Abstract
Mechanical forces are important signals for cell response and development, but detailed molecular mechanisms of force sensing are largely unexplored. The cytoskeletal protein filamin is a key connecting element between the cytoskeleton and transmembrane complexes such as integrins or the von Willebrand receptor glycoprotein Ib. Here, we show using single-molecule mechanical measurements that the recently reported Ig domain pair 20–21 of human filamin A acts as an autoinhibited force-activatable mechanosensor. We developed a mechanical single-molecule competition assay that allows online observation of binding events of target peptides in solution to the strained domain pair. We find that filamin force sensing is a highly dynamic process occurring in rapid equilibrium that increases the affinity to the target peptides by up to a factor of 17 between 2 and 5 pN. The equilibrium mechanism we find here can offer a general scheme for cellular force sensing.
Keywords: optical tweezers, mechanosensing
Sensing of signals is key for every cell to adapt and react to changing environmental conditions. Such signals can be chemical, optical, or even electrical in the case of nerve cells. In recent years, mechanical forces have been identified as important signals for cell development and cytokinesis. Mechanical signals have crucial implications for processes such as stem cell differentiation (1) or the remodeling of cardiac muscle tissue (2). Even though the sensing of mechanical forces has been postulated in various contexts, molecular mechanisms of force sensing have remained to a large degree elusive.
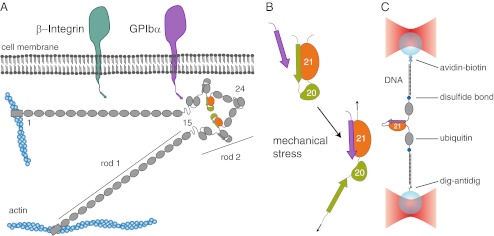
The cytoskeletal protein filamin is a dimeric actin cross-linker that acts as a signaling hub for various proteins (3). Two prominent examples for interaction partners are integrins (4–6) or glycoprotein Ib (GPIb), a constituent of a transmembrane complex found in platelets that binds to von Willebrand factor during blood clotting (7–9). Both the filamin–integrin and the filamin–GPIb interactions have been shown to be exposed to mechanical forces in living cells (10, 11). The filamin monomer consists of an N-terminal actin-binding domain and a sequence of 24 Ig domain repeats that form the filamin rod (Fig. 1A). Within the rod 2 region (Ig domains 16–23; Fig. 1A), filamin can interact with transmembrane protein complexes, thus establishing a mechanical connection between the cytoskeleton and the extracellular side. The major interaction sites within rod 2 are the odd-numbered domains, specifically domains 17, 19, and 21. The transmembrane interaction partners bind to these domains through a terminal peptide sequence that forms an additional β-strand extending the β-sheet structure of the domain (12, 13) (Fig. 1B).
Fig. 1.
Interaction of filamin with transmembrane proteins. (A) Schematics of the domain arrangement of filamin and its interaction with the transmembrane proteins integrin and GPIbα. Filamin consists of 24 Ig domains that dimerize at domain 24 and carry an N-terminal actin-binding domain. In its rod 2 region (Ig domains 16–24), domains 16–21 (FLNa16-21) arrange in pairs that contain binding sites for integrins (dark green) and GPIbα (purple). The domain pair FLNa20-21 is highlighted in orange and green. (B) Schematics of the autoinhibition mechanism of the domain pair FLNa20-21. The A-strand of FLNa20 binds to FLNa21, thus autoinhibiting its interaction with the transmembrane proteins (upper cartoon). It has been hypothesized that mechanical stress releases the autoinhibition and promotes interaction with the targets (lower cartoon). (C) Schematics of the single-molecule assay using a double-beam optical trap (see text and SI Materials and Methods).
In contrast to the linear arrangement of domains found in many structural proteins like titin, fibronectin, or rod 1 of filamin, the Ig domains 18–21 of filamin’s rod 2 are arranged in pairs (14, 15). In those pairs, the A-strand of the even-numbered domains is not integrated into the domain structure but binds to the subsequent odd-numbered domain, thus inhibiting the interaction of this domain with its peptide ligand (Fig. 1A and Fig. S1). It has been proposed that this special arrangement into domain pairs effects a force-sensing mechanism in filamin (Fig. 1B) (14, 16). Recent in vitro studies of mechanically strained filamin–cross-linked actin networks have provided further support for a potential force-sensing role of filamin (17).
In the present study, we have designed and performed single-molecule mechanical measurements that provide direct evidence for a force-sensing mechanism of human filamin A. We show quantitatively, at the single-molecule level, how force increases the binding of interaction partners by shifting the conformational equilibrium of the autoinhibited filamin domain pair 20–21 (FLNa20-21).
Results
Filamin Domain 21 Interaction with Different Tethered Target Peptides.
In a first set of experiments, we investigated the uninhibited mechanical binding strength of domain 21 to different interaction partners: the C-terminal peptide of GPIb’s α-chain (GPIbα), the integrin β7 cytoplasmic tail peptide (ITβ7), as well as the filamin-interacting peptide of the integrin regulator migfilin (Mig) (18, 19). To this end, we fused the respective interacting peptides directly to the N terminus of an isolated domain 21 of filamin (FLNa21) with a 6-aa residues spacer allowing the necessary flexibility (SI Materials and Methods). Force application to the molecular constructs was achieved by tethering the protein termini to 180-nm-long double-stranded DNA molecules through disulfide bonds. The ends of the DNA were functionalized with biotin and digoxigenin, respectively. This allowed attachment to 1-μm-diameter silica beads that were manipulated in a dual beam optical trap (Fig. 1C; for details, see SI Materials and Methods) (20–22).
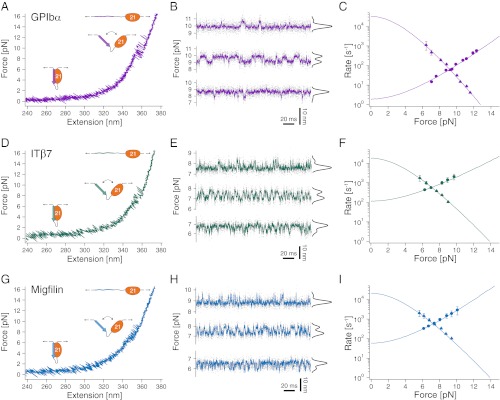
Upon stretching the DNA–protein construct, the force rises steeply beyond extensions of 320 nm, when the DNA is almost fully elongated (Fig. 2A). At forces between 7 and 12 pN, rapid near-equilibrium fluctuations of binding and unbinding of the GPIbα peptide can be observed. The contour length gain during this transition (ΔL = 12.5 ± 0.5 nm) is in excellent agreement with the expected length gain if the 26 aa residues of the bound peptide detach and are fully stretched (Table S1). To obtain a precise analysis of the force-dependent binding/unbinding kinetics of the peptide, we performed measurements where the trap centers were held at a constant separation, hence imposing a constant average force bias on the fluctuating molecule. Because only the separation of the trapping potentials is held constant, the force varies between the closed and open conformation. We define force bias as the mean of the force acting on the closed/bound and open/unbound state. This allowed us to observe fluctuations between bound (high force) and unbound (low force) states for several minutes at different biasing forces (Fig. 2B). At low forces, the molecule dwells predominantly in the bound state with few rapid excursions into the unbound state (Fig. 2B, bottom trace). With increasing force, the equilibrium (Fig. 2B, black population distributions) shifts to the unbound state together with altered kinetics (Fig. 2B, top two traces). The force-dependent population shift provides a direct measure for the free energy of binding in the absence of load of 10.6 kBT (Fig. S2 and SI Materials and Methods). A dwell time analysis using a hidden Markov model (HMM) (22, 23) yields the force-dependent kinetics of GPIbα binding/unbinding (Fig. 2C). Unbinding rates (circles) increase with force, whereas binding rates (triangles) drop with force. For extrapolation of the binding/unbinding branches to zero force, we used a model that considers the compliance of the unfolded peptide leading to a curvature in the branches (21). The slopes of the binding/unbinding branches reflect the distance of the transition state from the unbound and bound state, respectively. The sum of those distances (12.3 nm, measured in contour length) as obtained from the fits, is very close to the total contour length of the peptide (12.5 nm), which supports the validity of the analysis. The fast binding rate at zero load of 34,000 s−1 reflects the effectively high concentration of the ligand due to tethering. For the unbinding rate, we find 1.9 s−1 (Table S2).
Fig. 2.
Interaction of filamin with tethered peptides. (A) Force extension trace of the interaction between the target peptide of GPIbα and FLNa21 (GPIbα-FLNa21). At forces around 10 pN, the target peptide rapidly fluctuates between bound and unbound conformations and stays permanently unbound at higher loads (>12 pN). (B) Equilibrium traces obtained at three different biasing forces. At high loads (top trace), the probability (black histogram) is shifted to the unbound state, whereas at decreasing loads, the bound state becomes more and more populated (bottom two traces). (C) Opening (circles) and closing (triangles) rates as a function of force. The solid line is an extrapolation of the rates to zero-load taking into account the compliance of all mechanical elements in the construct (SI Materials and Methods). (D) Force extension trace of the interaction between the target peptide of β7-integrin and FLNa21 (ITβ7-FLNa21). (E) Equilibrium traces of ITβ7-FLNa21 obtained at three different biasing forces (compare B). (F) Opening (circles) and closing (triangles) rates as a function of force (compare C). (G) Force extension trace of the interaction between the target peptide of migfilin and FLNa21 (Mig-FLNa21). (H) Equilibrium traces of Mig-FLNa21 obtained at three different biasing forces (compare B and E). (I) Opening (circles) and closing (triangles) rates as a function of force (compare C and F).
In comparison with GPIbα, the ITβ7 ligand exhibits a significantly lower unbinding force (Fig. 2D) as well as free energy of binding (5.8 kBT; Fig. S2). The major difference in kinetics (Fig. 2 E and F) results from a much faster unbinding rate of 120 s−1 at zero load, whereas the binding rates are similar to the ones of GPIbα (Table S2). Since the unbinding lengths of the tethered ligands and hence the effective concentrations for both peptides are identical, the similar binding rates indicate that rebinding speed is not influenced by details of the sequence. This is further supported by our data of migfilin (Fig. 2 G–I) that exhibits very similar binding rates. The unbinding rates and the binding free energy of migfilin (6.5 kBT) are in between those of GPIbα and ITβ7 (Fig. S2). GPIbα exhibits by far the strongest binding to filamin of all peptides investigated. This finding is particularly important because platelet integrity critically depends on the mechanical strength of this bond once attached to von Willebrand factor during blood clotting (11). It has been shown that GPIbα binds von Willebrand factor on the extracellular side with forces similar to those we find for intracellular filamin binding (24). This could provide a stable mechanism to transmit force from the extracellular space to the actin cytoskeleton. The higher free energies of binding and interaction forces of migfilin relative to ITβ7 are consistent with migfilin’s role in competing off the cytoplasmic tail of β-integrins from filamin during talin-mediated integrin activation (18, 19).
Single-Molecule Mechanical Competition Assay: FLNa21 Interaction with Ligands in Solution.
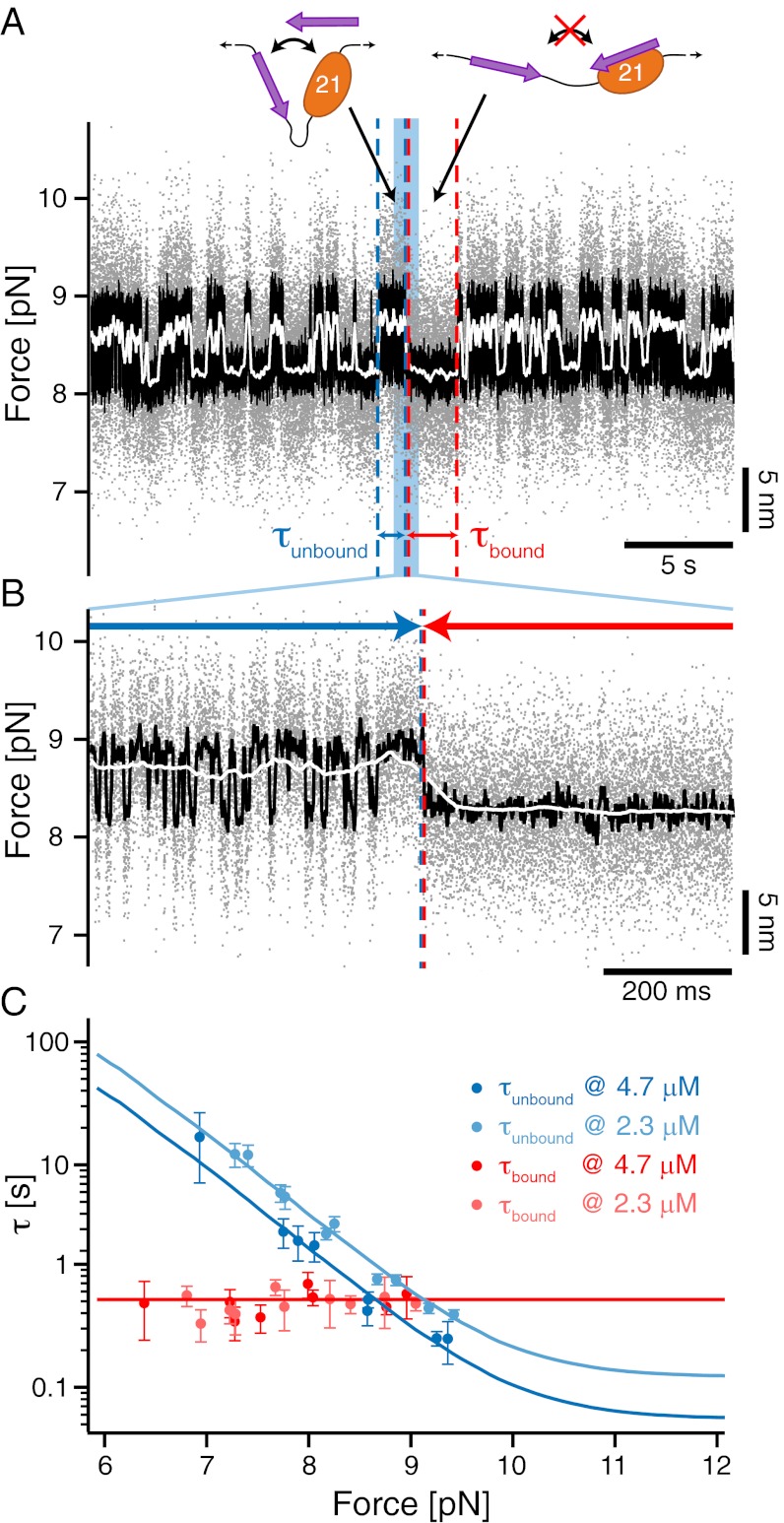
In their physiological context, the membrane protein tails are not tethered to the filamin domains. Even though our assay gives valuable information about the unbinding kinetics under load, measuring the free binding and unbinding rates of the ligands requires a different experimental approach. Fluorescent single-molecule methods cannot be used at the high concentrations necessary for observing binding (up to 100 μM) due to the associated high background. We therefore developed a mechanical single-molecule assay that allows direct observation of binding/unbinding events of freely diffusing ligands at those concentrations. To this end, we observed the opening and closing fluctuations of a tethered GPIbα-FLNa21 construct in the presence of freely diffusing GPIbα peptide in solution (Fig. 3A). Under load, the tethered ligand will constantly fluctuate between bound and unbound states as shown above (Fig. 3A, left cartoon). If the construct is in an open conformation, there will be a competition between the tethered ligand rebinding and a free ligand binding from solution. In the latter case (Fig. 3A, right cartoon), rebinding of the tethered ligand will be suppressed and the opening/closing fluctuations should vanish until the bound ligand spontaneously dissociates. A sample trace is shown in Fig. 3A where fluctuating states (high SD) can be clearly distinguished from mechanically quenched states (low SD). A zoom into this trace (Fig. 3B) shows that in the fluctuating state (Left), the construct undergoes rapid opening and closing transitions, whereas in the quenched state (Right), the construct dwells in the open conformation (see Fig. S3 for sample traces at different forces). The dwell times in the fluctuating state (τunbound) as well as in the quenched state (τbound) now provide direct information about the binding kinetics of the ligand from solution. Although 1/τbound directly yields the off-rate, 1/τunbound depends on both the solution concentration of the ligand and the applied force as follows:
Fig. 3.
Single-molecule mechanical competition assay to study peptide binding from solution. (A) Time traces of opening and closing of the GPIbα-FLNa21 construct held at a force bias of 8.5 pN in the presence of 2.3 μM GPIbα peptide in solution. The colored traces correspond to the 20-kHz data (gray), moving average filtered with 2.5 ms (black) and 50 ms (white) time window. Apparent high-SD regions with lifetimes 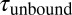 in the black and gray traces are interrupted by low-SD regions with lifetimes
in the black and gray traces are interrupted by low-SD regions with lifetimes 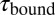 . (B) A zoom into the blue region shows the transition between a high-SD (rapid opening and closing cycles of the tethered construct) and low-SD region (blocked fluctuations due to competitive peptide binding from solution). (C) Dependence of the bound and unbound lifetimes as a function of applied force and solution concentration. As expected for binding from solution,
. (B) A zoom into the blue region shows the transition between a high-SD (rapid opening and closing cycles of the tethered construct) and low-SD region (blocked fluctuations due to competitive peptide binding from solution). (C) Dependence of the bound and unbound lifetimes as a function of applied force and solution concentration. As expected for binding from solution, 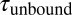 depends on the opening probability of the tethered construct and hence the applied force, as well as the solution concentration, whereas
depends on the opening probability of the tethered construct and hence the applied force, as well as the solution concentration, whereas 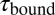 is independent (see text and SI Materials and Methods).
is independent (see text and SI Materials and Methods).
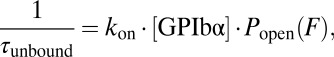 |
where 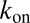 is the pseudo first-order on-rate,
is the pseudo first-order on-rate, 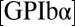 is the concentration of ligand, and
is the concentration of ligand, and  is the forcedependent probability for the tethered ligand to be in the open conformation (Fig. S2). We measured τbound and τunbound as a function of force and at two different ligand concentrations (Fig. 3C). As expected, τunbound decreases with force, because the probability for finding the tethered construct in an open conformation increases. Moreover, τunbound decreases with increasing solution concentration. From the fits to the concentration-dependent τunbound curves we obtain a pseudo first-order on-rate for GPIbα binding of kon = 3.7 (μM*s)−1. It is important to note that the shape of the fit curves is predetermined by
is the forcedependent probability for the tethered ligand to be in the open conformation (Fig. S2). We measured τbound and τunbound as a function of force and at two different ligand concentrations (Fig. 3C). As expected, τunbound decreases with force, because the probability for finding the tethered construct in an open conformation increases. Moreover, τunbound decreases with increasing solution concentration. From the fits to the concentration-dependent τunbound curves we obtain a pseudo first-order on-rate for GPIbα binding of kon = 3.7 (μM*s)−1. It is important to note that the shape of the fit curves is predetermined by  , which is not a fit parameter but measured directly (Fig. S2). The off-rate (1/τbound) is independent of concentration as well as applied force and we find
, which is not a fit parameter but measured directly (Fig. S2). The off-rate (1/τbound) is independent of concentration as well as applied force and we find 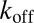 = 1.8 s−1. We note that this value is very close to the zero-force value that we extrapolated from the tethered construct in Fig. 2C. This again confirms the validity of the extrapolation methods used above.
= 1.8 s−1. We note that this value is very close to the zero-force value that we extrapolated from the tethered construct in Fig. 2C. This again confirms the validity of the extrapolation methods used above.
The new competition assay now allows to determine the force-free on- and off-rates for all three different peptides (Table S3 and Fig. S4 A and B). Even though the binding kinetics of those ligands have not been measured before, the equilibrium binding constants (KD) of the three peptides calculated from the kinetics agree well with bulk measurements (Table S3) (25). It is interesting to note that with KD values in the micromolar range and above, as well as lifetimes below a second, all those interactions are very dynamic.
Mechanical Response of the Autoinhibition of FLNa20-21.
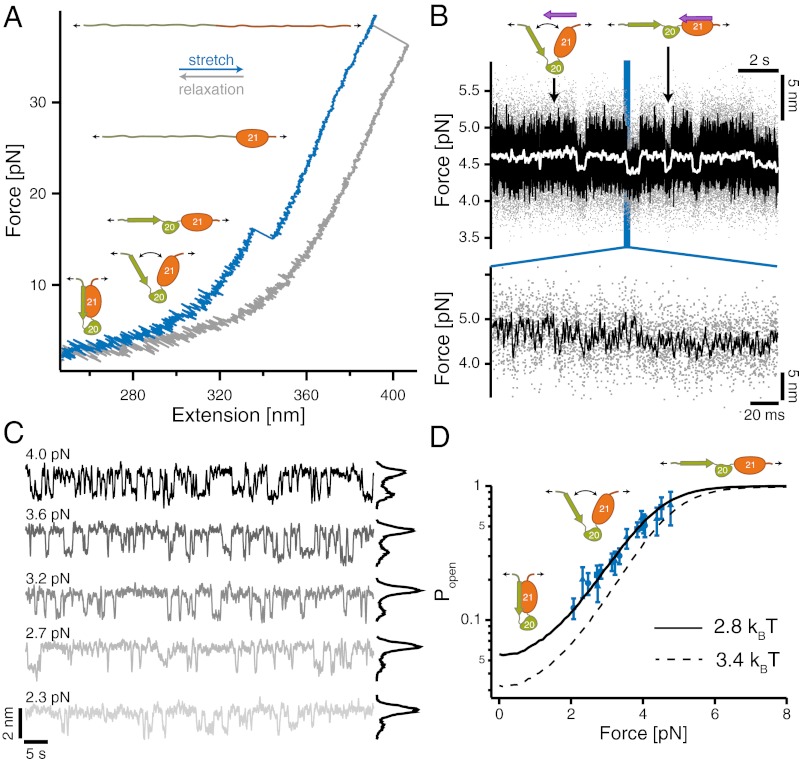
In a next set of experiments, we used the single-molecule mechanical competition assay to investigate the force-sensing mechanism of the domain pair FLNa20-21 (16). Can we see an autoinhibited conformation of the FLNa20-21 domain pair that is relieved with increasing loads? A force vs. extension curve of this domain pair is shown in Fig. 4A. Upon stretching (blue trace), we observe two major unfolding peaks around 15 and 37 pN, respectively. The lower peak exhibits a contour length change of ΔL = 17.7 ± 0.3 nm, and the higher peak has a ΔL of 28.8 ± 0.5 nm. The lower force peak can be directly associated with unfolding of domain 20, whereas the high peak reflects unfolding of domain 21. This assignment is confirmed by measurements with the isolated domains 20 and 21 (Fig. S5). Compared with other Ig domains of filamin (26–28), domain 20 appears much more mechanically labile.
Fig. 4.
Filamin domain pairs act as a precisely tuned mechano-sensor. (A) Low-resolution stretch (blue) and relax (gray) trace of FLNa20-21. At forces around 15 pN, FLNa20 unfolds, and at higher loads exceeding 30 pN, FLNa21 unfolds. Opening of the domain pair and release of the autoinhibition is not visible at this experimental resolution. (B) (Upper) Competition assay of domain pair opening in the presence of 2.7 μM GPIbα peptide in solution observed at loads of 4.5 pN (color scheme as in Fig. 3A). High-SD regions (opening–closing fluctuations) are interrupted with low-SD regions where bound peptide blocks fluctuations. (Lower) Zoom into a transition region. (C) Force-dependent binding of peptide from solution observed at biasing forces from 2.3 to 4.0 pN. From low to high loads, the binding probability (black histograms) increases constantly. (D) Force-dependent gating characteristics of the force-sensing domain pair as obtained from the force and concentration-dependent dwell times of Fig. S4C (blue data points; black solid line shows global fit). The three symbols (triangle, square, and circle) denote three different solution concentrations. The dashed black line is an independent measure of the force-dependent opening probability as obtained from the HMM analysis of the fluctuating state where no ligand from solution is bound (Fig. S6B).
A clear signal for opening the autoinhibited conformation, in which the A-strand of domain 20 is attached to domain 21, cannot be observed in the low-resolution trace of Fig. 4A. The single-molecule mechanical competition assay described above (Fig. 3) now offers a possibility to study the force-dependent relief of autoinhibition and hence the force-sensing properties of the domain pair together with its force-dependent binding rate of ligands from solution. At a solution concentration of 2.7 μM GPIbα, a force-dependent change in the fluctuation pattern of the domain pair can be observed when held at a biasing force of 4.5 pN (Fig. 4B). Again, rapidly fluctuating high-SD regions are interrupted by quenched low-SD dwells. The quenched levels (e.g. Fig. 4B, right arrow) exhibit exactly the same lifetimes of GPIbα unbinding (τbound = 520 ms; see Fig. S4D) as in Fig. 3C. The high-SD regions in the high-bandwidth trace (left arrow) also reflect the same concentration dependence and pseudo first-order on-rate as before (Fig. S4C), which confirms the unbinding and rebinding of GPIbα peptides from solution. A zoom into the high-SD region (Fig. 4B, Inset) reveals that the molecular construct undergoes rapid transitions between a closed and an open form. Force directly affects the probability of GPIbα binding to the domain pair from solution. This can be seen in the strongly smoothed curves of Fig. 4C, where increasing load increases the number of binding events observed. It is important to note that the force conditions shown were measured using the same molecule. The length change we observe between the closed and open conformation is 1.4 nm at a biasing force of 3.2 pN. From this length change, we can calculate a total contour length difference between the closed and open conformation of 14.5 ± 1.1 nm. This value is very close to the calculated contour length gain of 16.8 nm of a conformational change where the A-strand of FLNa20 detaches from FLNa21 and the domain pair opens up, as suggested by the crystal structure (14). Combining the force and concentration-dependent binding kinetics of GPIbα to FLNa20-21 allows reconstruction of the force-dependent gating-characteristics of the domain pair. In Fig. 4D, three data sets at different peptide concentrations were normalized by their maximal peptide binding rates, giving a direct measurement of the force-dependent opening probabilities (for details, see SI Materials and Methods). The solid line represents the globally fitted opening probability with a force of half-maximal opening F1/2 = 3.9 pN and a total free energy for opening of 2.8 kBT. Independently, the force-dependent opening probability of FLNa20-21 was obtained (Fig. 4D, dashed line) from dwell time analysis of the unquenched regions using HMM analysis (Fig. S6 and SI Materials and Methods).
Discussion
The force-gating characteristic we find for FLNa20-21 is tuned to surprisingly low forces. For comparison, the putative force-sensing titin-kinase domain acting in the high-force environment of muscle was reported to undergo force activation at around 40 pN (15, 29). However, in vivo and in vitro studies of the adhesion forces of integrins to fibronectin as well as the direct force measurements within load-bearing focal adhesion of living cells have indicated that a relevant force scale within cells may be much lower: on the order of a few piconewtons (30–32). The force sensing of filamin seems to be tuned exactly to this force range. Another important aspect of the force sensing of filamin is its smooth opening characteristic. Unlike the common description of force-induced structural changes within proteins as all-or-none events far from equilibrium, we find that filamin shifts its opening probability in a force-dependent and gradual manner from closed to open as indicated in the curve of Fig. 4D. This ensures a precise and well-controlled gating curve even though the involved binding energies lie below 4 kBT. It has been argued that an autoinhibition of filamin seems unlikely in in vitro experiments because it could be shown that an excess of ligand alone is able to convert the filamin domain pairs from a closed to an open conformation (33, 34). However, this stood in apparent contrast to earlier reports that the isolated domain 21 binds stronger to ligands than the domain pairs (14) as well as to binding studies on strained filamin–cross-linked actin networks (17). Our results are now able to reconcile this contradiction. Opening of the domain pair under load is, unlike a mechanical switch, a gradual process that shifts the binding constants by up to a factor of 17, and thus modulates affinity in a force-dependent manner.
A further important aspect of the force transmission through filamin is the transient and dynamic nature of the bonds involved. We find that even the strongest interaction of GPIbα with rod 2 domains of filamin, which critically determines platelet integrity under shear flow forces (11), is transient with a lifetime of less than a second even in the absence of force.
How can a cell sustain mechanical loads applied over much longer timescales? A view of the complete force transmission network helps to resolve this issue. Filamin does not offer only a single binding site to transmembrane receptors, but instead rod 2 has three reported domain pairs (Fig. 5). Therefore, in a dimer of filamin, many interaction sites act in concert leading to a strong avidity effect. Because bond energies are additive, the lifetimes will increase exponentially, which leads to long-lasting multiple bonds. Interestingly, the binding to domain 17 is likely not autoinhibited (15). The constitutively active domain 17 may be important for transiently recruiting filamin to the membrane receptor. The subsequent build-up of mechanical tension can then relieve the autoinhibition of the other domains promoting strong anchoring. The necessity of multiple parallel bonds for stable anchoring is in accord with the idea that the multiple binding sites of filamin can also induce clustering of transmembrane receptors (10, 25). Alternatively, filamin interactions may be substituted or stabilized by other adaptor molecules. In the case of GPIbα, a candidate for such a molecule is 14-3-3ζ, which is required for firm adhesion, and one of the 14-3-3ζ binding sites on GPIbα overlaps with that of filamin (35). An analogous function could be taken by talin in the case of integrin-based adhesion (14, 36).
Fig. 5.
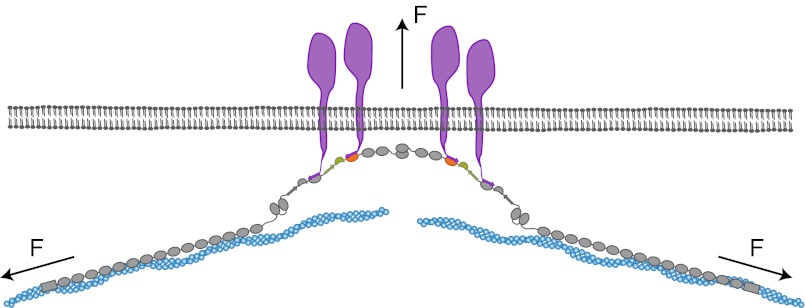
Model of the force-dependent binding and clustering of membrane receptors to filamin. Mechanical force in a strained cytoskeleton will lead to domain pair opening in the rod 2 segment of filamin, allowing the binding to membrane receptors. The simultaneous interaction with many domain pairs will significantly stabilize the cytoskeleton–membrane interaction and potentially induce clustering of receptors.
In conclusion, we have shown that the rod 2 domains of filamin act as force-sensing domains that react to the small cytoskeletal forces acting in cells. The dynamic equilibrium switching of this force sensor may provide a prototype for other force sensors found in living systems.
Materials and Methods
The domain pair 20–21 of human filamin A was genetically inserted between two ubiquitins with terminal cysteines that served as spacers. For the tethered peptide construct, domain 20 was replaced by the filamin-binding region of GPIbα/ITβ7/migfilin, linked to FLNa21 via an additional 6-aa glycine–serine spacer. To the terminal cysteines of the construct thiol-modified DNA handles of a length of 180 nm were attached. To create a dumbbell geometry, the other biotin/digoxigenin functionalized end was attached to micrometer-sized streptavidin/anti-digoxigenin silica beads (Fig. 1B). The beads were trapped in the foci of a custom-built dual beam optical tweezers setup and subjected to stretch-and-relax cycles or a constant force bias with fixed trap positions. All measurements were performed in PBS (10 mM phosphate buffer, 2.7 mM potassium chloride, and 137 mM sodium chloride, pH 7.4). For the single-molecule mechanical competition assay, peptides with the same sequence as the tethered ones were added into the solution.
Transitions between states were detected using a HMM analysis on the unfiltered raw data of the difference signal of the two traps. Complete descriptions of the methods used are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft Grant FOR 1352 and Sonderforschungsbereich 863 B7 (to M.R.) and Academy of Finland Grant 138327 (to J.Y.). M.R. acknowledges financial support through an instrument grant of the Institute for Advanced Study of Technische Universität München.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211274109/-/DCSupplemental.
References
- 1.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 2.Hoshijima M. Mechanical stress-strain sensors embedded in cardiac cytoskeleton: Z disk, titin, and associated structures. Am J Physiol Heart Circ Physiol. 2006;290(4):H1313–H1325. doi: 10.1152/ajpheart.00816.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura F, Stossel TP, Hartwig JH. The filamins: Organizers of cell structure and function. Cell Adhes Migr. 2011;5(2):160–169. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glogauer M, et al. The role of actin-binding protein 280 in integrin-dependent mechanoprotection. J Biol Chem. 1998;273(3):1689–1698. doi: 10.1074/jbc.273.3.1689. [DOI] [PubMed] [Google Scholar]
- 5.Gehler S, et al. Filamin A-beta1 integrin complex tunes epithelial cell response to matrix tension. Mol Biol Cell. 2009;20(14):3224–3238. doi: 10.1091/mbc.E08-12-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang P, Campbell ID. Integrin binding immunoglobulin type filamin domains have variable stability. Biochemistry. 2008;47(42):11055–11061. doi: 10.1021/bi8011466. [DOI] [PubMed] [Google Scholar]
- 7.Ruggeri ZM, Orje JN, Habermann R, Federici AB, Reininger AJ. Activation-independent platelet adhesion and aggregation under elevated shear stress. Blood. 2006;108(6):1903–1910. doi: 10.1182/blood-2006-04-011551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nesbitt WS, et al. A shear gradient-dependent platelet aggregation mechanism drives thrombus formation. Nat Med. 2009;15(6):665–673. doi: 10.1038/nm.1955. [DOI] [PubMed] [Google Scholar]
- 9.Huizinga EG, et al. Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain. Science. 2002;297(5584):1176–1179. doi: 10.1126/science.107355. [DOI] [PubMed] [Google Scholar]
- 10.Lynch CD, et al. Filamin depletion blocks endoplasmic spreading and destabilizes force-bearing adhesions. Mol Biol Cell. 2011;22(8):1263–1273. doi: 10.1091/mbc.E10-08-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cranmer SL, et al. High shear-dependent loss of membrane integrity and defective platelet adhesion following disruption of the GPIbα-filamin interaction. Blood. 2011;117(9):2718–2727. doi: 10.1182/blood-2010-07-296194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura F, et al. The structure of the GPIb-filamin A complex. Blood. 2006;107(5):1925–1932. doi: 10.1182/blood-2005-10-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiema T, et al. The molecular basis of filamin binding to integrins and competition with talin. Mol Cell. 2006;21(3):337–347. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Lad Y, et al. Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. EMBO J. 2007;26(17):3993–4004. doi: 10.1038/sj.emboj.7601827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heikkinen OK, et al. Atomic structures of two novel immunoglobulin-like domain pairs in the actin cross-linking protein filamin. J Biol Chem. 2009;284(37):25450–25458. doi: 10.1074/jbc.M109.019661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pentikäinen U, Ylänne J. The regulation mechanism for the auto-inhibition of binding of human filamin A to integrin. J Mol Biol. 2009;393(3):644–657. doi: 10.1016/j.jmb.2009.08.035. [DOI] [PubMed] [Google Scholar]
- 17.Ehrlicher AJ, Nakamura F, Hartwig JH, Weitz DA, Stossel TP. Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature. 2011;478(7368):260–263. doi: 10.1038/nature10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ithychanda SS, et al. Migfilin, a molecular switch in regulation of integrin activation. J Biol Chem. 2009;284(7):4713–4722. doi: 10.1074/jbc.M807719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das M, Ithychanda SS, Qin J, Plow EF. Migfilin and filamin as regulators of integrin activation in endothelial cells and neutrophils. PLoS One. 2011;6(10):e26355. doi: 10.1371/journal.pone.0026355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309(5743):2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 21.Gebhardt JCM, Bornschlögl T, Rief M. Full distance-resolved folding energy landscape of one single protein molecule. Proc Natl Acad Sci USA. 2010;107(5):2013–2018. doi: 10.1073/pnas.0909854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stigler J, Ziegler F, Gieseke A, Gebhardt JCM, Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334(6055):512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 23.Stigler J, Rief M. Hidden Markov analysis of trajectories in single-molecule experiments and the effects of missed events. Chemphyschem. 2012;13(4):1079–1086. doi: 10.1002/cphc.201100814. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Zhang C-Z, Zhang X, Springer TA. A mechanically stabilized receptor-ligand flex-bond important in the vasculature. Nature. 2010;466(7309):992–995. doi: 10.1038/nature09295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ithychanda SS, et al. Identification and characterization of multiple similar ligand-binding repeats in filamin: Implication on filamin-mediated receptor clustering and cross-talk. J Biol Chem. 2009;284(50):35113–35121. doi: 10.1074/jbc.M109.060954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuike S, Ito T, Yamazaki M. Mechanical unfolding of single filamin A (ABP-280) molecules detected by atomic force microscopy. FEBS Lett. 2001;498(1):72–75. doi: 10.1016/s0014-5793(01)02497-8. [DOI] [PubMed] [Google Scholar]
- 27.Schwaiger I, Kardinal A, Schleicher M, Noegel AA, Rief M. A mechanical unfolding intermediate in an actin-crosslinking protein. Nat Struct Mol Biol. 2004;11(1):81–85. doi: 10.1038/nsmb705. [DOI] [PubMed] [Google Scholar]
- 28.Chen H, et al. Differential mechanical stability of filamin A rod segments. Biophys J. 2011;101(5):1231–1237. doi: 10.1016/j.bpj.2011.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puchner EM, et al. Mechanoenzymatics of titin kinase. Proc Natl Acad Sci USA. 2008;105(36):13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang G, Giannone G, Critchley DR, Fukumoto E, Sheetz MP. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature. 2003;424(6946):334–337. doi: 10.1038/nature01805. [DOI] [PubMed] [Google Scholar]
- 31.del Rio A, et al. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323(5914):638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grashoff C, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466(7303):263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ithychanda SS, Qin J. Evidence for multisite ligand binding and stretching of filamin by integrin and migfilin. Biochemistry. 2011;50(20):4229–4231. doi: 10.1021/bi2003229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pentikäinen U, et al. Assembly of a filamin four-domain fragment and the influence of splicing variant-1 on the structure. J Biol Chem. 2011;286(30):26921–26930. doi: 10.1074/jbc.M110.195958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan Y, et al. Identification of a novel 14-3-3zeta binding site within the cytoplasmic domain of platelet glycoprotein Ibalpha that plays a key role in regulating the von Willebrand factor binding function of glycoprotein Ib-IX. Circ Res. 2009;105(12):1177–1185. doi: 10.1161/CIRCRESAHA.109.204669. [DOI] [PubMed] [Google Scholar]
- 36.Roca-Cusachs P, Gauthier NC, Del Rio A, Sheetz MP. Clustering of alpha(5)beta(1) integrins determines adhesion strength whereas alpha(v)beta(3) and talin enable mechanotransduction. Proc Natl Acad Sci USA. 2009;106(38):16245–16250. doi: 10.1073/pnas.0902818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.