| 19 | 39T | 65 | 4.7 | 447 | pos | neg | 2.2 | 2 | 0 |
| 20 | 40T | 67 | 3 | 434 | pos | neg | 6 | 3 | 0 |
Fig. 3.
Total and allele-specific copy number variations based on 300K MIP array platform analysis of paired Study 2 samples, plotted as a function of genomic location. A Summary box-plot of log2 copy number variations across all 20 normal samples and their matching 20 tumor samples. Red (increased copy number) and blue (decreased copy number) vertical lines indicate upper and lower quantile variations, colored by their median value; note different copy number scales for normal (−0.4 to +0.6) and tumor (−2 to +3) samples. Black dots indicate median values across all samples. B Genome-wide allele-specific copy number comparisons for the 9 HER2-positive tumor (T)–normal (N) sample pairs in which there was some genomic aberration detectable in the N sample, with 7 of these showing 17q12–21 HER2 copy number gains (# 2, 8, 9, 16, 17, 18, 20). Major (yellow) and minor (blue) allelic changes (scaled from 0 to 8 for all samples) are shown with estimated copy numbers >1.5 considered significant gains
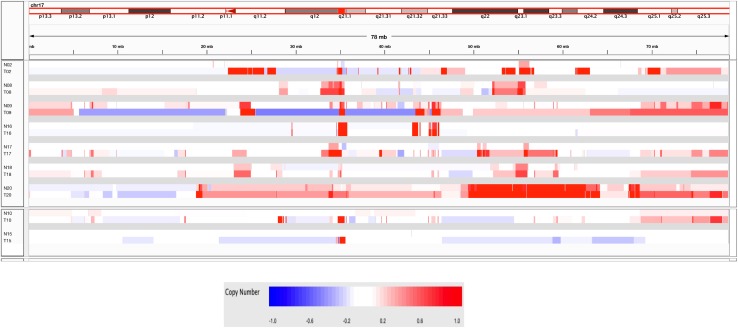
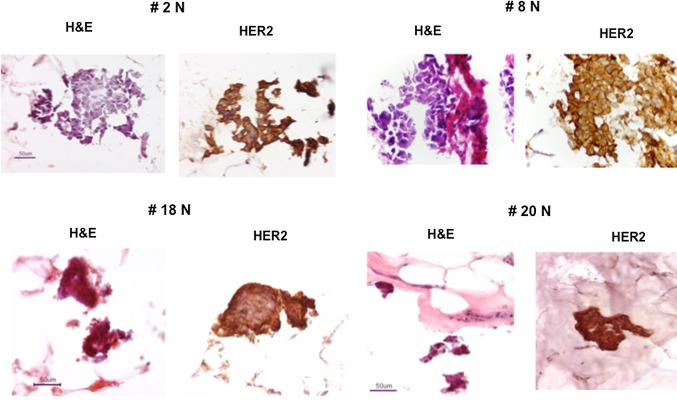
Of the nine normal samples exhibiting some evidence of genomic aberration (Fig. 3B), only two (# 10, 15) showed minimal but detectable copy number changes elsewhere besides the HER2 locus and chromosome 17, suggesting the absence of contaminating tumor cells and the possible presence of a field defect or cancerization in these two samples. Of note, microscopic examination of these normal samples revealed the presence of only normal-appearing mammary epithelium in both, with one (#10) showing the unusual absence of other stromal, fat, or skeletal muscle components (Table 2) suggesting that its twofold monoallelic gain at the telomere of 11p was present in that morphologically normal-appearing mammary epithelium. The remaining seven normal samples (35 % of Study 2 normals) showed genomic changes at loci similarly involved by their matching tumor samples (Fig. 3B), including monoallelic HER2 copy number gains and a general pattern of 17q, but not 17p, aberrations similar to their adjacent tumors, as illustrated in Fig. 4 and consistent with the possibility of occult tumor cells somewhere within the adjacent normal-appearing tissue sample. While 60 % of all the normal samples contained some epithelial components, three of the seven genomically aberrant normal samples contained normal stroma and fat but no microscopically detectable benign or malignant epithelial components (Table 2), suggesting that the cryosections flanking the larger core sample extracted for DNA were not adequate to capture contaminating tumor cells likely present in the core sample. In contrast, contaminating tumor cells were confirmed in four of the seven genomically aberrant normal samples (# 2, 8, 18, 20) which contained occasional 25–200 μm clusters of malignant, HER2-positive (IHC +3) cells, as illustrated in Fig. 5, with or without additional normal-appearing, HER2-negative (IHC +1 or +2) epithelial cells elsewhere in the same normal sample (Table 2).
Fig. 4.
IGV display comparing Study 2 chromosome 17 gains/losses and HER2/ERBB2 amplicon gains in the 7 T/N sample pairs for which increased 17q and HER2 copy numbers were seen the N samples (top heatmap panel), and the 2 other sample pairs in which the N samples showed normal chromosome 17 copy numbers but aberrant genomes elsewhere (bottom heatmap panel). Heatmaps show segmented overall copy number variations with red intensities indicating regions of copy number gain, blue intensities indicating regions of copy number loss, and white indicating no copy number change. The HER2/ERBB2 locus is indicated by the red rectangle on the ideogram (~35 mb)
Fig. 5.
Histologic (H&E) and IHC (+3 HER2 overexpression) images of scattered, occult tumor cell clusters (25–200 μ) identified in four HER2-positive cancer-adjacent normal samples (# 2N, 8N, 18N, 20N)
Discussion
Long-term (13–20 years) clinical follow-up of randomized trials of BCS have revealed in breast recurrence rates ranging from 6 to 20 %, even when negative surgical margins were as wide as 1–2 cm; these and other studies have suggested that up to half of such local recurrences might actually represent new primary cancers arising in mammary tissue at risk for developing cancer [8]. However, with most surgeons currently accepting a 2 mm or less free margin width as sufficient for complete breast cancer excision [8], such recurrence rates are also consistent with earlier mastectomy studies showing occult tumor cells commonly residing more than 2 cm beyond the microscopic tumor edge, potentially capable of invasive growth and regional spread [6]. We chose to study morphologically normal-appearing mammary tissue adjacent to HER2-positive breast cancers in part because this aggressive tumor phenotype is molecularly less heterogeneous than other breast cancer phenotypes yet likeliest to recur [7, 10], and because an earlier study had suggested that its adjacent mammary epithelium might be predisposed to monoallelic HER2 amplification [23]. Therefore, given the high resolution and sensitivity of MIP arrays, we expected to find genetic evidence of either field cancerization or contaminating tumor cells in up to 20 % of normal-appearing mammary tissue samples excised from 1 to 3 cm outside the tumor margins of HER2-positive breast cancers. Surprisingly, 16/32 (50 %) Study 1 and Study 2 sample pairs produced some evidence of genomic aberration within the adjacent normal tissue samples with 14/32 (44 %) of these showing significant copy number gains at the HER2 locus.
The observed single-parental origin of monoallelic amplicon haplotypes shared by informative tumor–normal sample pairs, as well as the commonly gained loci elsewhere on 17q in adjacent normal samples concordant with characteristic 17q gains previously described in HER2-positive breast tumors [31], provided compelling support for a preliminary conclusion that contaminating tumor cells probably explained the observed genomic aberrations in these HER2-positive cancer-adjacent normal samples. On the other hand, two sample pairs in Study 2 contained aberrations in their normal tissues that did not mirror their adjacent tumor genomes, suggesting possible field cancerization effects [11, 12], although such genetic field effects have not been described previously in association with HER2-positive breast cancers. Therefore, additional microscopic study of the multiple frozen sections flanking the DNA-extracted core normal samples from Study 2, blinded to the genomic results, was considered essential for histologic insight into the source of the observed genomic aberrations, understanding that absent microscopic evidence of contaminating tumor cells in these sections would not constitute proof of tumor cell absence in the larger DNA-extracted core samples. Indeed, upon histologic and genomic correlation, 4/7 normal samples with MIP array evidence of amplified HER2 gene copies also demonstrated isolated microscopic clusters of HER2-positive cancer cells easily missed depending on the number of frozen sections cut and evaluated. Sections flanking the other 3/7 normal samples showing obvious genomic aberrations and increased HER2 gene copies revealed normal mammary fat and stromal elements but no evidence of benign or malignant epithelium, leading to the presumption that occult tumor cell foci must have been confined to the larger DNA-extracted core sample. Importantly, of the 11 adjacent samples with entirely normal genomic profiles, 7 showed normal epithelial elements and 3 of these also contained some usual ductal hyperplasia (UDH), indicating that their normal genomic profiles were not likely false-negative findings due to the absence of adjacent breast epithelium. Notably, those three normals with UDH did not show any evidence of either increased HER2 gene copy number or HER2 overexpression by IHC (Table 2), suggesting that the UDH did not represent precursor lesions related to the adjacent HER2-positive invasive tumors. Finally, sections from the two normal samples with aberrant genomes but lacking increased HER2 gene copy numbers and appearing genomically dissimilar to their matching tumors, contained histologically normal epithelial elements, consistent with prior reports of field cancerization occurring in microscopically normal-appearing breast epithelium [11, 12, 14–16].
In sum, these findings suggest that using high-resolution genomic profiling, genetic aberrations commonly exist within morphologically normal breast tissue located well beyond what is commonly considered a clear surgical margin for BCS even for aggressive HER2-positive breast cancers, although the majority (55 %) of our Study 2 sample pairs revealed histologically and genetically normal tissue adjacent to the HER2-positive tumors. The genomic profiles and histologic study of our aberrant normal samples support the conclusion that tumor cell contamination rather than genetic field cancerization represents the likeliest explanation for clinically observed local recurrence rates up to 20 % following BCS despite post-operative breast radiotherapy. The observed frequency of genomically aberrant adjacent normal tissue in our two studies was consistent with earlier mastectomy studies showing frequent presence of isolated malignant cells well beyond tumor margins; and, as with many of these clinical studies, we found no correlation between our aberrant adjacent tissue genomic findings with other clinical characteristics of these HER2-positive tumors including age-at-diagnosis, size, nodal involvement, or hormone receptor expression. Importantly, this study did not address possibly common epigenetic field cancerization effects conducive to de novo tumorigenesis that would require DNA methylation, microRNA, and mRNA profiling for full characterization. As well, despite many recent molecular genetic approaches, there remain significant statistical challenges in trying to demonstrate the clonal relatedness of tumors arising in the same patient at distant sites or at different points in time [32–34]; therefore, even ignoring the technical limitation of characterizing minimal tumor DNA within a pool of normal germline DNA, we cannot absolutely conclude that the genomic aberrancies in our normal tissue samples equate to contaminating tumor cells derived from the adjacent primary tumors. In the future, it is possible that deep exome or whole genome sequencing of adjacent normal tissue samples will be able to better detect and differentiate genomic aberrations such as cancer-driving versus precursor mutations; and such studies are now underway within The Cancer Genome Analysis (TCGA) network [9]. The TCGA program, in particular, recognizes the growing need to employ surgically accessible normal organ tissues for in depth and accurate genomic profiling of breast and other cancers; and the present comparison of HER2-positive tumor and normal sample pairs indicates that great caution is needed in assuming the genomic normalcy of histologically benign appearing tissue within 1–3 cm of the invasive tumor margin.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplement Figure 1. Genome-wide allele-specific copy number variations from a 300 K MIP array platform analysis of the 11 HER2-positive tumor (T)-normal (N) sample pairs (# 1, 3, 4, 5, 6, 7, 11, 12, 13, 14, 19) in which each T sample demonstrates a HER2 amplicon at 17q12-21 but there are no genomic aberrations apparent in the matching N sample. Major (yellow) and minor (blue) allelic changes are shown, with estimated copy numbers > 1.5 considered significant gains. (TIFF 1520 kb)
Acknowledgments
This work was supported by National Institutes of Health grants U24-CA14358 (CCB), R21-CA155679 (CCB), P50-CA58207 (CCB, JWG), the Department of Defense grant W81XWH-11-1-0549-BC100597P1 (JWG), the U.S. Army Medical Research Acquisition Activity, 820 Chandler St., Fort Detrick, MD 21702-5014 is the awarding and administering acquisition office; and Hazel P. Munroe memorial funding (Buck Institute).We appreciate the valuable technical and bioinformatic assistance from Rachel Puckett (Buck Institute), Cathy Vitelli (Buck Institute Morphology & Imaging Core), Sandy DeVries (UCSF), Adam Olshen (UCSF), Victoria Carlton (Affymetrix) and Yuker Wang (Affymetrix).
Conflict of interest
The authors declare that they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
References
- 1.Carlson RW, Allred DC, Anderson BO, et al. Invasive breast cancer. J Natl Compr Cancer Netw. 2011;9:136–222. doi: 10.6004/jnccn.2011.0016. [DOI] [PubMed] [Google Scholar]
- 2.Shar JP, Rosen PP, Robbins GF. Pitfalls of local excision in the treatment of carcinoma of the breast. Surg Gynecol Obstet. 1973;136:721–725. [PubMed] [Google Scholar]
- 3.Morgenstern L, Friedmann NB. Breast cancer: the case against tylectomy. The factor of multicentricity. Prog Clin Cancer. 1978;76:113–122. [PubMed] [Google Scholar]
- 4.Rosen PP, Fracchia AA, Urban JA, Schottenfeld D, Robbins GF. “Residual” mammary carcinoma following simulated partial mastectomy. Cancer. 1975;35:739–747. doi: 10.1002/1097-0142(197503)35:3<739::AID-CNCR2820350329>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 5.Lagios MD, Westdahl PR, Rose MR. The concept and implications of multicentricity in breast carcinoma. Pathol Annu. 1981;16:83–102. [PubMed] [Google Scholar]
- 6.Holland R, Veling SHJ, Mravunac M, Hendriks JHCL. Histologic multifocality of Tis, T1–2 breast carcinomas—implications for clinical trials of breast conserving surgery. Cancer. 1985;56:979–990. doi: 10.1002/1097-0142(19850901)56:5<979::AID-CNCR2820560502>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 7.Morrow M, Harris JR, Schnitt SJ. Surgical margins in lumpectomy for breast cancer—bigger is not better. N Engl J Med. 2012;367:79–82. doi: 10.1056/NEJMsb1202521. [DOI] [PubMed] [Google Scholar]
- 8.Revesz E, Khan SA. What are safe margins of resection for invasive and in situ breast cancer? Oncology. 2011;25:890–895. [PubMed] [Google Scholar]
- 9.The Cancer Genome Atlas Research Network Comprehensive molecular portraits of human breast tumors. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowery AJ, Kell MR, Glynn RW, Kerin MJ, Sweeney KJ. Locoregional recurrence after breast cancer surgery: a systematic review by receptor phenotype. Breast Cancer Res Treat. 2012;133:831–841. doi: 10.1007/s10549-011-1891-6. [DOI] [PubMed] [Google Scholar]
- 11.Chai H, Brown RE. Field effect in cancer- an update. Ann Clin Lab Sci. 2009;39:331–337. [PubMed] [Google Scholar]
- 12.Heaphy CM, Griffith JK, Bisoffi M. Mammary field cancerization: molecular evidence and clinical importance. Breast Cancer Res Treat. 2009;118:229–239. doi: 10.1007/s10549-009-0504-0. [DOI] [PubMed] [Google Scholar]
- 13.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::AID-CNCR2820060515>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 14.Deng G, Lu Y, Zlotnikov G, Thor AD, Smith HS. Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science. 1996;274:2057–2059. doi: 10.1126/science.274.5295.2057. [DOI] [PubMed] [Google Scholar]
- 15.Trujillo KA, Heaphy CM, Mai M, et al. Markers of fibrosis and epithelial to mesenchymal transition demonstrate field cancerization in histologically normal tissue adjacent to breast tumors. Int J Cancer. 2011;129:1310–1321. doi: 10.1002/ijc.25788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roman-Perez E, Casbas-Hernandez P, Pirone JR, et al. Gene expression in extraturmoral microenvironment predicts clinical outcome in breast cancer patients. Breast Cancer Res. 2012;14:R51. doi: 10.1186/bcr3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardenbol P, Baner J, Jain M, et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 18.Hardenbol P, Yu F, Belmont J, et al. Highly multiplexed molecular inversion probe genotyping: over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 2005;15:269–275. doi: 10.1101/gr.3185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Moorhead M, Karlin-Neumann G, et al. Analysis of molecular inversion probe performance for allele copy number determination. Genome Biol. 2007;8:R246. doi: 10.1186/gb-2007-8-11-r246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Carlton VEH, Karlin-Neumann G, et al. High quality copy number and genotype data from FFPE samples using molecular inversion probe (MIP) microarrays. BMC Med Genomics. 2009;2:8. doi: 10.1186/1755-8794-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eppenberger-Castori S, Kueng W, Benz C, et al. Prognostic and predictive significance of ErbB-2 breast tumor levels measured by enzyme immunoassay. J Clin Oncol. 2001;19:645–656. doi: 10.1200/JCO.2001.19.3.645. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki J, Chen Y-Y, Scott GK, DeVries S, Chin K, Benz CC, Waldman FM, Hwang ES. Protein acetylation and histone deacetylase expression associated with malignant breast cancer progression. Clin Cancer Res. 2009;15:3163–3171. doi: 10.1158/1078-0432.CCR-08-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benz CC, Fedele V, Xu F, et al. Altered promoter usage characterizes monoallelic transcription arising with ERBB2 amplification in human breast cancers. Genes Chromosomes Cancer. 2006;45:983–994. doi: 10.1002/gcc.20364. [DOI] [PubMed] [Google Scholar]
- 24.Olshen AB, Es Venkatraman, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 25.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 26.Fridlyand J, Snijders AM, Ylstra B, et al. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 2006;6:96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fridlyand J, Dimitrov P (Release 2.10) aCGH: classes and functions for array comparative genomic hybridization data. http://www.bioconductor.org/packages/release/bioc/html/aCGH.html
- 28.Venkatraman ES, Olshen AB (Development 2.11) DNAcopy: a package for analyzing DNA copy data. http://www.bioconductor.org/packages/devel/bioc/html/DNAcopy.html
- 29.Sanborn JZ, Benz SC, Craft B, et al. The UCSC cancer genomics browser: update 2011. Nucleic Acids Res. 2011;39:D951–D959. doi: 10.1093/nar/gkq1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorvaldsdottir H, Robinson JT, Mesirov JP (2012) Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. doi:10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed]
- 31.Staaf J, Jonsson G, Ringner M, et al. High-resolution genomic and expression analyses of copy number alterations in HER2-amplified breast cancer. Breast Cancer Res. 2010;12:R25. doi: 10.1186/bcr2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlechter BL, Yang Q, Larson PS, et al. Quantitative DNA fingerprinting may distinguish new primary breast cancer from disease recurrence. J Clin Oncol. 2004;22:1830–1838. doi: 10.1200/JCO.2004.05.123. [DOI] [PubMed] [Google Scholar]
- 33.Bollet MA, Servant N, Neuvial P, et al. High-resolution mapping of DNA breakpoints to define true recurrences among ipsilateral breast cancers. J Natl Cancer Inst. 2008;100:48–58. doi: 10.1093/jnci/djm266. [DOI] [PubMed] [Google Scholar]
- 34.Ostrovnaya I, Begg CB. Testing clonal relatedness of tumors using array comparative genomic hybridization: a statistical challenge. Clin Cancer Res. 2010;16:1358–1367. doi: 10.1158/1078-0432.CCR-09-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Figure 1. Genome-wide allele-specific copy number variations from a 300 K MIP array platform analysis of the 11 HER2-positive tumor (T)-normal (N) sample pairs (# 1, 3, 4, 5, 6, 7, 11, 12, 13, 14, 19) in which each T sample demonstrates a HER2 amplicon at 17q12-21 but there are no genomic aberrations apparent in the matching N sample. Major (yellow) and minor (blue) allelic changes are shown, with estimated copy numbers > 1.5 considered significant gains. (TIFF 1520 kb)