Antibodies protect against lethal infection by bacteria or nonenveloped viruses such as adenovirus (1). They are secreted by plasma cells at the rate of thousands of molecules per second, and it has long been assumed that they function exclusively in the extracellular milieu. The recent discovery of a cytoplasmic process called antibody-dependent intracellular neutralization (ADIN) put an end to this dogma: virion-associated antibodies that fail to prevent virus entry into susceptible target cells have a second chance to block infection from within the target cell cytoplasm (2). In PNAS, Hauler et al. (3) generate a din of excitement by showing that the host AAA ATPase valosin-containing protein (VCP) cooperates with the E3 ubiquitin ligase tripartite motif-containing protein 21 (TRIM21) to dismantle the antibody-coated virion, thus permitting the proteasome access to the building block proteins of the virion capsid.
Generally, neutralizing antibodies are believed to block interaction of the virion with cognate cell surface receptors on susceptible target cells. This straightforward model has served well to guide basic immunology researchers as well as those developing prophylactic antiviral vaccination strategies. However, this model is not sufficient to explain a number of puzzling observations in the literature. In some cases, for example, antibodies block infection after virus is adsorbed to target cells (4). Despite the presence of multiple, functional binding sites for the cognate receptor on the surface of nonenveloped virion capsids a single antibody molecule per virion is sometimes sufficient to neutralize infectivity (5); this is inconsistent with the simple receptor blockade model. The IgG Fc is sometimes required for virus neutralization by antibody (6), lending further evidence that simple antigen recognition is not sufficient. The efficiency of virus neutralization by antibody is influenced by IFN, suggesting that unknown, innate immune cofactors are required for neutralization (7).
Perhaps the most enigmatic observation concerning neutralization of virus by antibody came from studies on TRIM21. This cytoplasmic host factor was isolated as an autoantigen using sera from patients with systemic lupus erythematosis and related disorders (8, 9). TRIM21 was later found to bind nonspecifically to IgG Fc in the yeast two-hybrid system (10). It was viewed as highly unlikely that this protein–protein interaction was physiologically relevant because TRIM21 could not have access to extracellular Ig. However, TRIM21 was then shown to have a binding affinity for IgG Fc as strong as that of any human Fc receptor (11). How could such a strong interaction be just an artifact?
An extraordinary series of TRIM21 experiments recently shed light on these unexplained observations. It was shown that, when adenovirus enters a susceptible target cell after incubation with antisera, the bound antibody is brought into the cell cytoplasm with the virion (2). TRIM21 was recruited to the antibody-coated virion in the cytoplasm, virion coat protein was degraded, and adenovirus infectivity was inhibited (Fig. 1). TRIM21 is a really interesting new gene (RING) E3 ubiquitin ligase that covalently attaches itself to K48-linked ubiquitin chains. The TRIM21 RING domain was required for ADIN, as was the proteasome (2). ADIN was blocked by a point mutation in the Fc domain of the antibody that disrupts interaction with TRIM21 (2).
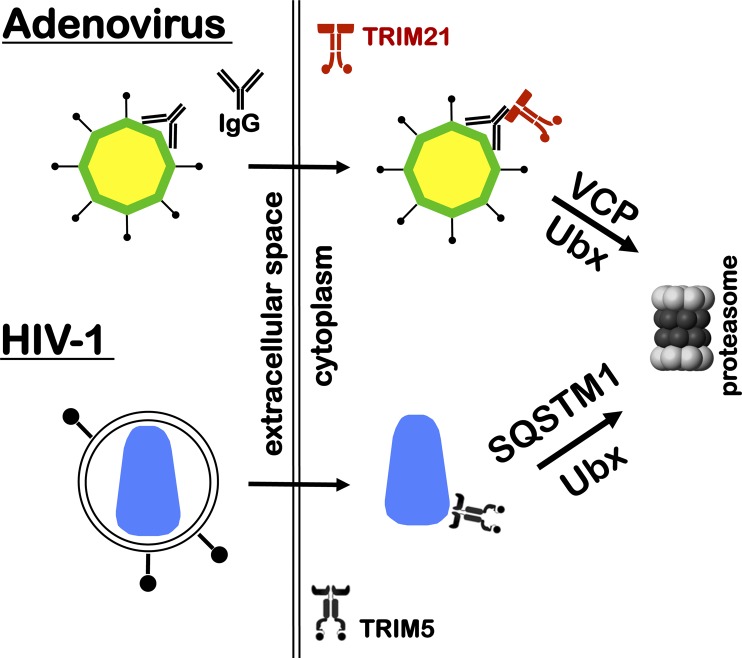
Fig. 1.
TRIM-mediated degradation of virion cores. If antibodies (IgG) bind to the adenovirus capsid (yellow and green octagon) but fail to prevent receptor binding and fusion, they hitch a ride into the cytoplasm with the invading capsid. Once in the cytoplasm the high-affinity Fc receptor TRIM21 (shown in red) binds the capsid-associated antibody and triggers capsid degradation by the proteasome. This host defense mechanism, ADIN, requires the TRIM21 RING domain, which has E3 ubiquitin ligase activity. As demonstrated by Hauler et al. (3), the ATPase VCP is also required, presumably to disassemble the capsid into “bite-size” pieces for the proteasome. Adenovirus is a nonenveloped virus. An analogous degradation pathway has been described for the enveloped virus, HIV-1 (blue conical capsid). Upon fusion of the HIV-1 virion membrane with a susceptible target cell, the capsid core is intercepted in the cytoplasm by the E3 ubiquitin ligase TRIM5. With assistance from p62/SQSTM1 (20), the capsid is degraded in a proteasome-dependent fashion.
Most of the observations mentioned above that were incompatible with the simple model for antibody neutralization can now be accounted for by ADIN, hinting that ADIN plays a role in normal physiology. Type 1 IFN increased TRIM21 protein levels, and this would explain how IFN directs more potent neutralization of virus by antibody (2). When TRIM21 levels were low in the target cell, even saturating concentrations of antibody failed to block adenovirus infection (2). Conversely, high levels of TRIM21 permitted neutralization in the presence of fewer than two antibody molecules per virion (12). Ultimately, it will be important to determine whether TRIM21, like Bruton’s tyrosine kinase or antibodies themselves (1), is essential for an animal to survive a challenge with adenovirus.
The PNAS article by Hauler et al. (3) takes the TRIM21 story further by addressing the question of how a substrate as large as a virion could be degraded by the proteasome. The proteasome is 15 nM long, with a chamber less than 5 nM across. Adenovirus cores measure approximately 100 nM, much larger than known cellular substrates for proteasomes. The article shows that the AAA (ATPase associated with diverse cellular activities) VCP is required for ADIN. VCP forms a 16-nM hexameric ring, lacks protease activity, associates with polyubiquitin chains, and dissociates proteins from large cellular structures (13). Interestingly, VCP was not required for TRIM21-dependent degradation of transfected IgG Fc, although this protein was degraded by the proteasome. In other words, VCP is not required for TRIM21-dependent proteasome degradation of all substrates, only very large, multiprotein structures.
The authors did not detect ubiquitination of viral capsid or the associated Ig using purified components in vitro (3). Perhaps ubiquitination of these proteins only occurs within cells where these intermediates are too transient to be detected. Alternatively, TRIM21 autoubiquitination might be sufficient for delivery of the substrate to the proteasome. Another open question is how VCP is recruited to the adenovirus–IgG–TRIM21 complex. Does VCP bind directly to TRIM21, or does it bind to the ubiquitin molecules conjugated to TRIM21? Does VCP recruitment require ubiquitin-binding cofactors (13)? Additionally, it remains to be determined whether VCP brings the adenovirus–IgG–TRIM21 complex to the proteasome or whether VCP promotes disassembly of the capsid so that smaller subunits are accessible to proteasome-mediated degradation.
TRIM21 is one of ∼100 tripartite motif genes in the mammalian genome (14). Many of these RING finger E3 ubiquitin ligases have antiviral activity and play roles
VCP is not required for TRIM21-dependent proteasome degradation of all substrates, only very large, multiprotein structures.
in innate immunity (15). One of the best studied of these proteins is TRIM5, a restriction factor for HIV-1 and other retroviruses (16–18). Many parallels exist between TRIM21 and TRIM5. Both associate with viral capsids in the cytoplasm. In the case of TRIM5, the C-terminal PRYSPRY domain binds directly to the capsid as a multimer, forming a lattice that is complementary to the capsid lattice (19). In the case of TRIM21, the PRYSPRY domain binds the IgG that is associated with the capsid. Both proteins inhibit infectivity in a process that involves proteasomes and autoubiquitination with K48-linked ubiquitin chains. In both cases, recruitment to the proteasome requires a ubiquitin-binding, disassembly facilitator, VCP for TRIM21 (3) and p62/sequestosome-1/SQSTM1 for TRIM5 (20). Interestingly, mutations in either of these adaptor proteins are associated with neurogenerative diseases like frontotemporal dementia (21, 22).
In addition to acting as a capsid-specific restriction factor, TRIM5 is a pattern recognition receptor that activates innate immune signaling in response to the retroviral capsid lattice (23). It does so by synthesizing unattached K63-linked ubiquitin chains that activate TGF-beta-activated kinase 1 (TAK1, also known as MAP3K7). This activity is activated by TRIM5 multimerization on the capsid lattice. Purified TRIM21 can synthesize similar chains in vitro (23), so it seems likely that TRIM21 will also activate TAK1-dependent signaling upon association with antibody-coated adenovirus. This would serve to limit infection further by the induction of antiviral cytokines—perhaps including type 1 IFN—that directly inhibit viral replication and alert other components of the acquired immune to the presence of a lethal invader.
Footnotes
The author declares no conflict of interest.
See companion article on page 19733.
References
- 1.Moore ML, McKissic EL, Brown CC, Wilkinson JE, Spindler KR. Fatal disseminated mouse adenovirus type 1 infection in mice lacking B cells or Bruton’s tyrosine kinase. J Virol. 2004;78(11):5584–5590. doi: 10.1128/JVI.78.11.5584-5590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mallery DL, et al. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21) Proc Natl Acad Sci USA. 2010;107(46):19985–19990. doi: 10.1073/pnas.1014074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hauler F, Mallery DL, McEwan WA, Bidgood SR, James LC. AAA ATPase p97/VCP is essential for TRIM21-mediated virus neutralization. Proc Natl Acad Sci USA. 2012;109:19733–19738. doi: 10.1073/pnas.1210659109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vrijsen R, Mosser A, Boeyé A. Postabsorption neutralization of poliovirus. J Virol. 1993;67(6):3126–3133. doi: 10.1128/jvi.67.6.3126-3133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wohlfart C. Neutralization of adenoviruses: Kinetics, stoichiometry, and mechanisms. J Virol. 1988;62(7):2321–2328. doi: 10.1128/jvi.62.7.2321-2328.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emini EA, Ostapchuk P, Wimmer E. Bivalent attachment of antibody onto poliovirus leads to conformational alteration and neutralization. J Virol. 1983;48(2):547–550. doi: 10.1128/jvi.48.2.547-550.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burdeinick-Kerr R, Wind J, Griffin DE. Synergistic roles of antibody and interferon in noncytolytic clearance of Sindbis virus from different regions of the central nervous system. J Virol. 2007;81(11):5628–5636. doi: 10.1128/JVI.01152-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark G, Reichlin M, Tomasi TB., Jr Characterization of a soluble cytoplasmic antigen reactive with sera from patients with systemic lupus erythmatosus. J Immunol. 1969;102(1):117–122. [PubMed] [Google Scholar]
- 9.Chan EK, Hamel JC, Buyon JP, Tan EM. Molecular definition and sequence motifs of the 52-kD component of human SS-A/Ro autoantigen. J Clin Invest. 1991;87(1):68–76. doi: 10.1172/JCI115003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, Eversole T, Lee DJ, Sontheimer RD, Capra JD. Protein-protein interactions between native Ro52 and immunoglobulin G heavy chain. Scand J Immunol. 1999;49(6):620–628. doi: 10.1046/j.1365-3083.1999.00547.x. [DOI] [PubMed] [Google Scholar]
- 11.James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc Natl Acad Sci USA. 2007;104(15):6200–6205. doi: 10.1073/pnas.0609174104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McEwan WA, et al. Regulation of virus neutralization and the persistent fraction by TRIM21. J Virol. 2012;86(16):8482–8491. doi: 10.1128/JVI.00728-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14(2):117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 14.Han K, Lou DI, Sawyer SL. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet. 2011;7(12):e1002388. doi: 10.1371/journal.pgen.1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uchil PD, et al. TRIM protein mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J Virol. 2012 doi: 10.1128/JVI01804-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 17.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430(6999):569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 18.Grütter MG, Luban J. TRIM5 structure, HIV-1 capsid recognition, and innate immune signaling. Curr Opin Virol. 2012;2(2):142–150. doi: 10.1016/j.coviro.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganser-Pornillos BK, et al. Hexagonal assembly of a restricting TRIM5alpha protein. Proc Natl Acad Sci USA. 2011;108(2):534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Connor C, et al. p62/sequestosome-1 associates with and sustains the expression of retroviral restriction factor TRIM5alpha. J Virol. 2010;84(12):5997–6006. doi: 10.1128/JVI.02412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubino E, et al. For the TODEM Study Group SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology. 2012;79(15):1556–1562. doi: 10.1212/WNL.0b013e31826e25df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sieben A, et al. The genetics and neuropathology of frontotemporal lobar degeneration. Acta Neuropathol. 2012;124(3):353–372. doi: 10.1007/s00401-012-1029-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pertel T, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472(7343):361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]