Abstract
Domestication of wild boar (Sus scrofa) and subsequent selection have resulted in dramatic phenotypic changes in domestic pigs for a number of traits, including behavior, body composition, reproduction, and coat color. Here we have used whole-genome resequencing to reveal some of the loci that underlie phenotypic evolution in European domestic pigs. Selective sweep analyses revealed strong signatures of selection at three loci harboring quantitative trait loci that explain a considerable part of one of the most characteristic morphological changes in the domestic pig—the elongation of the back and an increased number of vertebrae. The three loci were associated with the NR6A1, PLAG1, and LCORL genes. The latter two have repeatedly been associated with loci controlling stature in other domestic animals and in humans. Most European domestic pigs are homozygous for the same haplotype at these three loci. We found an excess of derived nonsynonymous substitutions in domestic pigs, most likely reflecting both positive selection and relaxed purifying selection after domestication. Our analysis of structural variation revealed four duplications at the KIT locus that were exclusively present in white or white-spotted pigs, carrying the Dominant white, Patch, or Belt alleles. This discovery illustrates how structural changes have contributed to rapid phenotypic evolution in domestic animals and how alleles in domestic animals may evolve by the accumulation of multiple causative mutations as a response to strong directional selection.
Several subspecies of wild boars have contributed to the development of the domestic pig, and there is a considerable divergence time between them (on the order of 1 million y) (1–4). The domestication process has, to a large extent, been going on in parallel in Asia and Europe. For instance, black coat color in Asian and European domestic pigs is caused by independent missense mutations in the melanocortin 1 receptor (MC1R) that occurred on haplotypes originating from the Asian and European wild boar, respectively (5). Since animal breeding became more organized in the 18th century, the selection goals in pigs have evolved in response to demand. The early focus on selection for fatness was driven by demand for energy-rich food and tallow for candles. In contrast, there has been very strong selection for lean growth (high protein and low fat content) during the last 60 y, driven by the demand for reduced caloric intake in modern society. Several mutations with major effects on lean growth have already been identified, including a missense mutation in Ryanodine receptor 1 (RYR1) (6), a missense mutation in PRKAG3 encoding the gamma 3 subunit of AMP-activated protein kinase (7), and a single base change at a repressor binding site in intron 3 of insulin-like growth factor 2 (IGF2) (8).
Here we used the pig draft genome sequence (Sscrofa10.2) (4) and whole-genome resequencing to reveal loci that have been under selection during and since pig domestication. We searched for selective sweeps and genetic variants showing marked allele frequency differences between pig and wild boar populations. The results are based on the combined analyses of two datasets: (i) mate pair reads from eight different pools of pigs and wild boars sequenced to ∼5× coverage/pool, and (ii) paired-end fragment reads (100 + 100 bp) from 37 individual pigs and 11 wild boars, each sequenced to ∼10× coverage (Table 1).
Table 1.
Samples used for whole-genome shotgun sequencing
| Population | Type | n | Sex |
| Dataset i | |||
| Large White Uppsala | ED | 8 | F |
| Danish Landrace | ED | 15 | F |
| Danish Duroc | ED | 15 | F |
| Danish Hampshire | ED | 15 | F |
| F2 intercross | F2 | 14 | M/F |
| Large White Roslin | ED | 10 | F |
| Meishan | AD | 20 | F |
| European wild boar | EWB | 20 | M/F |
| Dataset ii | |||
| Large White | ED | 14 | 2 M/12 F |
| Hampshire | ED | 2 | 2 M |
| Pietrain | ED | 5 | 2 M/3 F |
| Duroc | ED | 4 | 4 M |
| Landrace | ED | 5 | 1 M/4 F |
| European wild boar | EWB | 6 | 4 M/2 F |
| Asian wild boar | AWB | 5 | 3 M/2 F |
| Meishan | AD | 4 | 2 M/2 F |
| Xiang | AD | 2 | 2 F |
| Jianquhai | AD | 1 | 1 F |
| Sus scrofa (Sumatra) | OG | 2 | 1 M/1 F |
| Sus barbatus | OG | 1 | 1 M |
| Sus verrucosus | OG | 1 | 1 M |
| Sus cebifrons | OG | 1 | 1 F |
| Sus celebensis | OG | 1 | 1 F |
| Phacochoerus africanus | OG | 1 | 1 F |
Dataset i consisted of pooled samples sequenced, SOLiD mate pair reads. Gap sizes were in the range 1,010–1,430 bp. Dataset ii consisted of samples sequenced individually, Illumina paired-end reads. ED, European domestic; AD, Asian domestic; EWB, European wild boar, AWB, Asian wild boar; OG, outgroup; F2, F2 progeny from a Large White/wild boar intercross.
Results
Selective Sweep Analysis.
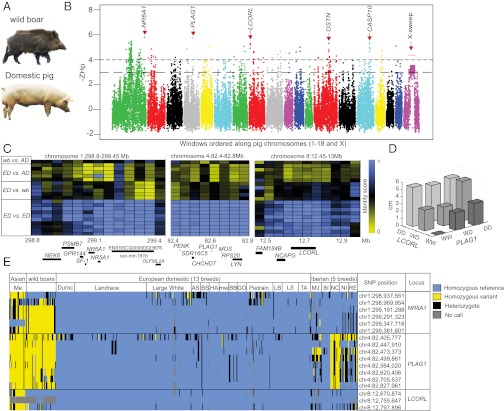
We analyzed the pooled sequences for selective sweeps in European domestic pigs by searching for genomic regions with excess homozygosity (9) (Fig. 1B, Fig. S1, and Table S1). A remarkable homozygosity was observed in a region spanning >50 Mb on chromosome X. Interestingly, Ma et al. (10) identified a region of ∼31 Mb on chromosome X with an extremely low rate of recombination in F1 hybrids between European and Asian pigs that overlaps the region of homozygosity detected here. However, the European wild boars shared the haplotype fixed in European domestic pigs, implying that this fixation predates domestication, and it was therefore not analyzed further in this study. In the genome-wide screen, 13 distinct loci showed a Z score of heterozygosity (ZHp) lower than −5, and 64 loci showed ZHp < −4 (Table S1). There was a striking correlation between putative sweep regions and well-established quantitative trait loci (QTL), exemplified by the melanocortin 4 receptor (MC4R) gene on chromosome 1 (position 178.5 Mb) underlying a QTL for feed intake and growth (11). Here we highlight our findings that three of the most convincing selective sweep candidates are colocalized with major QTLs, explaining a considerable portion of one of the most striking phenotypic changes during pig domestication—namely, elongation of the back and an increased number of vertebrae, as already noticed by Charles Darwin (12) (Fig. 1A). Wild boars have 19 vertebrae, whereas European domestic pigs intensively selected for meat production have 21–23 (13).
Fig. 1.
Summary of selective sweep analysis. (A) Striking phenotypic differences between a wild boar and a domestic pig. (B) Genome-wide Z score of heterozygosity (ZHp) plot. The y axis values are −ZHp, and the x axis shows positions of windows along each chromosome. Dotted lines indicate the −ZHp thresholds for inclusion (−4) and bridging/elongation (−3). Red arrows and names indicate loci discussed in the main text. For more comprehensive details on intersections of genes and sweeps, see Table S1 and Fig. S1. (C) Degree of haplotype sharing for the loci overlapping NR6A1, LCORL, and PLAG1 in pairwise comparisons. Boxes to the left indicate the comparison presented on that row (ED, European Domestic; wb, wild boar; AD, Asian Domestic). Heat map colors indicate identity scores. Locations of genes are indicated below the heat map. (D) Body length difference by genotypes at the LCORL and PLAG1 loci. Bar heights show the average length increase relative to homozygotes for the wild-type allele at both loci (WW, wild-type homozygotes; WD, heterozygotes; DD, domestic homozygotes). (E) Results from genotyping wild boars and a diverse panel of European domestic pigs for SNPs in the putative selective sweeps containing NR6A1, PLAG1, and LCORL. The plot is sorted by breeds but has not been clustered by individuals or by breeds. Me, Meishan; AS, Angler Sattelschwein; BS, British Saddleback; HA, Hampshire; mw, Middle White; BB, Bunte Bentheimer; GO, Gloucester Old Spot; LB, Large Black; LS, Linderödssvin; TA, Tamworth; MJ, Manchado de Jabugo; BI, Bisario; NC, Negro Canario; NI, Negro Iberico; RE, Retinto.
The strongest signature of selection (ZHp = −5.82) was observed for a locus on chromosome 1, which includes the NR6A1 (Nuclear Receptor 6 A1) gene (Fig. 1C). This region harbors a major QTL affecting the numbers of vertebrae in pigs, and a missense mutation in NR6A1 has been proposed to be causative (14). Two of the other convincing selective sweep candidates overlapped major QTLs for body length in our intercross between Large White pigs and wild boar (15, 16). One, located at 82.56–82.71 Mb on chromosome 4 (ZHp = −5.77), includes PLAG1 (pleomorphic adenoma gene 1; Fig. 1C); the other, located at 12.61–12.76 Mb on chromosome 8 (ZHp = −5.29), encompasses the entire coding region of ligand dependent nuclear receptor corepressor-like (LCORL; Fig. 1C). We genotyped the F2 population from our intercross (15) using informative markers in the LCORL and PLAG1 regions; the NR6A1 locus was not fully informative in this pedigree because both wild boar paternal grandsires were heterozygous for the “domestic” haplotype. The two QTLs associated with LCORL and PLAG1 acted additively with a combined effect of 5.3 cm in body length difference between the opposite homozygotes (Fig. 1D). Phylogenetic analysis of the NR6A1, LCORL, and PLAG1 loci using data from the individually sequenced pigs revealed a European origin of the swept haplotype for all three loci (Fig. S2).
To analyze the selective sweep candidates in more detail, we genotyped 384 SNPs from regions of the genome with highly significant effects in 418 individuals from a wide range of European domestic pig populations, 40 European wild boars, and 21 Asian domestic pigs. The results demonstrated strong signatures of selection at the NR6A1, LCORL, and PLAG1 loci across commercial lines of European domestic pigs used for meat production and in most local populations, except for Iberian pigs (Fig. 1E), which have been less intensely selected for bulk meat production. For all three loci, the most highly fixed SNP had a reference allele frequency of >0.99 in non-Iberian European domestic pigs.
Interestingly, LCORL has consistently been associated with human stature in genome-wide association studies (17), as well as with body size in dogs (18), cattle (19), and horses (20). The PLAG1 region has been associated with variation in height in humans (21) as well as with a major QTL for height in cattle (22). Although the same genes appear to contribute to variation in body length in pigs and humans, the estimated effects differ markedly. Together, LCORL and PLAG1 explained 18.4% of the residual variance in body length in our wild boar intercross. In contrast, 180 loci affecting variation in human height only explained 10% of the population variance (17). Our results imply that alleles with similarly large effects are also segregating in human populations, but that each such variant explains a small portion of the population variance.
Another particularly interesting candidate selective sweep locus is located on chromosome 13 (137.68–138.06 Mb) (ZHp = −5.32) and overlaps Osteocrin (OSTN), whose secreted protein product (OSTN) was first identified as an inhibitor of osteoblast differentiation (23). Shortly after its discovery, OSTN was rediscovered as “Musclin” in a screen for skeletal muscle-derived secretory factors (24), where OSTN expression levels were positively correlated with food intake in mice. Furthermore, it has been shown that insulin acts as a potent regulator of OSTN expression in murine myoblasts (24) and that OSTN is expressed at high levels in fast-twitch type IIb muscle fibers but at much lower levels in slow-twitch fibers (25). Interestingly, it is the fast-twitch type IIb muscle (white skeletal muscle) that is the most valuable part in pork production, and, for instance, the PRKAG3 gene underlying the RN muscle phenotype in pigs shows a similar differential expression between fast-twitch (white) and slow-twitch (red) muscle (26). Thus, the sweep overlapping OSTN present in domestic pigs may be related to selection for an altered body composition and/or altered skeletal development.
To validate the candidate selective sweeps identified in the pooled dataset, we proceeded to analyze the overlap between sweeps (33 candidate sweep windows, representing 18 loci) with SNPs showing extreme differences in genotype frequency in comparisons between individually sequenced domestic pigs and wild boars. We define these ”extreme SNPs” as those exhibiting derived allele frequencies of >0.9 and <0.1 in domesticated pigs and wild boars, respectively. Of 3,190 extreme SNPs, 254 were contained within candidate sweep windows (together encompassing 5.56 Mb of reference genome sequence). Thus, 8% of the extreme SNPs were present within candidate selective sweeps—the latter category taking up 0.2% of the genome. This overlap is much greater than would be expected by chance and supports our hypothesis that the identified selective sweeps are enriched for loci having undergone positive selection during domestication or during development of the modern domestic pig. The NR6A1, PLAG1, and LCORL loci contained 126, 9, and 53 of the 3,190 extreme SNPs, respectively.
Significant Excess of Derived Nonsynonymous Substitutions in Domestic Pigs.
We searched for mutations in coding sequence that have become fixed or nearly fixed in domestic pigs. We first looked for nonsense mutations as obvious candidates of functional significance that may have contributed to rapid evolution in domestic animals (27). However, none of the candidates (Table S2) occurred at a high frequency in several pig breeds, and we found no compelling evidence that any of these has been under positive selection. We conclude that gene inactivation has not played a prominent role during pig domestication, consistent with the results from a similar screen in chickens (9).
We then screened the individual sequence data for the presence of synonymous and nonsynonymous substitutions that showed a marked allele frequency difference between European domestic pigs and wild boars (>80% in one group, <20% in the other); we included both Asian (n = 5) and European wild boars (n = 6) to increase the resolution of this analysis. We also used outgroup species (Phacochoerus qfricanus, Sus cebifrons, Sus barbatus, Sus celebensis, and Sus verrucosus; Table 1) to deduce if the major allele in domestic pigs was ancestral or derived. The logic is that a derived allele may have occurred recently and increased in frequency due to positive selection. If the difference in allele frequency between populations is caused by genetic drift, the likelihood that a derived allele predominates among domestic pigs should be the same for synonymous and nonsynonymous substitutions. We found an excess of derived synonymous alleles in the European domestic pig group (Table 2), as expected because it is more genetically homogenous than the Asian/European wild boar group. However, the excess of derived alleles was much more pronounced for nonsynonymous than for synonymous substitutions (P = 0.00016; Fisher’s exact test), implying different selection pressures in wild and domestic pigs. Both positive selection for favorable mutations and relaxed purifying selection in domestic pigs may contribute to this difference. We argue that positive selection is an important component because if there is relaxed selection at many loci in pigs, we expect that nonsense mutations would be common, which is not the case. However, only 3 (NR6A1, CCT8L2, and MLL3) of the 72 derived nonsynonymous substitutions (Table S3) were colocalized with putative sweep regions detected in this study, implying that most of these substitutions have not increased in frequency due to recent sweeps. Furthermore, the poor overlap suggests that most of the sweeps reported in this study do not reflect selection for favorable mutations in protein-coding genes.
Table 2.
Derived nucleotide substitutions showing marked allele frequency differences between wild boars and domestic pigs
| Population |
||
| Substitution | Domestic pig | Wild boar |
| Nonsynonymous | 72 | 6 |
| Synonymous | 87 | 37 |
Values indicate the number of derived substitutions in which the frequency of the ancestral allele is >0.80 in the indicated population and <0.20 in the other.
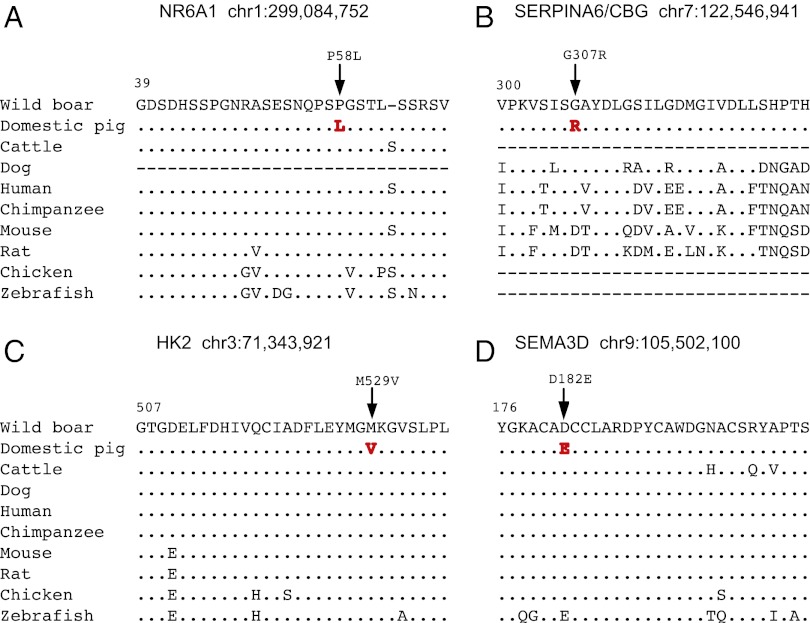
The 72 derived nonsynonymous substitutions (Table S3) approaching fixation in domestic pigs should be considered as candidate mutations underlying phenotypic differences between pigs and wild boars. Interestingly, previous reports indicate that two of these constitute quantitative trait nucleotides (28). The missense mutation (Pro192Leu) in NR6A1 (Fig. 2A) alters the binding affinity of NR6A1 to its coreceptors and has been proposed to be the causative mutation for the QTL affecting number of vertebrae (14) that is colocalized with one of the major sweeps detected in the present study (Fig. 1). Similarly, the missense mutation Gly307Arg in SERPINA6 (also known as CBG, corticosteroid-binding globulin; Fig. 2B) has been shown to affect cortisol-binding capacity and proposed to underlie a pleiotropic QTL affecting serum cortisol levels, fat deposition, and muscle content (29). The remaining 70 candidate causative mutations will require further research. We propose that many of these will be functionally important with support from SIFT (30)/PolyPhen-2 (31) analyses that classified as many as 32/72 substitutions as potentially damaging. For instance, most domestic pigs are homozygous for the missense mutation M529V in HK2 encoding hexokinase 2 (Fig. 2C), a key enzyme for glucose metabolism. Another particularly interesting example is the missense mutation D182E in SEMA3D encoding Semaphorin 3D (Fig. 2D). Semaphorins are known as axon guidance molecules of importance for neuronal development but may also have other biological functions (32).
Fig. 2.
Excess of derived nonsynonymous substitutions in domestic pigs. Multispecies alignment of four proteins for which domestic pigs are fixed or close to fixation for a derived amino acid substitution, NR6A1 (A), SERPINA6/CBG (B), HK2 (C), and SEMA3D (D). Positions beside protein names indicate the genomic coordinate of missense mutations. Dots indicate identities to the master sequence and dashes indicate missing data.
Copy Number Variation in the Pig Genome.
We used the mate-pair reads from the pooled samples to screen for deletions and duplications that showed large allele frequency differences between wild and domestic pigs and detected a large number of deletions and duplications with strong statistical support (Table S4). We intersected identified structural variants with our list of putative selective sweeps, and there was no statistically significant overlap between the two datasets, implying that the majority of copy number variants (CNVs) behave as neutral markers. However, we identified an 8-kb duplication within one of the major sweeps, in an intron of Caspase 10 (CASP10) on chromosome 15 (Fig. S9 and Table S4). This duplication occurred at a high frequency in all lines of domestic pigs used for the pooled sequencing but was rare or absent among wild boars.
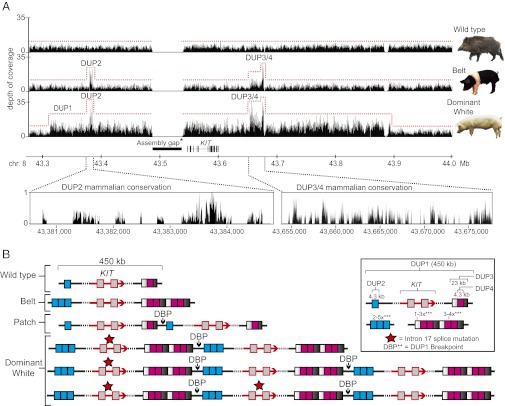
The most obvious structural variants with functional significance were detected at the KIT locus already known to control white spotting in pigs (33). KIT is a tyrosine kinase receptor, and normal KIT signaling is required for development and survival of neural crest-derived melanoblasts (34). Three major KIT variants have been described in pigs (Fig. S3): Dominant white (completely white; Fig. 3A), Patch (partially white), and Belt (white belt across forelegs; Fig. 3A). Dominant white is associated with a 450-kb duplication encompassing the entire KIT gene and a splice mutation causing exon skipping in at least one of the KIT copies; Patch has only the 450-kb duplication, whereas no causative mutation has been identified for Belt (33).
Fig. 3.
Evolution of white spotting alleles at the KIT locus in pigs. (A) Sequencing depth of coverage for the wild boar- (wild-type), Hampshire- (Belt), and Landrace- (Dominant White) pools demonstrating the presence of three duplications in addition to the previously described DUP1. *Further information on the assembly gap is presented in Fig. S8. Below are magnifications showing sequence conservation in 35 mammals for the DUP2, DUP3, and DUP4 regions. (B) Schematic presentation of our interpretation of porcine KIT alleles. Together, the splice site mutation and the variable copy numbers of the four duplications create great haplotype diversity. **The DUP1 breakpoint (DBP) was precisely defined by Giuffra et al. (49). ***Range of DUP2–4 copy numbers per allele was estimated from quantitative PCR (qPCR) of Belted pigs.
In the screen for CNVs using pooled samples, we confirmed the presence of the large 450-kb duplication (DUP1 in this study) in Dominant white pigs (Fig. 3A); the reference sequence (Sscrofa10.2) is from a Duroc pig and includes a single KIT copy. In Hampshire pigs (Belt phenotype), we detected a 4.3-kb duplication (DUP2) located ∼100 kb upstream of KIT and a 23-kb duplication (DUP3) ∼100 kb downstream of KIT, which in turn contained a fourth ∼4.3-kb duplication (DUP4) not present on wild-type chromosomes (Fig. 3A). A closer examination showed that DUP2–4 was also present in Dominant white pigs (Fig. 3A). We explored the distribution of DUP1–4 across a broad sample of pigs and wild boars using whole-genome sequencing data from individual samples (Fig. S4) as well as TaqMan assays (Fig. S5). The four duplications were exclusively present in pigs showing different white spotting patterns. DUP2–4 was present in Belted pigs representing four different breeds; a few of these pigs lacked DUP3, but all carried DUP2 and DUP4. All tested pigs with the Patch allele carried DUP1 but not DUP2–4 (Fig. S5). Finally, all Dominant white pigs representing five breeds carried DUP1–4.
This study has revealed an additional complexity at the KIT locus in pigs, and our new interpretation of porcine KIT alleles is summarized in Fig. 3B. Based on the complete concordance with white spotting phenotypes, we conclude that DUP2–4 is strongly associated with the presence of the Belt and propose that one or more of these duplications are required for manifestation of the Belt phenotype and contribute to the Dominant white phenotype. DUP2 is the strongest causative candidate because it overlaps with one of the most well-conserved noncoding regions located upstream of KIT (Fig. 3A). DUP2—which occurs as a single copy sequence in all tested wild-type alleles and in three to six copies in Belt and Dominant white alleles—may constitute a regulatory element that becomes stronger with copy number expansion, as recently demonstrated for a melanocyte-specific enhancer located within the duplication causing graying with age in horses (35).
Discussion
This work provides insights on the genetic basis for phenotypic evolution. We have shown that mutations in coding as well as noncoding sequences have contributed to the evolution of the domestic pig. Alleles with large phenotypic effects have played a significant role, and strong directional selection may result in the evolution of alleles/haplotypes that differ by multiple mutational steps from wild-type alleles. This study illustrates the strength of domestic animals as models for deciphering complex phenotype–genotype relationships. For instance, the reason why we can show that the Dominant white allele involves at least three causative mutations is that the Belt and Patch alleles constitute intermediate forms with distinct phenotypic effects. In a comparison between two different species—e.g., human and chimpanzee—it would be exceedingly difficult to unravel if multiple causative mutations in the same gene contributed to a phenotypic difference.
The origin of most of the nucleotide diversity present in domestic pigs predates domestication. The 10,000 y that have passed since pig domestication is simply not long enough to build up a genome-wide nucleotide diversity of ∼0.1%. A consequence of this recent divergence is that random genome sequences from wild boars and domestic pigs are very similar, and it is impossible to tell them apart. It is only at those loci under strong selection where one finds distinct sequence differences. One such locus is the MC1R, where a screen of 31 European domestic pig populations and 19 Chinese pig populations revealed that only one of these populations (Hungarian Mangalica) carried an MC1R wild-type allele (5). IGF2 is an example of a gene affected by a more recent selection pressure, in which the majority of European domestic pigs intensively selected for lean growth carry a specific IGF2 allele causing increased muscle growth and reduced fat depositions (8). IGF2 would have been a perfect positive control in our selective sweep analysis, but unfortunately this gene is in one of the gaps in the current assembly of the pig genome. The present study has revealed three more loci (NR6A1, PLAG1, and LCORL) in which the majority of European domestic pigs are homozygous for the same haplotype (Fig. 1E). For one of these (NR6A1), a strong candidate mutation (Pro192Leu) has already been proposed (14), and the results of the present study support this hypothesis. Further work is required to reveal the causative mutations associated with the PLAG1 and LCORL loci. An examination of our sequence data for these regions did not reveal any obvious candidate mutation in coding sequences, suggesting that they most likely represent regulatory mutations or mutations in noncoding RNA.
The aim of this study was to reveal loci under strong selection during pig domestication. Convincing associations were detected between putative sweeps and structural changes and the striking phenotypic differences between wild boars and domestic pigs as regards body length and coat color. The strong sweep signals associated with QTLs for body length most likely reflect the fact that the screen using pooled sequencing was based on pig populations intensively selected for meat production. Coat color is clearly one of the traits that changed early during domestication (5). However, we were not able to pinpoint sweeps, coding changes, or structural variants that are likely to explain the striking differences between wild and domestic pigs with regard to behavior or reproductive traits. There are several possible reasons for a lack of power in our attempts to identify such loci. Most importantly, there is a lack of genetic data showing the location of QTLs explaining differences for these interesting traits between wild and domestic pigs. Thus, we were not able to overlap our sweep signals with well-established QTLs as we could do for body length. Furthermore, we have primarily screened for fixed differences (hard sweeps), and it is an open question to which extent fixed differences are underlying a complex trait like behavior or whether behavioral differences between wild and domestic pigs instead are due to shifted allele frequencies at a large number of loci each with a minute phenotypic effect—which would make it very challenging to reveal such loci. In this respect, a recent study showing that a nonsense mutation in DMRT3 has a major impact on gait control in horses is encouraging because it demonstrates that alleles with large effects on a complex trait like the pattern of locomotion may be detected in domestic animals (36). Perhaps the best candidates for mutations affecting behavior in pigs found in the present study were the derived missense mutations in SEMA3D (Fig. 2D and Table S3) and SEMA3E (Table S3) that both encode axon guidance molecules.
Emerging data show that structural changes have contributed to rapid evolution in domestic animals, and most of these structural changes constitute cis-acting regulatory mutations (37). To our knowledge, the porcine KIT locus is the most striking example as it involves multiple structural changes. We propose that at least two out of the four KIT duplications exclusively present in white and white spotted pigs are causative. The porcine KIT locus also illustrates the evolution of alleles in domestic animals under strong positive selection. Dominant white is the most dominant KIT allele described in any species, and it is still fully viable in the homozygous condition, whereas KIT loss-of-function mutations causing pigmentation effects are lethal or sublethal (34). The reason is that Dominant white is not a single mutation effect but a haplotype effect caused by the combined action of at least two duplications and a splice mutation (Fig. 3B). The haplotype diversity at the porcine KIT locus is bewildering because DUP1–4 all show copy number variation, and the number of KIT copies carrying the splice mutation varies among haplotypes (Fig. 3B). The phenotypic effect of a dynamic locus like KIT in pigs may be grossly underestimated in a standard genome-wide association study, which is unlikely to capture such complex genotype–phenotype relationships. Haplotype effects due to multiple sequence polymorphisms affecting the structure or expression of a single gene may be common in many species and are probably one reason why it is so difficult to reveal the causative mutations at loci underlying complex traits and disorders, even when the genetic association is restricted to a very specific region as is the case in human genome-wide association studies.
Materials and Methods
Whole-Genome Resequencing of Pools.
For each population (Table 1), DNA samples from 8–20 individuals were pooled in equimolar quantities that were used to generate mate-pair sequencing libraries (mean insert sizes of ∼1.3 kb). The libraries were sequenced by using a SOLiD v.4 instrument (Life Technologies) according to the manufacturer’s instructions. The reads were mapped as paired reads to the porcine reference genome assembly Sscrofa10.2 by using BioScope (1.3.1) (Life Technologies), with a local mapping strategy that accepted 6 mismatches and 100 hits per read. This procedure resulted in an average read depth (RD) of ∼5× per breed pool. SNPs were called using the Bioscope SNP caller diBayes (Life Technologies) by first calling SNPs in each pool individually using a Bayesian approach. To select the values of the most important parameters, we proceeded to run a SNP detection analysis with only the libraries of the population that yielded most reads, the Uppsala Large White. We called SNPs with a low stringency (where a SNP can be called even though only a single observation of the nonreference allele is seen) to evaluate recall rate. For that purpose, we used the Illumina Porcine SNP60 chip (i.e., 60K SNP chip) (38) and mapped the probes to the genome assembly. We then studied the proportion of overlapped calls, varying the threshold of alignment-length/read-length ratio (ARR) and the minimum mapping/pairing quality value (MQV) parameters. We set MQV to 20 and ARR to 0.80 because the coverage at MQV of 10 and 15 may have been inflated due to spurious alignment, and the distribution seems to stabilize at QV = 20. ARR does not seem to be a decisive parameter in our study because it gives very similar results at 0.80 and 0.90. However, 0.80 seemed to yield a small increase in proportion of probe coverage. Finally, to exploit the full power of our libraries, we concatenated all of them to detect and produce the porcine set of SNPs presented herein (7,354,658 SNPs). The total RD of this analysis was reduced from an average of 30× to 25× at the MQV chosen, and the genome coverage decreased from 99% to 87% after discarding uncovered gap space. This last analysis was run with “call.stringency” setting to high. This flag is recommended for data in our coverage range to increase stringency of detection by requiring alleles to be seen in both strands and reduce artifacts at the expense of a reduced sensitivity to detect SNPs. We also used the Bioscope parameter “het.skip.high.coverage,” recommended for whole-genome resequencing applications, to avoid calling SNPs when the coverage of a position is too high compared with the median of the coverage distribution of all positions.
The resulting SNP set was further filtered by requiring at least three independent reads supporting the variant allele and also at least one reference allele in the union of reads from all sequenced pools. We avoided triallelic SNPs by discarding sites where the most commonly observed variant allele constituted <80% of the total sum of all variants. Following these filters, 6,792,483 SNPs remained, and these were included in the sweep analysis.
Sequencing of Individual Pigs.
A total of 37 domestic pigs, 11 wild boars, and 7 other suids were sequenced by using Illumina HiSeq technology. Illumina (v. 1.3–1.7) formatted fastq files, with sequence reads between 60 (Illumina GA2, part of the data for S. verrucosus and S. celebensis) and 100 bp (Illumina GA2 and HiSeq2000), were subject to quality trimming before sequence alignment. The trimming strategy involved a 3-bp sliding window, running from 5′ to 3′, with sequence data upstream being discarded if the 3-bp window average quality dropped to <13 (i.e., average error probability equal to 0.05). Only sequences 45 bp or longer were retained. In addition, sequences with mates of <45 bp after trimming were discarded. During trimming, quality scores were recoded to follow the Sanger fastq format to standardize downstream processing.
Sequences were aligned against the Sscrofa10.2 reference sequence by using Mosaik 1.1.0017 (http://bioinformatics.bc.edu/marthlab/Mosaik). Alignment was performed by using a hash size of 15, with a maximum of 10 matches retained, and 7% maximum mismatch score, for all pig populations and outgroup species. Alignment files were then sorted by using the Mosaiksort function, which entails removing ambiguously mapped reads that are either orphaned or fall outside a computed insert-size distribution. Alignment archives were converted to BAM format (39) by using the Mosaiktext function. Processing of BAM files, such as merging of alignments archives pertaining the same individual, were conducted by using SAMtools v. 1.12a (39).
Variant allele calling was performed per individual by using the pileup function in SAMtools, and variations were initially filtered to have a minimum quality of 50 for indels and 20 for SNPs. In addition, all variants showing higher than 3× average read density, estimated from the number of raw sequence reads, were also discarded to remove false-positive variant calling originating from off-site mapping as much as possible. This procedure yielded high-quality variants for 55 pigs, wild boar, and outgroup species.
To obtain genotype calls for all polymorphic sites identified across 55 individuals, every individual was examined for the genotype call for each of the sites found to be polymorphic, including the species-specific differences. Sequence depth, SNP, and consensus quality were retrieved for these sites by using the SAMtools pileup function. These de facto genotype calls were filtered based on sequence depth (genotype of each particular individual retained if the depth ranged between four and twice the average genome-wide depth of that individual). Further filtering was performed on SNP and consensus quality (for homozygotes, either a SNP or consensus quality of >20 was used, and for heterozygotes, both consensus and SNP qualities of >20 were applied). All indels were removed. After filtering, genotype calls were established for a total of 66,668,635 single nucleotide positions in the genome.
Sweep Analysis.
The selective sweep screen was performed by using the sequenced DNA pools. At each identified SNP position, we counted the numbers of reference and variant allele reads observed in each breed. We next slid along the genome using 50% overlapping windows of variable sizes (ranging from 10 to 550 kb) and plotted the distributions of SNP counts within these windows. After reviewing the distributions (Fig. S6), we concluded that 150 kb was the most appropriate window size, because this size yielded few windows with <20 SNPs (9), while retaining a theoretically appropriate length to detect smaller sweeps. For each 150-kb window along the reference genome, we calculated the pooled heterozygosity (Hp) and the ZHp as described (9) (Hp and ZHp distributions are presented in Fig. S7) and also collected other statistics such as pairwise and global FST and identity scores (i.e., haplotype similarities). Because sex chromosomes and autosomes are subjected to different selective pressures and have different effective population sizes, we decided to calculate the ZHp for autosomes and chromosome X separately. Windows with ZHp ≤ −4 were retained as candidate selective sweeps, and adjacent ZHp ≤ −4 regions were merged into the candidate sweep loci presented in Table S1. Single windows of ZHp ≤ −3 were allowed to bridge adjacent ≤ −4 windows in the merging step.
SNP Genotyping.
SNPs located in putative selective sweeps and other regions of interest identified in initial screens were used to design a VeraCode GoldenGate assay (Illumina) targeting 384 SNPs. A total of 418 individuals from the Pig Biodiversity panel (40), as well as from other sources, representing a wide range of European domestic pig populations, 40 European wild boars, and 21 Asian domestic pigs, were genotyped by using the standard protocol provided by Illumina. The GoldenGate assays were read by using a BeadXpress Reader (Illumina), and data were analyzed by using the GenomeStudio V2011.1 software (Genotyping module Version 1.9.4; Illumina).
Phylogenetic Analysis.
Sequence assemblies for the NR6A1, LCORL, and PLAG1 loci were extracted per individual according to their genomic coordinates from the BAM files generated from individually sequenced pigs using SAM tools (Version 1.12b) (39). Because the PLAG1 gene is small, an additional 10 kb of sequence data was obtained upstream and downstream of the gene. Phylogenetic analysis was performed by using RAxML (41), using 10 iterations, implementing a GTRGAMMA model, and with the African warthog as outgroup.
Coding Mutations.
To identify candidate nonsense mutations, we first used the software ANNOVAR (42) (Release 2012Feb23). We identified 106 SNPs in which one of the alleles created a stop codon. We next filtered these SNPs using spliced expressed sequence tags (ESTs) retrieved from the University of California Santa Cruz Genome browser and mapped them to the porcine reference genome assembly (Sscrofa10.2) using the standalone version of BLAT (Basic Local Alignment Tool) (43). We retained the ones with the highest matching scores and no gaps. We manually reviewed each of the sites for validity of gene model and SNP calls. After reviewing, we retained 12 of the 106 SNPs as candidate nonsense mutations listed in Table S2. A complicating factor in this analysis was that the reference genome is from a domestic pig, which means that if a nonsense mutation has become fixed in the domestic pig, it is likely that the corresponding gene model may be wrong. To remove this bias, we developed an alternative approach where we screened the initial dataset of SNPs (6.25 million) to search for nonsense SNPs within any of the six ORFs by selecting 90 bp (30 codons) upstream and downstream to the stop-causing site. We next performed a BLAST search for human homologs using Homo sapiens GRCh37.61. After manual review, we retained 23 candidate nonsense mutations having human homologs. However, we did not find EST support for any of these candidates.
SNPs with Extreme Allele Frequency Differences Between Domestic Pigs and Wild Boars.
The procedure entailed grouping of 11 wild boar vs. 30 European domesticated pigs that were all individually sequenced (Table 1). For each SNP discovered in the entire pool of animals, the direction (i.e., derived or ancestral) of the alleles was determined by comparison with five outgroup species, four other species in the genus Sus, and the African warthog (Table 1). Only SNPs where all five outgroup species agreed on the direction of the SNP were considered, with the criteria that the SNP needed to be scored in at least two of the species. Furthermore, only SNPs that had a genotype scored for at least 50% in both the wild boar and European domestic group were considered. SNPs with an extreme difference in ancestral allele frequency between the wild boar and European domestic group (>80% in one group, <20% in the other) were subsequently analyzed by using ANNOVAR (42) to investigate locations in genes and potential coding properties.
Structural Changes.
We screened the pooled samples for structural variants using the software CNV-seq (44), CNVnator (45), and a custom script for identifying fixed deletions (FD) described in Rubin et al. (9). We scanned the genome to identify CNVs using RDs from each domestic pig population in pairwise comparisons relative to the wild boar using CNV-seq. To gain further support for identified candidate CNVs and deletions, we used CNVnator to slide 100 bp along each chromosome in each pool to compare observed RD with the GC-matched RD of the same chromosome in the same pool. Our final set of structural variants was derived from intersections of CNV-seq determined CNVs with CNVnator-duplications and correspondingly intersections of FD with CNVnator deletions. The genomic distribution of the final set of candidate structural variants is presented together with intersections between structural variants and candidate selective sweeps in Fig. S9.
Genomic Copy Number PCR Analyses.
TaqMan primers and probes were designed by using standard parameters in Primer3Plus (46) (Table S5). PCR products were tested for secondary structures by using mfold (http://mfold.rna.albany.edu/?q=mfold). Target probes were 5′ labeled with 6-FAM and 3′ labeled with the minor groove binder nonfluorescent quencher. The reference probe ESR1 was 5′ labeled with VIC and 3′ labeled with TAMRA (ABI). TaqMan reactions were performed by using Gene Expression MasterMix (Life Technologies) containing 10 ng of genomic DNA, 800 nM each primer, and 250 nM probe in a total volume of 10 µL. Primer/probe performances were evaluated in both simplex and duplex with the control primer/probe set by using a 5-point standard curve with a 10× dilution factor and the criteria described (47). Genomic qPCR assays were performed by using individuals representing Dominant white, Belted, Patched and wild-type colored pig breeds from the Pig Biodiversity panel (40) (Fig. S5). For each qPCR assay and DNA sample, Ct values from a minimum of three runs were used to estimate genomic copy number of the duplications in relation to the single copy locus ESR1 by using the ΔΔCt methodology (48). Mean copy numbers were then calculated for each breed.
Supplementary Material
Acknowledgments
Pig samples were provided by the European Community PigBioDiv2 Project QLK5-CT-2002-01059. Silvia D'Andrea and Elisabetta Giuffra provided samples from Cinta Sinese pigs. We thank Uppsala Genome Center for performing SOLiD sequencing, the SNP genotyping platform of SciLifeLab Uppsala, and UPPMAX for providing computational and storage resources. The study was supported by the Swedish Foundation for Strategic Research, Formas, Swedish Research Council and European Research Council under the European Community's Seventh Framework Programme (FP7/2007-2013)/European Research Council Grant Agreement 249894. A.L.A. was supported by a Biotechnology and Biological Sciences Research Council Institute Strategic Programme grant.
Footnotes
The authors declare no conflict of interest.
Data deposition: The Illumina and SOLiD data have been deposited in the National Center for Biotechnology Information Sequence Read Archive, http://www.ncbi.nlm.nih.gov/sra) [accession nos. ERP001813 (Illumina) and SRA057461 (SOLiD)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217149109/-/DCSupplemental.
References
- 1.Giuffra E, et al. The origin of the domestic pig: Independent domestication and subsequent introgression. Genetics. 2000;154(4):1785–1791. doi: 10.1093/genetics/154.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kijas JMH, Andersson L. A phylogenetic study of the origin of the domestic pig estimated from the near-complete mtDNA genome. J Mol Evol. 2001;52(3):302–308. doi: 10.1007/s002390010158. [DOI] [PubMed] [Google Scholar]
- 3.Larson G, et al. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science. 2005;307(5715):1618–1621. doi: 10.1126/science.1106927. [DOI] [PubMed] [Google Scholar]
- 4.Groenen MAM, et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 2012;491(7424):393–398. doi: 10.1038/nature11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang M, Larson G, Ribeiro HS, Li N, Andersson L. Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 2009;5(1):e1000341. doi: 10.1371/journal.pgen.1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujii J, et al. Identification of a mutation in porcine ryanodine receptor associated with malignant hyperthermia. Science. 1991;253(5018):448–451. doi: 10.1126/science.1862346. [DOI] [PubMed] [Google Scholar]
- 7.Milan D, et al. A mutation in PRKAG3 associated with excess glycogen content in pig skeletal muscle. Science. 2000;288(5469):1248–1251. doi: 10.1126/science.288.5469.1248. [DOI] [PubMed] [Google Scholar]
- 8.Van Laere AS, et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 2003;425(6960):832–836. doi: 10.1038/nature02064. [DOI] [PubMed] [Google Scholar]
- 9.Rubin C-J, et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464(7288):587–591. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- 10.Ma J, et al. Recombinational landscape of porcine X chromosome and individual variation in female meiotic recombination associated with haplotypes of Chinese pigs. BMC Genomics. 2010;11:159. doi: 10.1186/1471-2164-11-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim KS, Larsen N, Short T, Plastow G, Rothschild MF. A missense variant of the porcine melanocortin-4 receptor (MC4R) gene is associated with fatness, growth, and feed intake traits. Mamm Genome. 2000;11(2):131–135. doi: 10.1007/s003350010025. [DOI] [PubMed] [Google Scholar]
- 12.Darwin C. The Variation of Animals and Plants Under Domestication. London: John Murray; 1868. [Google Scholar]
- 13.King JWB, Roberts RC. Carcass length in the bacon pig: Its association with vertebrae numbers and prediction from radiographs of the young pig. Anim Prod. 1960;2:59–65. [Google Scholar]
- 14.Mikawa S, et al. Fine mapping of a swine quantitative trait locus for number of vertebrae and analysis of an orphan nuclear receptor, germ cell nuclear factor (NR6A1) Genome Res. 2007;17(5):586–593. doi: 10.1101/gr.6085507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersson L, et al. Genetic mapping of quantitative trait loci for growth and fatness in pigs. Science. 1994;263(5154):1771–1774. doi: 10.1126/science.8134840. [DOI] [PubMed] [Google Scholar]
- 16.Andersson-Eklund L, et al. Mapping quantitative trait loci for carcass and meat quality traits in a wild boar x Large White intercross. J Anim Sci. 1998;76(3):694–700. doi: 10.2527/1998.763694x. [DOI] [PubMed] [Google Scholar]
- 17.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaysse A, et al. LUPA Consortium Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet. 2011;7(10):e1002316. doi: 10.1371/journal.pgen.1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pryce JE, Hayes BJ, Bolormaa S, Goddard ME. Polymorphic regions affecting human height also control stature in cattle. Genetics. 2011;187(3):981–984. doi: 10.1534/genetics.110.123943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Signer-Hasler H, et al. A genome-wide association study reveals loci influencing height and other conformation traits in horses. PLoS ONE. 2012;7(5):e37282. doi: 10.1371/journal.pone.0037282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gudbjartsson DF, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40(5):609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 22.Karim L, et al. Variants modulating the expression of a chromosome domain encompassing PLAG1 influence bovine stature. Nat Genet. 2011;43(5):405–413. doi: 10.1038/ng.814. [DOI] [PubMed] [Google Scholar]
- 23.Thomas G, et al. Osteocrin, a novel bone-specific secreted protein that modulates the osteoblast phenotype. J Biol Chem. 2003;278(50):50563–50571. doi: 10.1074/jbc.M307310200. [DOI] [PubMed] [Google Scholar]
- 24.Nishizawa H, et al. Musclin, a novel skeletal muscle-derived secretory factor. J Biol Chem. 2004;279(19):19391–19395. doi: 10.1074/jbc.C400066200. [DOI] [PubMed] [Google Scholar]
- 25.Banzet S, et al. Musclin gene expression is strongly related to fast-glycolytic phenotype. Biochem Biophys Res Commun. 2007;353(3):713–718. doi: 10.1016/j.bbrc.2006.12.074. [DOI] [PubMed] [Google Scholar]
- 26.Mahlapuu M, et al. Expression profiles representing the γ-subunit isoforms of AMP-activated protein kinase suggest a major role for γ3 in white skeletal muscle fibers of mammals. Am J Physiol Endocrinol Metab. 2004;286:E194–E200. doi: 10.1152/ajpendo.00147.2003. [DOI] [PubMed] [Google Scholar]
- 27.Olson MV. When less is more: Gene loss as an engine of evolutionary change. Am J Hum Genet. 1999;64(1):18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackay TFC. Quantitative trait loci in Drosophila. Nat Rev Genet. 2001;2(1):11–20. doi: 10.1038/35047544. [DOI] [PubMed] [Google Scholar]
- 29.Guyonnet-Dupérat V, et al. Functional implication of an Arg307Gly substitution in corticosteroid-binding globulin, a candidate gene for a quantitative trait locus associated with cortisol variability and obesity in pig. Genetics. 2006;173(4):2143–2149. doi: 10.1534/genetics.105.053983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sim NL, et al. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40(Web Server issue):W452-7. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ton QV, Kathryn Iovine M. Semaphorin3d mediates Cx43-dependent phenotypes during fin regeneration. Dev Biol. 2012;366(2):195–203. doi: 10.1016/j.ydbio.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersson L, Plastow G. Molecular genetics of coat colour variation. In: Rothschild MF, Ruvinsky A, editors. The Genetics of the Pig. Oxon, UK: CAB International; 2011. pp. 38–50. [Google Scholar]
- 34.Chabot B, Stephenson DA, Chapman VM, Besmer P, Bernstein A. The proto-oncogene c-kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature. 1988;335(6185):88–89. doi: 10.1038/335088a0. [DOI] [PubMed] [Google Scholar]
- 35.Sundström E, et al. Identification of a melanocyte-specific, MITF-dependent regulatory element in the intronic duplication causing hair greying and melanoma in horses. Pigment Cell Melanoma Res. 2012;25:28–36. doi: 10.1111/j.1755-148X.2011.00902.x. [DOI] [PubMed] [Google Scholar]
- 36.Andersson LS, et al. Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature. 2012;488(7413):642–646. doi: 10.1038/nature11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersson L. How selective sweeps in domestic animals provide new insight into biological mechanisms. J Intern Med. 2012;271(1):1–14. doi: 10.1111/j.1365-2796.2011.02450.x. [DOI] [PubMed] [Google Scholar]
- 38.Ramos AM, et al. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS ONE. 2009;4(8):e6524. doi: 10.1371/journal.pone.0006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.SanCristobal M, et al. Genetic diversity within and between European pig breeds using microsatellite markers. Anim Genet. 2006;37(3):189–198. doi: 10.1111/j.1365-2052.2005.01385.x. [DOI] [PubMed] [Google Scholar]
- 41.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 42.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kent WJ. BLAT–The BLAST-Like Alignment Tool. Genome Res. 2002;4:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie C, Tammi MT. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics. 2009;10:80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21(6):974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Untergasser A, et al. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35(Web Server issue)(suppl 2):W71-4. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishii T, Sootome H, Shan L, Yamashita K. Validation of universal conditions for duplex quantitative reverse transcription polymerase chain reaction assays. Anal Biochem. 2007;362(2):201–212. doi: 10.1016/j.ab.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 48.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 49.Giuffra E, et al. A large duplication associated with dominant white color in pigs originated by homologous recombination between LINE elements flanking KIT. Mamm Genome. 2002;13(10):569–577. doi: 10.1007/s00335-002-2184-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.