Abstract
Capture and isolation of flowing cells and particulates from body fluids has enormous implications in diagnosis, monitoring, and drug testing, yet monovalent adhesion molecules used for this purpose result in inefficient cell capture and difficulty in retrieving the captured cells. Inspired by marine creatures that present long tentacles containing multiple adhesive domains to effectively capture flowing food particulates, we developed a platform approach to capture and isolate cells using a 3D DNA network comprising repeating adhesive aptamer domains that extend over tens of micrometers into the solution. The DNA network was synthesized from a microfluidic surface by rolling circle amplification where critical parameters, including DNA graft density, length, and sequence, could readily be tailored. Using an aptamer that binds to protein tyrosine kinase-7 (PTK7) that is overexpressed on many human cancer cells, we demonstrate that the 3D DNA network significantly enhances the capture efficiency of lymphoblast CCRF-CEM cells over monovalent aptamers and antibodies, yet maintains a high purity of the captured cells. When incorporated in a herringbone microfluidic device, the 3D DNA network not only possessed significantly higher capture efficiency than monovalent aptamers and antibodies, but also outperformed previously reported cell-capture microfluidic devices at high flow rates. This work suggests that 3D DNA networks may have broad implications for detection and isolation of cells and other bioparticles.
Keywords: circulating tumor cells, multivalency, point-of-care, cell sorting, microfluidics
Capture and isolation of flowing cells and particulates from a biological fluid (i.e., peripheral blood), including tumor cells, bacteria, viruses, and exosomes, is important for disease diagnosis, monitoring therapy, elucidating biology, and developing new drugs (1–4). Conventional technologies are limited to multistep, complex, and inefficient methods, such as immunomagnetic enrichment or flow cytometric cell sorting (1, 2, 5). The more recent microfluidic devices that use monovalent capture agents, including antibodies (6) and nucleic acid aptamers (7–9), represent a promising approach to capture circulating cells with potentially greater simplicity, sensitivity and throughput (10, 11). However, such monovalent adhesion ligands that extend only a few nanometers into the solution fail to capture large-sized entities, such as cells (∼10–30 μm) under high shear stress, which renders the assay inefficient, time-consuming, and unable to rapidly process large volumes of blood. Thus, the existing devices for cell capture are typically designed with low channel height (7) and with complex surface topologies [e.g., microposts (6), herringbone grooves (12), or 3D structures (13–16)]. Even in these geometries, efficient cell separation requires low shear stress conditions to maximize cell-surface contact (6). Moreover, cell separation devices typically fail to gently release captured cells, which limits observation of the cells to the device substrate and presents significant challenges for subsequent molecular analysis to elucidate new biology and screen for effective chemotherapeutic drugs (6).
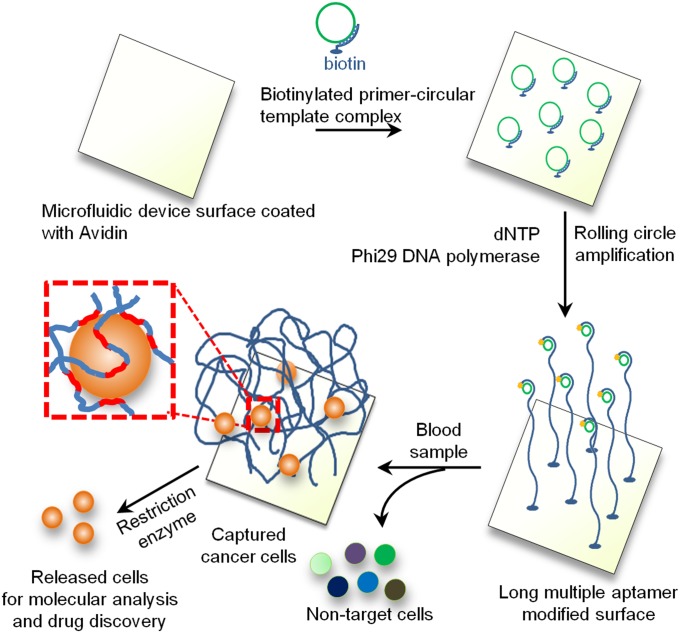
Many creatures in nature, such as jellyfish and sea cucumbers, have evolved long tentacles or arms to effectively capture flowing food particles and prey (17, 18). A common feature of these tentacles is that they are long, extend into the flow, and contain repeating units of adhesive moieties (e.g., mucus), which maximizes the contact with flowing targets and therefore the capture efficiency (17, 18). Inspired by nature, we herein present a unique method to effectively capture and isolate flowing entities in peripheral blood including cancer cells using a microfluidic chip tethered with long (tens to hundreds of microns) DNA molecules containing multiple target-binding aptamers (typically hundreds of repeating aptamer units/strand). As shown in Fig. 1, we hypothesized that the aptamer-decorated DNA strands would enable effective capture by binding to cell surface markers in a cooperative manner to enhance avidity, leading to higher cell capture efficiency. Moreover, the extension of DNA strands into the 3D space could increase the accessibility and frequency of interactions to permit cell capture under high flow rates. This effect may be especially important as the deformability of the cells induces repulsive hydrodynamic forces (19) that may limit cell-surface interactions at higher shear stresses. Finally, the DNA strands can be cleaved by restriction enzymes, which permits the captured cells to be easily released for molecular analysis.
Fig. 1.
Isolation and detection of cancer cells in whole blood using long multivalent DNA aptamer-based microfluidic device. The zoomed-in box illustrates a captured cell is bound by several long DNA molecules via multiple aptamer domains (red color).
Results
Long Rolling Circle Amplification Products Containing Multiple Aptamers Bind to Target Cells Specifically and More Effectively than Monovalent Aptamers.
Long DNA strands are produced by an isothermal, enzymatic polymerization method called rolling circle amplification (RCA) (20, 21), where DNA polymerase replicates the circular template many times to yield a single-stranded (ss) DNA product that typically comprises tens of thousands of nucleotides (Fig. S1A) corresponding to hundreds of nanometers to hundreds of microns in length (21–24). Importantly, the length of the RCA product can be simply tuned by the RCA reaction time (Fig. S1A). The RCA products contain repetitive sequence units that are complementary to the circular DNA template and therefore can be tailor-designed (Fig. S2A) (21). In our model system, we encoded in the circular template a previously isolated aptamer (25) specific to protein tyrosine kinase 7 (PTK7) (sequences presented in Table S1) that is expressed in certain leukemia cells [acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML)] (25), and in lung and colon cancers (26). Additionally, PTK7 is a valid cancer marker for phenotyping AML (27) (5–50% positive in ∼30% AML patient blood samples (n = 6) (Fig. S3). Immunostaining and flow cytometry studies of a representative AML patient sample demonstrated that PTK7 is positive on cells that also express CD34, a common AML cancer marker (28, 29). Use of a substrate functionalized with the PTK7 aptamer targeting rare leukemia cells in peripheral blood may be useful for detection of minimal residual disease (MRD). Preliminary studies strongly suggest that the presence of MRD correlates with increased relapse and poor survival (30). However, a large proportion of patients without detectable MRD, analyzed by traditional techniques such as flow cytometry, still relapse because of the rare, undetectable residual cancer cells (31). Therefore, simple technologies that improve the sensitivity of MRD detection are urgently needed to identify patients at high risk of relapse and progression, and to monitor and inform treatments.
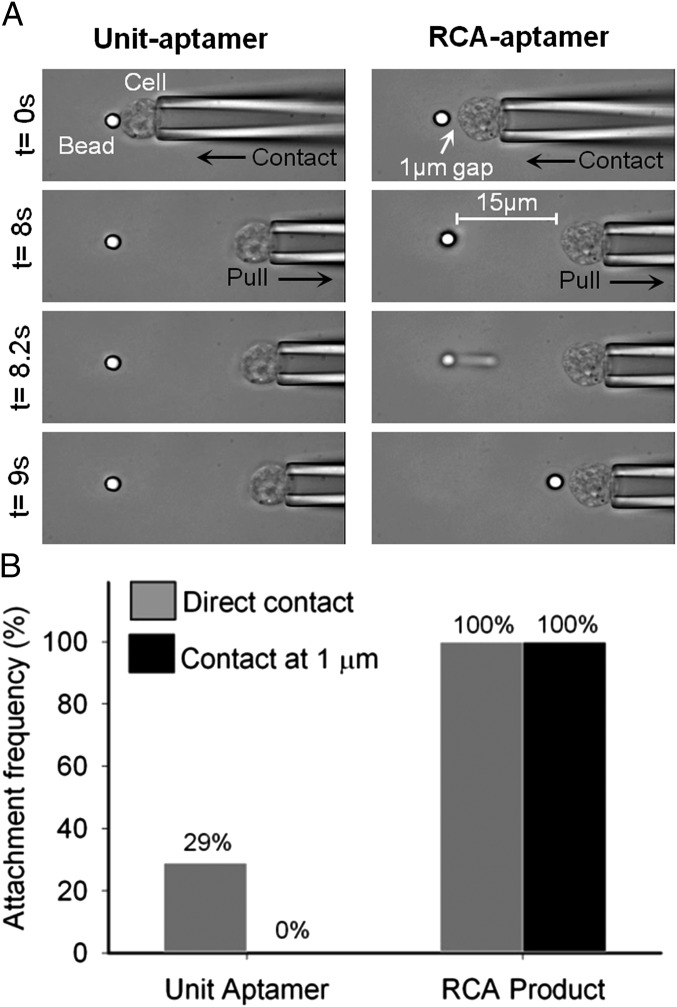
The specificity of the RCA product (hybridized to dye-conjugated polyA) (Fig. S2A) was demonstrated via flow cytometry by showing binding to PTK7 expressing CCRF-CEM (CCL-119 T-cell, human ALL) cells (7), whereas controls including the RCA product of the scrambled DNA sequence, unhybridized dye-conjugated polyA or Ramos cells (lacking PTK7), showed minimal fluorescence signals (Fig. S2B). Next, we directly compared the 3D DNA network formed by surface-immobilized RCA product and the Unit-aptamer with respect to bond strength, attachment probability, and capture distance though an optical trap experiment. Specifically, a CCRF-CEM cell held by a peizo-controlled micropipette was repeatedly brought in contact with indentation force of 3 pN (or in proximity) and then separated from an optically trapped microbead (trap force of 40 pN, loading rate of ∼58 pN/s) functionalized with RCA aptamer or unit-aptamer (Fig. 2 and Movie S1). We found that target cells bound more strongly to RCA-aptamers than to Unit-aptamers. For 100% of the tests (n = 7), the RCA-aptamer functionalized microbead exhibited strong adhesion to the cells that could not be disrupted by the maximum force exerted by the optical trap (i.e., bond strength > 40 pN). In contrast, 98% of the Unit-aptamer-bead/cell complexes were broken during separation (n = 97). Interestingly, in the case of RCA-aptamer-bead/cell experiment, long tethers (a few cell diameters) formed between the bead and cells that eventually pulled the bead out from the optical trap and then recoiled (Movie S1). Because cells were fixed in this experiment and therefore the cell membrane was nondeformable (32), we speculate that these long tethers are long DNA molecules that extend upon pulling. Further support for this speculation is provided by estimates of the force-extension curve of ssDNA. Single ssDNA molecules are capable of withstanding forces much higher than that of the optical trap (40 pN) (33); although the force-extension behavior of ssDNA is complex (33, 34), we can approximately estimate the recoil time using the freely jointed chain model with a Kuhn length of 1.5 nm obtained from experiments under similar salt conditions (35). In one of our experiments (Fig. 2), the bead recoiled from a distance of 24.5 μm from the cell to 5.8 μm in 95 ± 35 ms, and the corresponding recoil time estimated for a single ssDNA tether using the freely jointed chain model is ∼98 ms, in good agreement given the simplicity of the model. In comparison, a linear elastic spring model yields a recoil time of ∼24 ms. These estimates further suggest that an ssDNA molecule was tethered between the bead and the cell. We also found that RCA-aptamer beads exhibited a much higher probability [100% (7 of 7)] of binding to target cells than Unit-aptamer beads [29% (28 of 97)] when pressed into cells with a 3-pN indentation force. Strikingly, RCA-aptamer beads did not require direct contact (between the bead surface and cell as long RCA-aptamer extends far from the surface) to promote a binding event, whereas Unit-aptamer-bead complex formation required direct bead-cell contact. When the cell was approached to the microbead to a distance of ∼1 µm before retracting, it resulted in an attachment frequency of 100% (7 of 7) vs. 0% (0 of 50) for the RCA-aptamer vs. Unit-aptamer beads, respectively. Collectively, these results suggest that long DNA with multivalent aptamers bind to target cells with a higher efficiency that is likely the result of a combination of higher binding avidity (multivalency) and higher probability of association (large size and 3D conformation).
Fig. 2.
Single-cell force measurement revealed that the RCA-aptamer binds target cells more effectively than Unit-aptamers. (A) Representative images of individual CCRF-CEM cells held in a piezo-controlled micropipette that were brought (Left) into direct contact with an optically trapped Unit-aptamer coated bead or (Right) at a 1-μm distance from an RCA-aptamer coated bead. RCA-aptamers formed stronger bonds to the target cell than the maximum trap force (>40 pN and a force loading rate of ∼58 pN/s) as evidenced by the micropipette/cell pulling the bead out of the optical trap, whereas the Unit-aptamer system was unable to achieve this. Images were obtained with 40× oil immersion objective. (B) RCA-aptamer beads exhibited a much higher frequency of binding to target cells (100% for direct contact and 100% for 1-µm distance) than Unit-aptamer beads (29% for direct contact and 0% for a 1-µm distance).
Three-Dimensional DNA Networks Enable Efficient Capture of Cells Under Shear Flow.
We first developed a simple microfluidic 2D laminar flow device that enabled us to directly compare the capture performance between RCA-aptamer and Unit-aptamer under simple shear flow over a flat surface and to test cell capture using the 3D DNA network (Fig. S4). The RCA reaction was conducted using Phi29 DNA polymerase and surface-tethered primer/circular DNA template (Fig. 1 and Materials and Methods). Staining of the surface immobilized DNA strands with Sybr Green II permitted visualization of individual DNA strands by epi-fluorescence microscopy. We found that RCA products formed globular DNA networks on the surface with diameters ranging approximately from <500 nm to 5 µm (Fig. S5A). The density of these individual globular networks depended on the primer/circular template concentration. A graft density of ∼1 µm−2 was obtained when primer/circular template concentration was 2.5 µM, and high-resolution confocal imaging confirmed that the DNA networks extended as far as 20 µm in the solution and “wiggled” in 3D within the solution phase (Fig. S5 B and C and Movie S2).
After we confirmed the successful synthesis of RCA products on the microfluidic surface, we conducted a functional cell-capture assay under controlled dynamic flow conditions on the device (Materials and Methods). We first demonstrated that CCRF-CEM cells suspended in buffer could be captured specifically by the RCA-aptamer device (Fig. S6 and Movie S3). This finding was validated by control experiments that showed minimal cell capture with a scrambled RCA sequence or PTK7 negative Ramos cells. Additionally, examination with confocal microscopy (Fig. S6B) showed colocalization of cells with one or multiple long RCA products that appeared to be wrapped around the captured cells, suggesting that such long molecules bind to cells in a multivalent, cooperative manner. In a separate experiment, repeated pulses of high shear (∼20 dyn/cm2) were applied to the captured cells on the device substrate. Strikingly, in addition to firmly attached stationary cells, several captured cells were displaced during each pulse and then recoiled without detaching from the substrate (Fig. S6C and Movie S4). These recoils had different timescales and amplitudes ranging from sub- or multiple cell diameters to occasionally several tens and hundreds of microns, reflecting the potential diversity of RCA product length and elasticity. This result is consistent with the observations in the micropipette experiment and suggests that surface-immobilized RCA products are deformable, elastic, and can be used to maintain immobilized cells on a substrate in the presence of high shear.
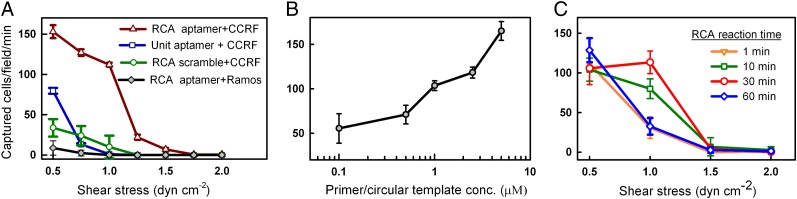
Remarkably, the multivalent DNA networks exhibited significantly greater cell capture efficiency compared with Unit-aptamers under flow conditions (∼113 ± sevenfold enhancement at 1 dyn/cm2) (Fig. 3A) and could capture cells at a much higher shear stress (up to 1.5 dyn/cm2). This finding is significant given that the substrate density of the Unit-aptamers (∼10–20 nm in size) is expected to be higher than that of the RCA products (∼10–100 µm in size). To our knowledge, monovalent antibody- or aptamer-immobilized substrates reported to date perform poorly in capturing cells at higher shear stress and are usually operated at shear rates of 0.4 dyn/cm2 or lower (1, 2, 6, 7). Thus, compared with existing approaches, use of RCA products offers a fresh approach to capture cells flowing across surfaces with potential to process larger sample volumes in a shorter time.
Fig. 3.
(A) RCA-aptamer device captured cells specifically and exhibited significantly greater cell capture efficiency compared with Unit-aptamers. Cell-capture performance is shown as a function of (B) graft density and (C) length of RCA products.
One important feature of RCA products is their engineering versatility. Specifically, the length and graft density of the RCA products on a substrate can be readily tuned by varying the time for RCA reaction and primer/circular template concentration. To maximize capture efficiency, we studied the effect of RCA product length and graft density. As anticipated, cell-capture performance improved at higher RCA product graft density (Fig. 3B). Furthermore, the cell capture efficiency increased as the RCA product length was increased through lengthening the RCA reaction time from 1 min to 30 min (Fig. 3C). However, the capture efficiency decreased when the RCA reaction was extended to 60 min. Although longer RCA products contain more aptamer units, extended reaction times likely introduces more intra- and intermolecular interactions, which may limit the accessibility of aptamers. Interestingly, for the aptamer-containing RCA products to efficiently capture flowing cells, a spacer domain between aptamers was required (in this work we used a polyT sequence). Experiments suggested that hybridization of the spacer domain with its complementary oligonucleotide between aptamer domains enabled the function of aptamer-decorated DNA strands likely by extending the RCA products, thereby minimizing nonspecific base-pairing with the aptamer domains.
Three-Dimensional DNA Networks Enable Selective Capture of Target Cells from Heterogeneous Mixtures.
A clinically useful substrate should capture target cells selectively from a mixture of nontarget cells (e.g., healthy blood cells). To assess the performance of the 3D DNA network-modified substrate, we first introduced a mixture of CCRF-CEM cells (target) and Ramos cells (nontarget) at a 1:1 ratio with a combined cell concentration of 106 cells/mL suspended in cell-capture buffer into the 2D laminar flow microfluidic device with the surface modified with the RCA DNA networks. The purity of the captured cells, which is dependent on the shear stress (i.e., higher shear stresses yield higher capture purity) (Fig. S7), was as high as 99% at a shear stress of 1.5 dyn/cm2. To demonstrate the versatility of our platform, we show that devices immobilized with 3D DNA networks containing the Ramos cell-binding aptamer (8) can capture Ramos cells selectively under dynamic flow conditions with a performance similar to the CCRF-CEM aptamer system (Fig. S8 A–C). After demonstrating selective capture from a mixture of two cell types, we examined the capture of the CCRF-CEM target cells from undiluted whole blood (Fig. S8 D–F). The simple, 2D laminar flow microfluidic device was easily able to detect the target cells spiked at ∼1,000 cells/mL in whole blood under a shear stress of 1.25 dyn/cm2 (141 ± 14 cells were captured from a total of 1,000 cells flown through in 1 mL) (Fig. S8F). Fluorescently labeled CCRF-CEM cells represented 84 ± 2% of the total captured cells (CCRF-CEM + white blood cells), indicating the ability for high-purity isolation of cells from whole blood. In contrast, existing cell-capture microfluidic devices typically have a capture purity of <14% for isolation of circulating epithelial cells at similar concentrations in blood (12). We attribute the high purity obtained in our system to the high shear stresses and the higher avidity because of multivalent interactions that was enabled by the unique 3D DNA network system [higher shear stresses yield higher capture purity (Fig. S7)].
Captured Cells Can Be Minimally Invasively Released from the Device by DNase I.
A major advantage of the DNA network-based capturing system compared with antibody-functionalized devices is that the DNA strands are accessible to molecules in solution and can be cleaved by nucleases (e.g., DNase I) (Fig. S1B) to release captured cancer cells for subsequent molecular characterization and drug discovery. When DNase I (100 U/mL in Hepes buffer) was continuously flowed into the device for 10 min at 37 °C, 68 ± 6% of the captured cells were released from the surface (Fig. S9A and Movie S5). The viability of the released cells was 66 ± 6%, compared with 87% ± 4% for the control cells (incubated with DNase I in solution), and both the released cells and control cells exhibited similar growth rates (Fig. S9B).
High-Throughput and High-Efficiency Capture of Cells by Functionalization of 3D DNA Networks in Herringbone Microfluidic Devices.
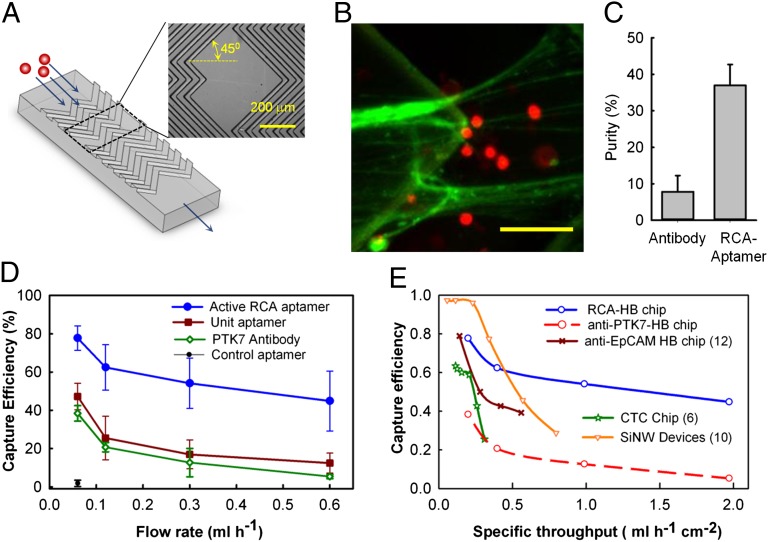
Although the laminar flow device was useful for systematically studying cell capture under controlled flow conditions, large device height (75 µm) with lack of any features to enhance cell-surface interactions resulted in a relatively low cell capture efficiency (∼20%) (Fig. S8F). To improve the cell capture efficiency and increase throughput, we integrated the RCA-aptamer system into a herringbone microfluidic device (HB-chip) (12) (Fig. 4A) that disrupts streamlines and therefore maximizes collisions between target cells and the surface. The cell-capture channel in the HB-chip was 800-µm wide, 45-µm high, with 40-µm deep grooves repeating over a total length of 38 mm. The surface of the HB-chips was modified with RCA-aptamer product and whole human blood spiked with CCRF-CEM cells at a concentration of ∼1,000 cells/mL was infused into the HB-chips at different flow rates (SI Materials and Methods). The RCA-aptamer–modified HB-chip demonstrated excellent capture efficiency (∼80%) at a flow rate of 60 µL/h (Fig. 4D). In contrast to the sharp drop-off in cell capture efficiency with flow rates reported in other devices, a cell capture efficiency of 50% was maintained even when the flow rate was increased to 600 µL/h. This capture efficiency was three- to fivefold higher than that for the Unit-aptamer and PTK7 antibody under the same conditions. Conversely, the RCA-aptamer surface enabled a 10-fold higher flow rate yet provided the same cell capture efficiency (50%) as the immobilized Unit-aptamer and the PTK7 antibody. Moreover, the purity of the captured cells on the RCA-aptamer HB-chip was five-times higher (∼38%) than that of the PTK7-antibody coated chip (∼8%) at a similar shear stress (Fig. 4C), which is in agreement with our earlier observations of higher capture purity for the RCA aptamer within the laminar flow system.
Fig. 4.
Microfluidic RCA-HB chip for cell capture. (A) Design of the HB-chip with grooves shown in the blowout. (B) Fluorescent image (false colored) of CCRF-CEM cells captured in the 3D DNA network on the herringbone surface. The RCA-product was stained with Sybr Green II (Green) and the cells were fluorescently tagged with CMTPX (Red). (Scale bar, 20 µm.) (C) Purity of captured CCRF-CEM cells spiked at a concentration of 1,000 cells/mL of whole blood and flowed at 120 µL/h (nominal shear stress of 1.5 dyn/cm2) through a HB-chip functionalized with RCA-aptamer or PTK7-antibody. (D) Capture efficiency of CEM-CCRF cells (1,000 cells/mL of whole blood) at different flow rates for HB-chip modified with RCA product, Unit-aptamer, PTK7-antibody, and scrambled RCA-aptamer (control). True mass balance was used to calculate efficiency values. The values shown are mean ± SD for n = 5 (RCA-aptamer) or n = 3 (other groups) independent experiments. (E) Comparison between the current RCA system and other major cell capture devices in terms of efficiency vs. specific throughput (flow rate over device footprint area). RCA HB-chip, PTK7 antibody HB-chip, EpCAM antibody HB-chip (12), CTC chip (6), and EpCAM antibody functionalized silicon nanowires incorporated in a microfluidic mixer (10) are shown.
Given that the cell-capture efficiency depends on the flow rate and device dimensions, we defined the specific throughput as the flow rate normalized by the active footprint of the device. This metric enabled direct comparison of the performance of the RCA HB-chip with the existing state-of-the-art cell-capture methods, with the caveat that the antibodies and target cells differ between studies. However, this comparison is useful to visualize trends in the capture efficiency as the flow rates are varied. The circulating tumor cell (CTC) chip (6), HB-chip with Epithelial cell adhesion molecule (EpCAM) antibody (12), and HB-chip grafted with anti-EpCAM functionalized silicon nanowires (SiNW-chip) (10) were compared against the RCA HB-chip. Although the capture efficiencies at lower specific throughputs were excellent for all of the devices (Fig. 4E), only the RCA HB-chip maintained high cell capture efficiency at high specific throughputs. Although the SiNW-chip does extend cell-capture molecules into the solution, its performance also drops off at high specific throughput. The combined effect of the increased valency and flexibility of the RCA-aptamer and the passive mixing because of the herringbone structure helps to maintain a consistent performance over a wide range of flow rates.
Discussion
Previous approaches to capture cells have focused on 2D or modified 2D substrates that are limited to monovalent adhesive domains extending tens of nanometers from the substrate. Although multivalent aptamers (i.e., dimer, trimer, and tetramers) have been reported (36–38), they have not been used for cell or particle capture. We have shown that use of multivalent RCA products for capturing cells is a highly versatile approach where the length and graft density of RCA products can be readily adjusted to optimize capture performance. Interestingly, marine creatures often have tentacles with different sizes that may have different optimal flow regimes that maximize capture of prey (39). Given that RCA products are tunable and exhibit a range of lengths, this may provide a unique advantage for efficient cell capture, although this theory requires further testing.
The use of a leukemia cell-targeting aptamer permits the device to be applicable for the detection and monitoring of MRD. The extent of MRD in the peripheral blood and bone marrow is highly predictive of relapse following therapy, where levels of leukemia cells greater than 0.01% of peripheral leukocytes (28) is considered to have a poor prognosis. In such a scenario, operating our device at a specific flow rate of 1 mL/h/cm2 would capture ∼500 leukemia cells/h/cm2 (assuming a sample concentration of leukemia cells at 0.01%). Thus, a device of a size of standard microscope slide (area ∼10 cm2) could collect more than 100 target cells within 2 min of operation. If the concentration of leukemia cells is reduced 10-fold, we would only require 20 min to capture the same number of cells.
Moreover, given that the repeating unit in the RCA product is complementary to the circular DNA template, one may tailor-design the sequence of the RCA product including, for example, the incorporation of a polyT spacer, which is important to reduce the nonspecific inter- and intramolecular DNA interactions and facilitate accessibility of aptamer domains to cells. Given that aptamers for any cell type can potentially be isolated by SELEX (40), substrates can be coated with a panel of different aptamers that collectively define a particular cancer cell phenotype and are capable of multiplex cancer cell detection.
The use of aptamers is particularly advantageous in this regard compared with antibody-based cell-capture assays because of its robustness and ability to release the captured cells. Although not all of the captured cells were released with the current device, the ability to release cells minimally invasively (8, 15, 41) represents a significant step toward studying the biology of these cells or screening for new drugs. One could envision that by encoding restriction sites for different restriction enzymes in the RCA product, one could release captured cells in a selective manner. It is interesting to note that although DNase I cleaves both long RCA-aptamer and short Unit-aptamer, it could only release the cells captured by the RCA-aptamer but not the Unit-aptamer. Compared with the Unit-aptamer, the RCA product provides increased spacing between the cell and substrate and increased number of cleavage sites. Furthermore, low background binding with high-capture purity allows processing of relatively large sample volumes over a small device footprint, thus effectively concentrating the captured cells. Therefore, use of our device to detect low concentrations of leukemia cells in blood may be useful to predict cancer relapses and to screen drugs for personalized medicine (42, 43), or it may allow larger volumes of blood to be processed to recover more CTCs for further analysis. We envision that this approach will have broad applications in the detection, isolation, enrichment, and sorting of rare cell types (e.g., cancer cells, CD4 T cells, stem/progenitor cells, and pathogens) for therapeutic and diagnosis applications.
Materials and Methods
Detailed methods are provided in SI Materials and Methods.
Reagents and Cells.
All DNA (Table S1) were obtained from Integrated DNA Technology. Cells were obtained from ATCC. Cell staining dyes and Sybr Green II were purchased from Invitrogen. Avidin, PBS, 3-Mercaptopropyl trimethoxysilane, and 4-Maleimidobutyric acid N-hydroxysuccinimide ester (GMBS) were purchased from Sigma. Neutravidin was purchased from Pierce. dNTP and Phi 29 DNA polymerase, and DNA T4 Ligation kit were purchased from Fermentas. Fresh whole-blood samples from healthy donors were obtained from Research Blood Components. Peripheral blood samples from patients were obtained from Brigham and Women’s Hospital with approval of Institutional Review Board Protocol # 2011-P-001624/1.
RCA Reaction on Microfluidic Chips.
The microfluidic chips were fabricated in PDMS (Polydimethoxysilane) by standard soft lithography techniques (see SI Materials and Methods for details). Avidin (1 mg/mL) was first introduced to the microfluidic device and incubated for 10 min, after which biotilylated preannealed circular template-primer complex (2.5 µM) was perfused and allowed to react for 15 min. Next, the RCA reaction mixture containing 0.3 mM dNTP and 0.5 µL Phi29 DNA polymerase in RCA reaction buffer was added. The reaction was incubated at 37 °C and following the RCA process, the channel was thoroughly washed. The HB-chips were fabricated in a similar manner with some modification in the protocol. See SI Materials and Methods for details.
Cell Capture and Release Assay.
Cells were infused from a vial using a pressure source that was calibrated to obtain a pressure-flow rate operational chart for the experimental system. The cell capture assay was performed by infusing fluorescently labeled target CCRF-CEM cells or control Ramos cells in cell capture buffer [1% (vol/vol) FCS in PBS] or whole human blood at desired concentrations.
To release the captured cells from the surface, freshly prepared DNase I solution comprising 100 U/mL DNase I in 10mM HEPE buffer (supplemented with 100 mM NaCl, 60 mM MgCl2, and 10 mM CaCl2) was continuously flown into the device for 10 min with the temperature of the stage maintained at 37 °C. Cells were collected at the outlet into a flat bottom 96-well plate containing growth medium. Viability of released cells was evaluated using LIVE/DEAD cell viability assay (Invitrogen). Details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by an International Human Frontier Science Program Organization postdoctoral fellowship (to W.Z.); the China Scholarship Council and National Natural Science Foundation of China Grants 51273159 and 51072159 (to D.G.); Program for New Century Excellent Talents in Universities (Chinese Ministry of Education) Grant NCET-08-0444 (2301G107aaa) (to D.G.); and National Institutes of Health Grants HL097172 and HL095722 (to J.M.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211234109/-/DCSupplemental.
References
- 1.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8(5):329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 2.Dharmasiri U, Witek MA, Adams AA, Soper SA. Microsystems for the capture of low-abundance cells. Annu Rev Anal Chem (Palo Alto Calif) 2010;3:409–431. doi: 10.1146/annurev.anchem.111808.073610. [DOI] [PubMed] [Google Scholar]
- 3.Galanzha EI, et al. In vivo magnetic enrichment and multiplex photoacoustic detection of circulating tumour cells. Nat Nanotechnol. 2009;4(12):855–860. doi: 10.1038/nnano.2009.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maheswaran S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359(4):366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riethdorf S, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res. 2007;13(3):920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 6.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450(7173):1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phillips JA, Xu Y, Xia Z, Fan ZH, Tan W. Enrichment of cancer cells using aptamers immobilized on a microfluidic channel. Anal Chem. 2009;81(3):1033–1039. doi: 10.1021/ac802092j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y, et al. Aptamer-based microfluidic device for enrichment, sorting, and detection of multiple cancer cells. Anal Chem. 2009;81(17):7436–7442. doi: 10.1021/ac9012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan Y, et al. Surface-immobilized aptamers for cancer cell isolation and microscopic cytology. Cancer Res. 2010;70(22):9371–9380. doi: 10.1158/0008-5472.CAN-10-0568. [DOI] [PubMed] [Google Scholar]
- 10.Wang S, et al. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew Chem Int Ed Engl. 2011;50(13):3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dharmasiri U, et al. High-throughput selection, enumeration, electrokinetic manipulation, and molecular profiling of low-abundance circulating tumor cells using a microfluidic system. Anal Chem. 2011;83(6):2301–2309. doi: 10.1021/ac103172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stott SL, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci USA. 2010;107(43):18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han W, Allio BA, Foster DG, King MR. Nanoparticle coatings for enhanced capture of flowing cells in microtubes. ACS Nano. 2010;4(1):174–180. doi: 10.1021/nn900442c. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, et al. Three-dimensional nanostructured substrates toward efficient capture of circulating tumor cells. Angew Chem Int Ed Engl. 2009;48(47):8970–8973. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, et al. Aptamer-mediated efficient capture and release of T lymphocytes on nanostructured surfaces. Adv Mater. 2011;23(38):4376–4380. doi: 10.1002/adma.201102435. [DOI] [PubMed] [Google Scholar]
- 16.Chen GD, Fachin F, Fernandez-Suarez M, Wardle BL, Toner M. Nanoporous elements in microfluidics for multiscale manipulation of bioparticles. Small. 2011;7(8):1061–1067. doi: 10.1002/smll.201002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruppert E, Barnes R. Invertebrate Zoology. Philadelphia, PA: Saunders College Publishing; 1994. 1056 pp. [Google Scholar]
- 18.Lambert P. Sea Cucumbers of British Columbia, Southeast Alaska, and Puget Sound. Vancouver, BC: UCB Press; 1997. 166 pp. [Google Scholar]
- 19.Coupier G, Kaoui B, Podgorski T, Misbah C. Noninertial lateral migration of vesicles in bounded Poiseuille flow. Phys Fluids. 2008;20(11):111702–111706. [Google Scholar]
- 20.Fire A, Xu SQ. Rolling replication of short DNA circles. Proc Natl Acad Sci USA. 1995;92(10):4641–4645. doi: 10.1073/pnas.92.10.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao W, Ali MM, Brook MA, Li Y. Rolling circle amplification: Applications in nanotechnology and biodetection with functional nucleic acids. Angew Chem Int Ed Engl. 2008;47(34):6330–6337. doi: 10.1002/anie.200705982. [DOI] [PubMed] [Google Scholar]
- 22.Cheglakov Z, Weizmann Y, Braunschweig AB, Wilner OI, Willner I. Increasing the complexity of periodic protein nanostructures by the rolling-circle-amplified synthesis of aptamers. Angew Chem Int Ed Engl. 2008;47(1):126–130. doi: 10.1002/anie.200703688. [DOI] [PubMed] [Google Scholar]
- 23.Tian Y, He Y, Mao C. Cascade signal amplification for DNA detection. ChemBioChem. 2006;7(12):1862–1864. doi: 10.1002/cbic.200600336. [DOI] [PubMed] [Google Scholar]
- 24.Zhao W, Gao Y, Kandadai SA, Brook MA, Li Y. DNA polymerization on gold nanoparticles through rolling circle amplification: Towards novel scaffolds for three-dimensional periodic nanoassemblies. Angew Chem Int Ed Engl. 2006;45(15):2409–2413. doi: 10.1002/anie.200600061. [DOI] [PubMed] [Google Scholar]
- 25.Shangguan D, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci USA. 2006;103(32):11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng L, et al. Silencing of PTK7 in colon cancer cells: Caspase-10-dependent apoptosis via mitochondrial pathway. PLoS ONE. 2010;5(11):e14018. doi: 10.1371/journal.pone.0014018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prebet T, et al. The cell polarity PTK7 receptor acts as a modulator of the chemotherapeutic response in acute myeloid leukemia and impairs clinical outcome. Blood. 2010;116(13):2315–2323. doi: 10.1182/blood-2010-01-262352. [DOI] [PubMed] [Google Scholar]
- 28.Campana D. Minimal residual disease studies in acute leukemia. Am J Clin Pathol. 2004;122(Suppl):S47–S57. doi: 10.1309/YJP8CEQ1E7RYP3MF. [DOI] [PubMed] [Google Scholar]
- 29.Dworzak MN, et al. Austrian Berlin-Frankfurt-Münster Study Group Prognostic significance and modalities of flow cytometric minimal residual disease detection in childhood acute lymphoblastic leukemia. Blood. 2002;99(6):1952–1958. doi: 10.1182/blood.v99.6.1952. [DOI] [PubMed] [Google Scholar]
- 30.San Miguel JF, et al. Immunophenotyping investigation of minimal residual disease is a useful approach for predicting relapse in acute myeloid leukemia patients. Blood. 1997;90(6):2465–2470. [PubMed] [Google Scholar]
- 31.Radich J, Ladne P, Gooley T. Polymerase chain reaction-based detection of minimal residual disease in acute lymphoblastic leukemia predicts relapse after allogeneic BMT. Biol Blood Marrow Transplant. 1995;1(1):24–31. [PubMed] [Google Scholar]
- 32.Vassar PS, Hards JM, Brooks DE, Hagenberger B, Seaman GV. Physicochemical effects of aldehydes on the human erythrocyte. J Cell Biol. 1972;53(3):809–818. doi: 10.1083/jcb.53.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar S, Mishra G. Stretching single stranded DNA. Soft Matter. 2011;7(10):4595–4605. [Google Scholar]
- 34.Dessinges MN, et al. Stretching single stranded DNA, a model polyelectrolyte. Phys Rev Lett. 2002;89(24):248102. doi: 10.1103/PhysRevLett.89.248102. [DOI] [PubMed] [Google Scholar]
- 35.Smith SB, Cui YJ, Bustamante C. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271(5250):795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 36.Kim Y, Cao Z, Tan W. Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proc Natl Acad Sci USA. 2008;105(15):5664–5669. doi: 10.1073/pnas.0711803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, Yan H, Liu Y, Chang Y. Targeted cell-cell interactions by DNA nanoscaffold-templated multivalent bispecific aptamers. Small. 2011;7(12):1673–1682. doi: 10.1002/smll.201002292. [DOI] [PubMed] [Google Scholar]
- 38.Mallikaratchy PR, et al. A multivalent DNA aptamer specific for the B-cell receptor on human lymphoma and leukemia. Nucleic Acids Res. 2011;39(6):2458–2469. doi: 10.1093/nar/gkq996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anthony K. Prey capture by the sea anemone Metridium senile (L.): Effects of body size, flow regime, and upstream neighbors. Biol Bull. 1997;192(1):73–86. doi: 10.2307/1542577. [DOI] [PubMed] [Google Scholar]
- 40.Fang X, Tan W. Aptamers generated from cell-SELEX for molecular medicine: A chemical biology approach. Acc Chem Res. 2010;43(1):48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plouffe BD, Brown MA, Iyer RK, Radisic M, Murthy SK. Controlled capture and release of cardiac fibroblasts using peptide-functionalized alginate gels in microfluidic channels. Lab Chip. 2009;9(11):1507–1510. doi: 10.1039/b823523f. [DOI] [PubMed] [Google Scholar]
- 42.Kang H, et al. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood. 2010;115(7):1394–1405. doi: 10.1182/blood-2009-05-218560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jmili N, et al. Flow cytometry evaluation of minimal residual disease in acute lymphoblastic leukaemia type B. The Open Leukemia Journal. 2010;3:47–54. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.