Abstract
Humans host an intestinal population of microbes—collectively referred to as the gut microbiome—which encode the carbohydrate active enzymes, or CAZymes, that are absent from the human genome. These CAZymes help to extract energy from recalcitrant polysaccharides. The question then arises as to if and how the microbiome adapts to new carbohydrate sources when modern humans change eating habits. Recent metagenome analysis of microbiomes from healthy American, Japanese, and Spanish populations identified putative CAZymes obtained by horizontal gene transfer from marine bacteria, which suggested that human gut bacteria evolved to degrade algal carbohydrates—for example, consumed in form of sushi. We approached this hypothesis by studying such a polysaccharide utilization locus (PUL) obtained by horizontal gene transfer by the gut bacterium Bacteroides plebeius. Transcriptomic and growth experiments revealed that the PUL responds to the polysaccharide porphyran from red algae, enabling growth on this carbohydrate but not related substrates like agarose and carrageenan. The X-ray crystallographic and biochemical analysis of two proteins encoded by this PUL, BACPLE_01689 and BACPLE_01693, showed that they are β-porphyranases belonging to glycoside hydrolase families 16 and 86, respectively. The product complex of the GH86 at 1.3 Å resolution highlights the molecular details of porphyran hydrolysis by this new porphyranase. Combined, these data establish experimental support for the argument that CAZymes and associated genes obtained from extrinsic microbes add new catabolic functions to the human gut microbiome.
Two phyla dominate the bacterial community of the adult distal gut in humans and in other mammals: the Firmicutes and the Bacteroidetes (1). Metagenomic studies and experiments with isolated strains revealed that members of the genus Bacteroides are broadly adapted to the degradation of host glycans and terrestrial plant material (2). This adaptation is exemplified by the enzyme systems devoted to starch, pectin, hemicelluloses, and other plant carbohydrates as well as host glycans (2, 3); the wide variety of genes encoding enzymes for the degradation of these carbohydrates dominate carbohydrate active enzyme, or CAZyme, arsenals in human gut metagenome datasets. Hence, the evolution of this carbohydrate catabolizing machinery is key for host and microbiome mutualism. An important question, which has thus far only been addressed by bioinformatic approaches, regards how members of this community evolve—for instance, by horizontal gene transfer (HGT) (4, 5)—to process new carbohydrate resources (6).
In Bacteroidetes, the genes encoding CAZymes that target a specific carbohydrate or related groups of carbohydrates are often found in gene clusters termed polysaccharide utilization loci (PUL) (7). These systems use a generally conserved strategy to sense, degrade, bind, and import carbohydrates encountered in diverse environments like the gut, ocean, and soil (7, 8). The CAZymes (www.cazy.org) (9) cleave high-molecular-weight glycans into oligosaccharides that can be bound by the Sus-like proteins, imported into the bacteria, and ultimately catabolized (10). Using a bioinformatic approach, we recently identified a putative PUL in the gut bacterium Bacteroides plebeius; the PUL contains possible CAZymes encoding genes that appear to have been acquired by HGT from marine microbes and which may target carbohydrates from red seaweeds (4).
The major matrix polysaccharides in the cell walls of red algae—which are the most common dietary red seaweed polysaccharides consumed by humans and present in many processed foods (11, 12)—are carrageenans (13), agars, and porphyran (14), and all contain sulfate esters that are absent in terrestrial plants. Furthermore, the sugar backbones can contain unique monosaccharides, such as the 3,6-anhydro-d-galactose present in the carrageenan of Chondrus crispus (Irish moss) and the 3,6-anhydro-l-galactose (LA) found in agars of Gelidium and Gracilaria spp. In both cases, the 3,6-anhydro-galactose is α-1,3 linked to d-galactose (G) and the resulting disaccharide is connected with β-1,4 linkages (SI Appendix, Fig. S1). Porphyran from Porphyra spp. belongs to the agar-family of polysaccharides, although the majority of the LA is replaced by l-galactose-6-sulfate (L6S), and C-6 methylations of G are frequent (14). The unique structural properties of these red algal galactans requires a distinct set of enzymes that are predominantly encoded in genomes of marine microbes and are far less frequent or absent in bacteria that break down terrestrial polysaccharides (4, 8). Thus, genetic and functional tracking of CAZymes with these rare specificities holds the potential to dissect carbohydrate resource partitioning by human gut microbes. For example, putative agarases, porphyranases, and alginate lyases have recently been identified in the intestinal microbiomes of Japanese (4), Spanish (15), and American (16) people, suggesting that their gut microbes may have adapted to novel algal carbohydrates in the modern diet. However, all of these previous studies focused on bioinformatic analysis and lacked functional characterizations of gut microbes and their enzymes, raising the question of whether these HGT events conferred active pathways to the human microbiome or simply introduced cryptic genes.
In this study, we used seaweed polysaccharides as substrates and established that a horizontally acquired integrative and conjugative element (ICE) expanded the catabolic repertoire of B. plebeius allowing it to use porphyran. Furthermore, through functional screening with additional algal substrates, two other gut Bacteroides, B. uniformis NP1 and B. thetaiotaomicron VPI-3731, were identified, which grew on agar and on carrageenan, contrasting previous reports that suggested such activities are lacking in intestinal microbes (17). Using B. plebeius as a model organism, we show that when the bacterium was grown using porphyran as a sole carbon source, the genes of the PUL were up-regulated. Consistent with this result is the presence of active β-porphyranases, the crystal structures of which are presented, including a structural analysis of a family 86 glycoside hydrolase. Our results show that gut bacteria catabolize red seaweed galactans, a feature that may influence health (18) and refines our understanding of the evolutionary plasticity of the human gut microbiome.
Results
Screening of Gut Bacteria for Growth on Red Algal Galactans.
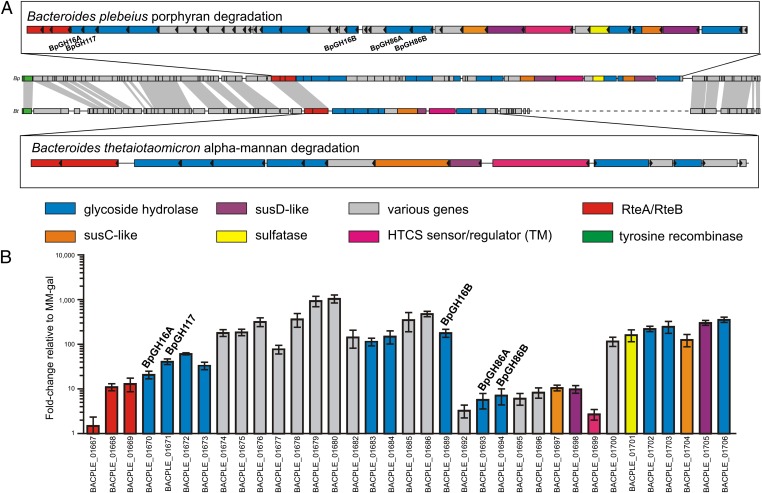
B. plebeius contains a putative porphyran degrading PUL covering up to 40 ORFs from BACPLE_01667 to BACPLE_01706. Within this set, the products of two genes (BACPLE_01670 and BACPLE_01689) share high amino acid sequence identity with GH16 homologs from marine seaweed-degrading microbes; these homologs have previously demonstrated activity on porphyran. This finding suggested that agarose, porphyran, or even carrageenan may be nutrients for this gut microbe. Thus, we used these three galactans as substrates and observed that B. plebeius grew specifically on porphyran (SI Appendix, Fig. S2A). To test whether this result was an isolated case or if these common algal food galactans are catabolized by other human gut microbes, we screened an additional collection of 291 human gut Bacteroidetes (see SI Appendix for details) and identified several carrageenolytic and agarolytic bacteria. One isolate, B. uniformis NP1, grew on agarose (SI Appendix, Fig. S2B) and showed reduced growth on porphyran, whereas another, B. thetaiotaomicron VPI-3731, exhibited growth specificity for carrageenan (SI Appendix, Fig. S2C). Together, these growth experiments showed that human gut bacteria catabolize seaweed carbohydrates and that each of these strains is specific for a certain type of red algal galactan. To further understand the molecular mechanism of the degradation of such galactans, we focused on the PUL of B. plebeius for which the whole genome sequence is available.
B. plebeius PUL Is Activated by Porphyran.
The 40 genes that comprise the putative B. plebeius PUL include 12 putative glycoside hydrolases, 2 SusC/SusD-like protein pairs (encoded by BACPLE_01697–01698 and BACPLE_01704–01705), and a hybrid two-component system (HTCS) sensor/regulator (ref. 19; BACPLE_01699) (Fig. 1A and SI Appendix, Table S2). Using quantitative PCR (qPCR), the expression of 35 genes in the PUL was examined in response to porphyran as the only carbohydrate source in the growth medium (growth in galactose served as a reference). Compared with galactose, porphyran promoted expression of all 35 examined genes (Fig. 1B). A notable feature is the mosaic profile in which the expression of genes BACPLE_01692–01699, measured by fold-change analysis, appears lower. We quantified transcript abundance of the low-fold-change susC/susD genes (BACPLE_01697–01698) and compared them to the high-fold-change susC/susD genes (BACPLE_01704–01705), using genomic DNA that contained equal copies of each gene as a standard. Our results indicated that the susC/susD genes (BACPLE_01697–10698) that exhibited low fold change had 16- and 18-fold higher basal expression, respectively, in MM-galactose, suggesting that these genes (BACPLE_01692–01699) are actually expressed at a higher level in the absence of porphyran.
Fig. 1.
Expression of genes when B. plebeius is grown in porphyran. (A) Synteny between the adjacent genomic region surrounding the porphyran PUL to a fungal α-mannan–specific PUL in B. thetaiotaomicron. (B) The expression of 35 genes within the PUL was monitored by RT-PCR upon incubation with porphyran compared with the control incubated with galactose. Five additional genes ranging in size from 126 to 489 bp (BACPLE_01681, 01687, 01688, 01690, and 01691) are present within this locus but were not probed by qPCR. Data in B are mean and SD of independent biological replicates.
B. plebeius PUL Is Contained in an ICE.
Notably, the final three genes (BACPLE_01667–01669) in the PUL encode products that are homologous to RteA and RteB, which are two-component sensor histidine kinase and response regulator proteins, respectively (note that the BACPLE_01667/01668 ORFs each encode part of an RteB homolog that is split by a frameshift mutation that was validated by resequencing). RteA and RteB are important to initiate the excision and mobilization of Bacteroides conjugative transposons that carry antibiotic-resistance genes (20). Examination of the genomic region surrounding these porphyran-inducible genes further suggested that the PUL is contained within an ICE, which includes genes that encode mobilization proteins and an integrase tyrosine recombinase that lies immediately adjacent to a tRNAlys (Fig. 1A). We identified a pair of identical 18-bp direct repeats, one of which overlaps the 3′ end of tRNAlys and the other located 107,705 bp upstream, suggesting that this location is the integration site at which this putative ICE entered the B. plebeius genome.
The HGT notion is further supported by the identification of a related element in the genome of B. thetaiotaomicron VPI-5482. In contrast to the porphyran ICE, the element contained in B. thetaiotaomicron harbors a PUL involved in fungal α-mannan degradation (21), but it is located amid ICE genes that are homologous between both species, including homologs of RteA and RteB (genes connected by gray bars in Fig. 1A). Thus, these two ICEs have very similar organization, but contain different PUL cargo. Unlike the B. plebeius ICE, the one in B. thetaiotaomicron is located adjacent to a tRNAphe, suggesting that this location is its integration site. Similar to B. plebeius, there is a pair of 22-bp direct repeat sequences that are located 90,450 bp apart that define the “left” and “right” ends of these putative ICEs, which share homologous and syntenic genetic organization. Comparison of the surrounding genomic regions in B. thetaiotaomicron VPI-5482 and two other sequenced B. thetaiotaomicron strains revealed the precise absence of all 90,450 bp that are located between the direct repeats. Finally, using PCR with primers that flank the putative ICE insertion sites in both species, we were able to validate excision of these elements. Sequencing of the recombined amplification products confirmed that the 18- and 22-bp direct repeat sequences do indeed mediate recombination (SI Appendix, Fig. S3). Despite the fact that expression of each PUL activates transcription of its associated rteA and rteB genes—a feature that suggests these two elements may be transferred in response to their cognate substrate—we were unable to detect increased excision or circularization of either element in response to porphyran or α-mannan. An attempt to transfer the B. plebeius ICE into B. thetaiotaomicron was unsuccessful (SI Appendix), suggesting that this element is either no longer mobile or that we have not yet identified appropriate conditions for high-frequency excision and subsequent transfer.
B. plebeius PUL Contains an Active Agarase and Active Porphyranases.
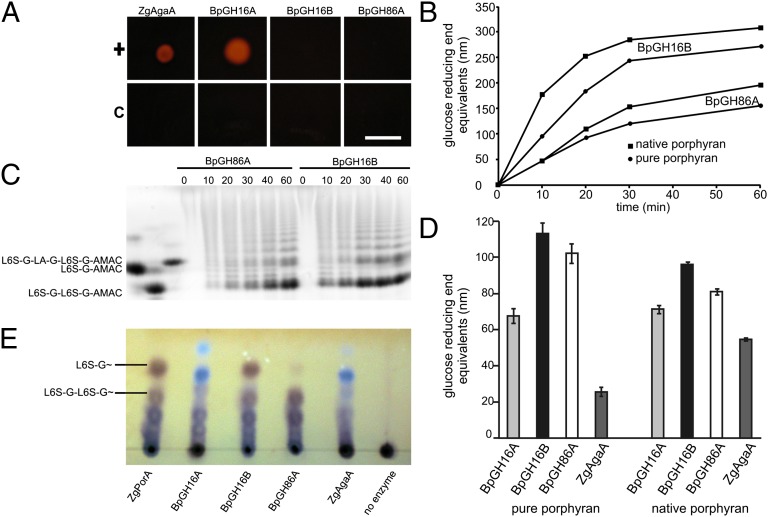
Of the 12 putative glycoside hydrolase encoding genes in the B. plebeius porphyran PUL, 4 are predicted to have endo-polysaccharolytic activity and may thus initiate porphyran degradation. Two family 16 glycoside hydrolases have been identified: BpGH16A (BACPLE_01670) and BpGH16B (BACPLE_01689), with putative agarolytic and porphyranolytic activity, respectively. The two family 86 glycoside hydrolases, BpGH86A (BACPLE_01693) and BpGH86B (BACPLE_01694), have unknown specificity, yet all characterized enzymes in family GH86 have been β-agarases, which suggested a similar function in B. plebeius. To analyze these putative agarases and porphyranases, we cloned the gene fragments encoding their catalytic domains, produced them in Escherichia coli, and examined their activities.
BpGH16A, BpGH16B, and BpGH86A were assessed for agarolytic and porphyranolytic activity, whereas BpGH86B could not be expressed. BpGH16A was active on solid agarose, supporting its predicted agarolytic function, whereas BpGH16B and also BpGH86A were inactive on this substrate (Fig. 2A); thus, we also tested their activity on native and pure porphyran (Fig. 2B). Although agarases possess comparable activities on native porphyran, they show significantly lower activity on pure porphyran, which allows one to differentiate between porphyranases and agarases (4). Both BpGH16B and BpGH86A were active on native and pure porphyran preparations (Fig. 2B), whereas BpGH16A showed a significantly lower level of activity on pure and native porphyran preparations (Fig. 2D), comparable to the β-agarase ZgAgaA from Zobellia galactanivorans (22).
Fig. 2.
B. plebeius contains two endo-acting porphyranases, BpGH16B and BpGH86A, and an agarase, BpGH16A. (A) Activity test on agar plate revealed similar agarase activity of BpGH16A to the previously characterized ZgAgaA from Z. galactanivorans (positive control). BpGH16B and GH86A were inactive on this substrate. Heat-inactivated enzymes served as control. (Scale bar: 1 cm.) (B) Relative activity of BpGH16B and BpGH86A with porphyran as substrate measured by reducing sugar assays showed porphyranolytic activity. (C) The reaction products of BpGH16B and BpGH86A, incubated with native porphyran, were analyzed by fluorophore-assisted PACE showing a ladder-like pattern typical for endo-acting glycoside hydrolases. (D) Comparative hydrolysis of native and pure porphyran by B. plebeius enzymes and ZgAgaA showing that BpGH16B and BpGH86A have higher activity than the agarases. (E) TLC analysis of degradation products reveals that BpGH16B releases similar reaction products to the previously characterized ZgPorA (positive control), which releases L6S-G∼ and L6S-G-L6S-G∼ as major products (14), whereas BpGH86A releases mainly the larger oligosaccharide L6S-G-L6S-G∼. Notably, these products are not released by the previously characterized agarase ZgAgaA (negative control). BpGH16A has an agarase-like reaction pattern similar to ZgAgaA. Data in D are mean and SD of independent enzymatic replicates.
For product profiling, porphyran was treated with BpGH16B or BpGH86A, and the reaction products were labeled with the fluorophore 2-aminoacridone (AMAC) and analyzed by carbohydrate polyacrylamide gel electrophoresis (PACE). Both BpGH86A and BpGH16A successively degraded the high-molecular-weight polysaccharide into smaller oligosaccharides of random size, apparent in the ladder-type pattern that is typical for endo-acting glycoside hydrolases (Fig. 2C). One well-resolved degradation product from both enzymes corresponded to a band that had the same mobility as an AMAC-labeled tetra-oligosaccharide standard derived from porphyran (L6S-G-L6S-G-AMAC), which is consistent with cleavage of the β-glycosidic linkage (for sugar residue nomenclature, see SI Appendix, Fig. S1). We further used TLC to analyze the reaction products of the different enzymes (Fig. 2E). BpGH16B showed a reaction profile similar to the β-porphyranase ZgPorA from Z. galactanivorans, with L6S-G∼ as a major end product (14). BpGH86A produced predominantly larger oligosaccharides, the smallest of which was the tetra-oligosaccharide L6S-G-L6S-G∼. Consistent with its agarolytic activity, BpGH16A had a different reaction profile than the porphyranases, yet a similar reaction profile to the β-agarase ZgAgaA; the major reaction products showed the typical blue color of neoagarooligosaccharides as observed with the resorcinol staining procedure (23). Combined, these results support the assignment of BpGH16A as a β-agarase, whereas BpGH16B and BpGH86A are β-porphyranases.
Crystal Structure of the Porphyranase BpGH86A.
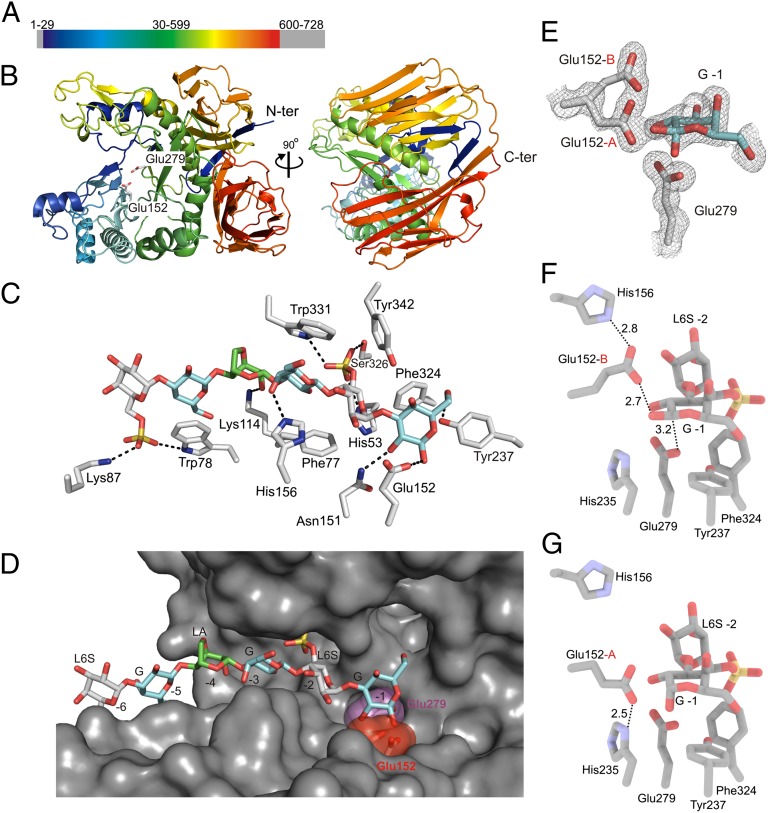
After establishing that BpGH86A is a porphyranase, we determined its structure to 1.3-Å resolution. The structure of BpGH86A, comprising the catalytic domain (residues 25–599) (Fig. 3A), revealed a multidomain architecture consisting of an N-terminal (β/α)8 barrel and two C-terminal β-sandwich domains, which align via their convex faces to the exterior of the (β/α)8 barrel and connect with two N-terminal β-strands that become part of these C-terminal β-sandwich domains (Fig. 3B; data collection and refinement statistics are summarized in SI Appendix, Table S1). The (β/α)8 barrel fold, or TIM-barrel, is found in glycoside hydrolases belonging to clans GH-A,D,H,K, and it generally forms a closed ring structure (24), the center of which harbors the catalytic residues. In BpGH86A, the distal part of the barrel lacks the helix α2 in the outer ring and parts of the surface loops, which form the rim in the canonical (β/α)8 barrel fold. The absence of this distal α-helix and associated loops shapes the structure of the enzyme into an open toroid, further accentuating an already pronounced cleft of ∼40-Å length (SI Appendix, Fig. S4A), which is longer than in BpGH16B (∼32 Å; SI Appendix, Fig. S5A) and consistent with the longer oligosaccharides produced by BpGH86A.
Fig. 3.
The structure of BpGH86A features a toroid-TIM barrel with an extended substrate binding cleft and two accessory β-sandwich domains and the complex with porphyran/agarose hexasaccharide supports a retaining mechanism. (A) The primary structure of BpGH86A consists of an N-terminal signal peptide, the central part which contains the catalytic TIM barrel domain of family GH86 and a C-terminal domain of unknown function. (B) Cartoon plot of the catalytic domain, color ramped from blue (N terminus) to red (C terminus) and with a view centered on the active site with the two putative catalytic residues and of the back of the enzyme where the two N-terminal β-sheets complement the C-terminal β-sandwich domains. (C) Stick representation showing the hydrophobic, ionic, and hydrogen-bond interactions between enzyme and the bound hexasaccharide. (D) Surface representation with the hexasaccharide bound to the − subsites with the putative nucleophile in magenta and the putative acid/base colored with a red surface. (E) The electron densities of the G in subsite −1, the acid/base glutamate in its double conformation, and the nucleophile are shown in a gray mesh as a refined maximum likelihood/σa-weighted 2Fo − Fc map contoured at 1σ (0.19 e/Å3). (F) The conformation of the catalytic acid/base residue (Glu-152-A) that engages the β-anomer of the g in subsite −1 is depicted. (G) The conformer of the acid/base glutamate that interacts with the α-anomer is presented.
Product Complex of BpGH86A with a Hybrid Porphyran-Agarose-Oligosaccharide.
The crystal structure revealed unambiguous electron density for the six sugar rings of an oligosaccharide bound in the pronounced cleft of the enzyme. The electron density showed that this bound oligosaccharide, likely a product from the hydrolysis of the porphyran added during crystallization, was a hybrid-hexamer of structure L6S-G-LA-G-L6S-G∼ occupying six minus subsites (25) (Fig. 3C and SI Appendix, Fig. S3B). A series of hydrogen bonds and hydrophobic interactions with the enzyme stabilized the carbohydrate.
The reducing end of the oligosaccharide is a G residue that fits into a subsite whose architecture, namely, the presence of the putative catalytic residues Glu-152 and -279 (see below), suggest it is the −1 subsite. Cleavage at this site of the polysaccharide is consistent with the classification of BpGH86A as a β-galactanase that cleaves the β-1,4 glycosidic bond in porphyran (Fig. 3C). In this −1 subsite, the sugar ring is situated above and stabilized by the hydrophobic platform formed by the residues Tyr-237 and Phe-324, and its C6–OH projects toward a small pocket in the active site (Fig. 3 C and D). This pocket may accommodate the methyl substitutions that are frequently found on the C6 of G residues in porphyran; such substitutions inhibit productive binding in the GH16 porphyranases because they lack such a pocket (SI Appendix, Fig. S5 E and F) (14).
The L6S bound in subsite −2 is surrounded by His-53, Phe-77, Phe-324, and Tyr-342, and these side chains form the cavity that harbors the sugar ring and the sulfate substitution on C6 (Fig. 3C). This sulfate group points toward the interior of the substrate binding cleft and is accommodated in a pocket similar to BpGH16B (SI Appendix, Fig. S5F) and previously described GH16 β-porphyranases (4). Furthermore, His-53, located below the sugar ring, stabilizes the sulfate group by an ionic interaction.
The G unit bound in subsite −3 is clamped between Tyr-331 and Phe-77, which align with their β- and α-faces, respectively, with the sugar ring. The axial hydroxyl group on C4 of the sugar makes a hydrogen bond to His-114. Compared with the three sugar rings that are bound in subsites −1, −2, and −3, the LA in subsite −4 is far less engaged by the enzyme and makes a single hydrogen bond with Lys-76. This bicyclical sugar is in the 4C1 conformation compared with the 1C4 conformation of the L6S. This arrangement leads to all equatorial glycosidic bonds between the LA and adjacent G sugars and an almost coplanar arrangement of these three sugar rings, highlighting the conformational and chemical heterogeneity introduced by varying anhydro- or C6 sulfate modifications in natural agars and carrageenans (SI Appendix, Fig. S1). The paucity of interactions in the −4 subsite allows binding of either L6S or LA, and therefore hydrolysis close to interspersed LA-G motifs in porphyran. However, because the bound hexasaccharide was not further degraded to L6S-G∼ and LA-G-L6S-G∼ or L6S-G-LA-G∼, we assume that BpGH86A does not accept LA in subsites −2 or +1. The G residue bound in subsite −5 stacks with its β-face to Trp-78, and the L6S bound in subsite −6 is stabilized by the side chains of Trp-83 and the Lys-87, which form an ion bridge to the sulfate group on C6 (Fig. 3C).
Active-Site Architecture of BpGH86A Supports a Retaining Catalytic Mechanism.
The G in subsite −1 is present in an undistorted 4C1 conformation, and both the β- and α-anomer have been modeled at the C1 atom (Fig. 3E). The G interacts with the putative acid/base residue Glu-152, which was modeled in a double conformation. One conformation makes a hydrogen bond to the equatorial hydroxyl group (Fig. 3F), and the other conformation coincides with the axial hydroxyl group at the C1 of the sugar ring (Fig. 3G).
Hydrogen bonds with two histidine residues stabilize each of the two conformations of Glu-152. His-235 forms a hydrogen bond (2.8 Å) to the conformer of Glu-152 that approaches the equatorial hydroxyl; in this position, Glu-152 is appropriately positioned to protonate the β-1,4 glycosidic bond (Fig. 3F), whereas Glu-279 attacks the anomeric carbon from below at a distance of 3.2 Å, supporting its role as the catalytic nucleophile. Thus, His-235 may reduce the pKa of Glu-152, which becomes poised to act as an acid for protonation of the glycosidic bond. The second conformer of Glu-152 further approaches the C1 with its now axial C1–OH and comes to rest almost at place of the equatorial hydroxyl group of the β-anomer (Fig. 3G). This conformer is stabilized by a hydrogen bond to His-235 (2.5 Å), and we suggest that this second conformation may protect the C1 of the bound intermediate from the back reaction with the aglycone in a transglycosylation reaction. Thus, the deglycosylation step may be initiated when Glu-152 turns back to its interaction with His-235 and now as a base activates the incoming water that attacks at C1 and releases the glycon. It is further possible that the dynamic range of interactions with both histidines modulates pKa cycling of the Glu-152 during the retaining mechanism, according to its dual roles as an acid and a base (26). However, as an alternative or complementing scenario, we propose that the role of this interaction, between Glu-152 and His-235, may be a molecular ratchet favoring hydrolysis over transglycosylation (27), which may be beneficial in cases where strong binding between products and enzyme are encountered.
Discussion
The collective group of CAZymes in the human gut microbiome is large and diverse, allowing it to extract energy and carbon from a wide variety of carbohydrates. Using B. plebeius as a model microbiome species that possesses a rare catalytic ability, we were able to associate a 40-gene PUL with the catabolism of porphyran, a structurally unique algal polysaccharide. The biochemical analysis revealed that the encoded agarase (BpGH16A) and porphyranases (BpGH86A, BpGH16B) concertedly degrade porphyran, consistent with the heterogeneous nature of this galactan.
The porphyran PUL displayed a mosaic expression profile in which its central part, including genes BACPLE_01692–01699, showed lower relative induction by porphyran because of their higher basal expression in galactose. The high basal expression of these genes, even in the absence of their polysaccharide substrate, suggested the production of “surveillance” levels of their respective enzymes. This common theme in gut Bacteroides delivers small amounts of each PUL-encoded system to initially sense and begin the degradation of a specific polysaccharide (21). In particular, we postulate that BpGH86A, located within the surveillance set of genes, with its longer substrate binding cleft compared with GH16 porphyranases, may play a key role by performing the initial production of larger oligosaccharides, allowing the HTCS protein and Sus-like system to sense and respond to their presence (2). The release of oligosaccharides from porphyran, and not agarose, which failed to induce growth of the bacterium, subsequently triggers the production of the porphyranases (BpGH16B), agarases [BpGH16A, BpGH117 (28)], and additional CAZymes and accessory proteins [sulfatase (29)] that can further deconstruct these larger oligosaccharides (21).
The acquisition of the B. plebeius porphyran PUL is likely to have originated from a marine Bacteroidete because similar gene clusters are present in oceanic species (8, 30–32). Because the B. plebeius PUL is harbored at a similar location on an otherwise very similar ICE that is present in B. thetaiotaomicron, this finding suggests that HGT via conjugative elements is the mechanism of transfer and that similar ICE vehicles are capable of acquiring new cargo (20). The observation that both the B. plebeius and B. thetaiotaomicron elements were observed to excise and circularize from the genome via site-specific recombination events supports our conclusion that we have identified the extent of these ICEs involved in PUL transfer. The apparently low excision rate, which was only detected with 36–70 additional cycles of PCR, suggests that these two example ICEs do not undergo efficient excision in the growth conditions tested, which included the cognate substrates that trigger expression of the RteA/B homologs. The frameshift mutation in the B. plebeius RteB homolog indicates that this ICE has lost the ability to excise and mobilize by the same mechanism as that of Bacteroides conjugative transposons with similar structure (20).
The degradation of seaweeds by intestinal microbes may have health implications because red algal galactans, including porphyran, have been associated with a wide range of pharmacological activities, such as antiviral, anticancer, anti-inflammatory, antioxidative, and anticoagulative effects (see ref. 33 for review). These activities largely depend on the molecular weight of the polymer (34) and the amount of sulfations along the polysaccharide chain, both factors which are modulated with specific microbial CAZymes and sulfatases (29). Indeed, health implications and concerns have been associated with the low-molecular-weight degradation products of carrageenan, which elicited ulcerative colitis in animal models (11, 35–37). Our screening for red algal galactan degrading gut microbes revealed B. thetaiotaomicron VPI-3731, which presented strong growth on carrageenan and must therefore contain one or more carrageenases. Future studies are needed to characterize the enzymes involved in these red algae degrading pathways and to test whether they are distributed in human gut microbiomes and form harmful oligosaccharides in vivo.
Overall, this work demonstrates that B. plebeius, a member of the gut microbiome, has an active PUL that degrades the marine red algal polysaccharide porphyran. This finding supports the hypothesis that the gut microbiome coevolves with host diet through HGT from extrinsic microbes enabling the catabolism of new carbohydrates.
Materials and Methods
Gut isolates were grown as described (21), and for qPCR of B. plebeius PUL genes, B. plebeius DSM 17135 was grown in triplicate on either galactose or porphyran as a sole carbon source, and cells were harvested during exponential growth (A600 values of 0.41, 0.42, 0.43 for galactose and 0.57, 0.56, 0.57 for porphyran). The catalytic domains of the porphyranases and agarase were cloned from B. plebeius DSM 17135 genomic DNA (28). Crystallization screenings were carried out in sitting-drop experiments by using commercial screens and optimizations in hanging-drop setups by grid screen expansions around initially successful hits. Biochemical characterization of the enzymes was carried out as described (4, 14). The structure of BpGH16B was solved by molecular replacement, and the structure of BpGH86A was solved by the single-wavelength anomalous dispersion method. The data collection and refinement statistics are listed in SI Appendix, Table S1. (For additional details, see SI Appendix, SI Materials and Methods.)
Supplementary Material
Acknowledgments
We thank Dr. Mirjam Czjzek for recombinant agarases and porphyranases from Z. galactanivorans and for the photograph of C. crispus. We thank the Stanford Synchrotron Radiation Lightsource Beamline 9-2 staff and also the Canadian Light Source Canadian Macromolecular Crystallography Facility staff. This work was supported by a Natural Sciences and Engineering Research Council of Canada Discovery Grant. The Canadian Light Source is supported by the Natural Sciences and Engineering Research Council of Canada, the National Research Council Canada, the Canadian Institutes of Health Research, the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan. J.-H.H. was supported by an European Molecular Biology Organization Long Term Fellowship. Work performed in the E.C.M. laboratory was supported by National Institutes of Health Career Development Grant K01DK084214. A.B.B. is a Canada Research Chair in Molecular Interactions and a Michael Smith Foundation for Health Research Career Scholar.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4AW7 and 4AWD).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211002109/-/DCSupplemental.
References
- 1.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martens EC, et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9(12):e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonnenburg ED, et al. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141(7):1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hehemann J-H, et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464(7290):908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 5.Smillie CS, et al. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480(7376):241–244. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 6.Sonnenburg JL. Microbiology: Genetic pot luck. Nature. 2010;464(7290):837–838. doi: 10.1038/464837a. [DOI] [PubMed] [Google Scholar]
- 7.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Complex glycan catabolism by the human gut microbiota: The Bacteroidetes Sus-like paradigm. J Biol Chem. 2009;284(37):24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas F, Hehemann J-H, Rebuffet E, Czjzek M, Michel G. Environmental and gut bacteroidetes: The food connection. Front Microbiol. 2011;2:93. doi: 10.3389/fmicb.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shipman JA, Berleman JE, Salyers AA. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J Bacteriol. 2000;182(19):5365–5372. doi: 10.1128/jb.182.19.5365-5372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michel C, Macfarlane GT. Digestive fates of soluble polysaccharides from marine macroalgae: Involvement of the colonic microflora and physiological consequences for the host. J Appl Bacteriol. 1996;80(4):349–369. doi: 10.1111/j.1365-2672.1996.tb03230.x. [DOI] [PubMed] [Google Scholar]
- 12.Bixler HJ. Recent developments in manufacturing and marketing carrageenan. Hydrobiologia. 1996;326-327:35–57. [Google Scholar]
- 13.Michel G, Nyval-Collen P, Barbeyron T, Czjzek M, Helbert W. Bioconversion of red seaweed galactans: A focus on bacterial agarases and carrageenases. Appl Microbiol Biotechnol. 2006;71(1):23–33. doi: 10.1007/s00253-006-0377-7. [DOI] [PubMed] [Google Scholar]
- 14.Correc G, Hehemann J-H, Czjzek M, Helbert W. Structural analysis of the degradation products of porphyran digested by Zobellia galactanivorans [beta]-porphyranase A. Carbohydr Polym. 2011;83:277–283. [Google Scholar]
- 15.Rebuffet E, et al. Discovery and structural characterization of a novel glycosidase family of marine origin. Environ Microbiol. 2011;13(5):1253–1270. doi: 10.1111/j.1462-2920.2011.02426.x. [DOI] [PubMed] [Google Scholar]
- 16.Cantarel BL, Lombard V, Henrissat B. Complex carbohydrate utilization by the healthy human microbiome. PLoS ONE. 2012;7(6):e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salyers AA, West SE, Vercellotti JR, Wilkins TD. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl Environ Microbiol. 1977;34(5):529–533. doi: 10.1128/aem.34.5.529-533.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tobacman JK. Review of harmful gastrointestinal effects of carrageenan in animal experiments. Environ Health Perspect. 2001;109(10):983–994. doi: 10.1289/ehp.01109983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowe EC, Baslé A, Czjzek M, Firbank SJ, Bolam DN. A scissor blade-like closing mechanism implicated in transmembrane signaling in a Bacteroides hybrid two-component system. Proc Natl Acad Sci USA. 2012;109(19):7298–7303. doi: 10.1073/pnas.1200479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon K, Shoemaker NB, Gardner JF, Salyers AA. Regulation of excision genes of the Bacteroides conjugative transposon CTnDOT. J Bacteriol. 2005;187(16):5732–5741. doi: 10.1128/JB.187.16.5732-5741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4(5):447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allouch J, et al. The three-dimensional structures of two beta-agarases. J Biol Chem. 2003;278(47):47171–47180. doi: 10.1074/jbc.M308313200. [DOI] [PubMed] [Google Scholar]
- 23.Duckworth M, Yaphe W. Thin-layer chromatographic analysis of enzymic hydrolysates of agar. J Chromatogr A. 1970;49(3):482–487. doi: 10.1016/s0021-9673(00)93663-x. [DOI] [PubMed] [Google Scholar]
- 24.Henrissat B, et al. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc Natl Acad Sci USA. 1996;93(11):5674. doi: 10.1073/pnas.93.11.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies GJ, Wilson KS, Henrissat B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem J. 1997;321(Pt 2):557–559. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McIntosh LP, et al. The pKa of the general acid/base carboxyl group of a glycosidase cycles during catalysis: A 13C-NMR study of bacillus circulans xylanase. Biochemistry. 1996;35(31):9958–9966. doi: 10.1021/bi9613234. [DOI] [PubMed] [Google Scholar]
- 27.Baumann MJ, et al. Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: Biological implications for cell wall metabolism. Plant Cell. 2007;19(6):1947–1963. doi: 10.1105/tpc.107.051391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hehemann J-H, Smyth L, Yadav A, Vocadlo DJ, Boraston AB. Analysis of keystone enzyme in Agar hydrolysis provides insight into the degradation (of a polysaccharide from) red seaweeds. J Biol Chem. 2012;287(17):13985–13995. doi: 10.1074/jbc.M112.345645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjdia A, Martens EC, Gordon JI, Berteau O. Sulfatases and a radical S-adenosyl-L-methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont, Bacteroides thetaiotaomicron. J Biol Chem. 2011;286(29):25973–25982. doi: 10.1074/jbc.M111.228841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong Z, et al. Sequence analysis of a 101-kilobase plasmid required for agar degradation by a Microscilla isolate. Appl Environ Microbiol. 2001;67(12):5771–5779. doi: 10.1128/AEM.67.12.5771-5779.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teeling H, et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336(6081):608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 32.Bauer M, et al. Whole genome analysis of the marine Bacteroidetes ‘Gramella forsetii’ reveals adaptations to degradation of polymeric organic matter. Environ Microbiol. 2006;8(12):2201–2213. doi: 10.1111/j.1462-2920.2006.01152.x. [DOI] [PubMed] [Google Scholar]
- 33.Holdt S, Kraan S. Bioactive compounds in seaweed: Functional food applications and legislation. J Appl Phycol. 2011;23:543–597. [Google Scholar]
- 34.Hatada Y, Ohta Y, Horikoshi K. Hyperproduction and application of α-agarase to enzymatic enhancement of antioxidant activity of porphyran. J Agric Food Chem. 2006;54(26):9895–9900. doi: 10.1021/jf0613684. [DOI] [PubMed] [Google Scholar]
- 35.Marcus AJ, Marcus SN, Marcus R, Watt J. Rapid production of ulcerative disease of the colon in newly-weaned guinea-pigs by degraded carrageenan. J Pharm Pharmacol. 1989;41(6):423–426. doi: 10.1111/j.2042-7158.1989.tb06493.x. [DOI] [PubMed] [Google Scholar]
- 36.Carthew P. Safety of carrageenan in foods. Environ Health Perspect. 2002;110(4):A176–A177. doi: 10.1289/ehp.110-a176a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhattacharyya S, et al. Carrageenan-induced innate immune response is modified by enzymes that hydrolyze distinct galactosidic bonds. J Nutr Biochem. 2010;21(10):906–913. doi: 10.1016/j.jnutbio.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.