Abstract
Recurrent missense mutations in the RNA polymerase II Mediator subunit MED12 are associated with X-linked intellectual disability (XLID) and multiple congenital anomalies, including craniofacial, musculoskeletal, and behavioral defects in humans with FG (or Opitz-Kaveggia) and Lujan syndromes. However, the molecular mechanism(s) underlying these phenotypes is poorly understood. Here we report that MED12 mutations R961W and N1007S causing FG and Lujan syndromes, respectively, disrupt a Mediator-imposed constraint on GLI3-dependent Sonic Hedgehog (SHH) signaling. We show that the FG/R961W and Lujan/N1007S mutations disrupt the gene-specific association of MED12 with a second Mediator subunit, CDK8, identified herein to be a suppressor of GLI3 transactivation activity. In FG/R961W and Lujan/N1007S patient-derived cells, we document enhanced SHH pathway activation and GLI3-target gene induction coincident with impaired recruitment of CDK8 onto promoters of GLI3-target genes, but not non–GLI3-target genes. Together, these findings suggest that dysregulated GLI3-dependent SHH signaling contributes to phenotypes of individuals with FG and Lujan syndromes and further reveal a basis for the gene-specific manifestation of pathogenic mutations in a global transcriptional coregulator.
X-linked intellectual disability (XLID) affects 1–2 of every 1,000 males and accounts for ∼10% of all intellectual disability (ID) (1). Approximately one-third of XLID cases are associated with sufficiently coincident somatic, neurobehavioral, or metabolic features to permit diagnostic designation and are therefore classified as “syndromal” in nature. To date, more than 140 syndromal XLID conditions have been identified, almost half of which are attributable to mutations in single genes (2). For most of these XLID genes, the underlying mechanisms responsible for the broad spectrum of clinical phenotypes arising from their mutation have not been established. In this regard, our interest has centered on two related XLID disorders, FG (or Opitz-Kaveggia) syndrome and Lujan (or Lujan-Fryns) syndrome. In addition to ID, FG and Lujan syndromes share several overlapping clinical manifestations, including agenesis/dysgenesis of the corpus callosum, macrocephaly, hypotonia, craniofacial dysmorphisms, seizures, and behavioral disturbances (3, 4). Although neither condition was originally considered in the differential diagnosis of the other, these two syndromes were recently found to be allelic, arising from different missense mutations in the Xq13 gene encoding MED12, a subunit of the RNA polymerase II transcriptional Mediator (3–5).
Mediator is a conserved multiprotein interface between gene-specific transcription factors and RNA polymerase II (6). In this capacity, Mediator serves to channel regulatory signals from activator and repressor proteins to affect changes in gene expression programs that control diverse physiological processes including cell growth and homeostasis, development, and differentiation (7). Recent structural and comparative genomics analyses support a deep evolutionary origin for a four-module Mediator complex among eukaryotes (8, 9). In addition to an evolutionarily conserved core composed of “head” and “middle” modules that dock with RNA polymerase II, Mediator also includes more evolutionarily divergent “tail” and “kinase” modules through which most activators and repressors target Mediator. The kinase module, comprising MED12, MED13, CDK8, and cyclin C, has been ascribed both activating and repressing functions and exists in variable association with Mediator, implying that its regulatory role is restricted to a subset of RNA polymerase II transcribed genes.
Within the Mediator kinase module, MED12 is a critical transducer of regulatory information conveyed by signal-activated transcription factors linked to diverse developmental pathways, including the EGF, Notch, Wnt, and Hedgehog pathways (10–12). Furthermore, MED12 has been implicated in vertebrate neural development, and genetic variation in MED12 is associated with neuoropsychiatric illness, including schizophrenia and psychoses, in humans (5, 13). However, the underlying basis by which genetic disruption of MED12 elicits the broad spectrum of clinical phenotypes observed in FG and Lujan syndromes remains unknown. Notably, several phenotypes associated with FG and/or Lujan syndromes including macrocephaly, corpus callosal defects, hypertolerism, syndactyly, and cognitive impairment, overlap with a subset of those variously appearing in Greig cephalopolysyndactyly syndrome (GCPS) and/or Pallister-Hall syndrome (PHS) arising from mutations in GLI3, a Sonic Hedgehog (SHH) signaling effector and direct interaction target of MED12 (12, 14).
The SHH signaling pathway is a preeminent developmental pathway essential for proper embryonic pattern formation and adult tissue homeostasis (15). Accordingly, dysregulation of SHH signaling has been linked to both congenital anomalies and cancer (16). The physiological and pathological manifestations of SHH signaling arise from the specification of unique gene expression programs dependent upon key nuclear effectors of the GLI transcription factor family. In vertebrates, three GLI family members, GLI1–3, subsume the functions of a single ancestral GLI homolog, Cubitus interruptus (Ci), originally identified in invertebrates (17). Whereas GLI1 and GLI2 function primarily as activators, GLI3 more closely resembles Ci as a functionally bipartite regulator with the ability to directly repress and activate a common set of target genes in the absence and presence of SHH, respectively (18). In the absence of SHH, its transmembrane receptor Patched (PTCH) inhibits the SHH transducer Smoothened (SMO), a transmembrane G-protein–coupled receptor (15). This inhibition promotes phosporylation-dependent proteolytic processing of GLI3 to an N-terminal repressor (19). Binding of SHH to PTCH liberates SMO from inhibition, triggering the accumulation and translocation of full-length GLI3 into the nucleus whereupon it is phosphorylated and converted to a labile transcriptional activator (19). SHH thus dictates the transcriptional output of a cell by altering the ratio of GLI3 repressor to GLI activator species.
Whereas the GLI3 repressor is known to exert a broad and essential function in development by antagonizing SHH activity, emerging studies have also established important developmental roles for a GLI3 activator function in proper patterning of the ventral spinal cord, brain, limb digits, inner ear, and sclerotome (18, 20–23). Nonetheless, relatively few studies have focused on the mechanism by which GLI3 stimulates target gene transcription in the nucleus. In this regard, we previously showed that SHH-activated GLI3, through a unique C-terminal transactivation domain (MED12/Mediator-binding domain, MBD) physically targets both Mediator and the histone acetyltransferase CBP to activate transcription (12). Furthermore, we found that the GLI3 MBD targets Mediator through both an unidentified subunit(s), likely required for MBD-directed Mediator recruitment onto GLI3-target genes, and MED12 within the kinase module. The GLI3–MED12 interaction was shown to be functionally important, because dominant negative disruption of this interaction by overexpression of the GLI3-binding domain on MED12 (PQL domain) inhibited GLI3 transactivation in response to SHH. Surprisingly, however, RNAi-mediated MED12 depletion enhanced GLI3 transactivation induced by SHH, implicating MED12/Mediator in suppression of GLI3 transactivation activity. Based on these findings, we proposed a model in which SHH-activated GLI3, through its MBD, targets the MED12 interface in Mediator to reverse a Mediator-imposed constraint on GLI3 MBD transactivation activity (12). Given the physical and functional interaction between GLI3 and MED12 coupled with the fact that mutations in each of these interacting proteins elicit congenital anomaly syndromes with overlapping phenotypes, we hypothesized that pathogenic mutations in MED12 leading to FG and Lujan syndromes elicit dysregulated GLI3-dependent SHH signaling. Herein we confirm this prediction with important implications for the pathology of MED12-associated syndromal XLID disorders.
Results
FG and Lujan Mutations in MED12 Disrupt a Mediator-Imposed Constraint on GLI3-Dependent Shh Signaling.
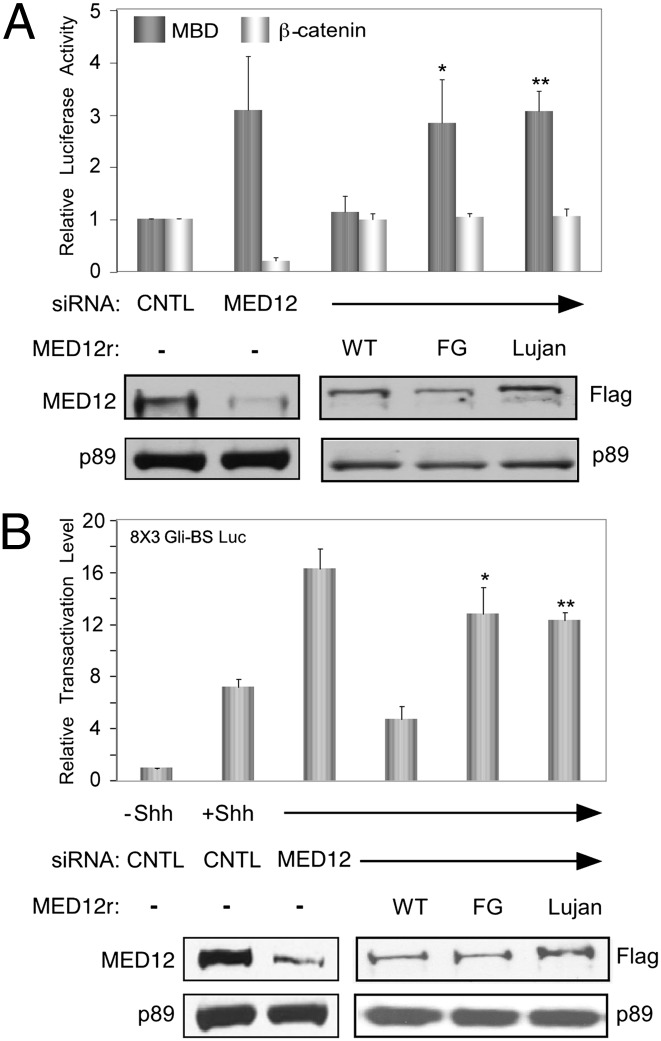
Our prior work revealed that an essential function of the GLI3 MBD in gene activation induced by Shh is to physically bind the MED12 interface in Mediator to functionally reverse a MED12/Mediator-imposed constraint on its transactivation activity (12). Notably, the isolated GLI3 MBD, when fused to a heterologous (i.e., Gal4) DNA-binding domain, will drive high levels of activated transcription in the absence of Shh through an identical mechanism (12), because the principal function of Shh is to promote the nuclear accumulation of a full-length GLI3 activator carrying its C-terminal MBD. This Gal4–MBD-dependent transactivation assay thus offers a facile means to assess the functional consequences of the FG and Lujan mutations in MED12 on GLI3 transactivator function. Therefore, to examine the impact of the FG/R961W and Lujan/N1007S mutations in MED12 on GLI3 MBD-driven transactivation, we initially compared siRNA-resistant WT MED12 (MED12r) and its corresponding FG and Lujan mutant derivatives for their respective abilities to suppress enhanced GLI3 MBD activity observed in cells depleted of endogenous MED12 by RNAi. As seen previously (12), siRNA-mediated MED12 depletion enhanced Gal4–MBD-driven transactivation from a Gal4-responsive reporter gene due to removal of a MED12-dependent constraint on the GLI3 MBD (Fig. 1A). Strikingly, whereas MED12r was fully competent to reverse this effect, both the FG and Lujan mutant derivatives were severely compromised in this ability (Fig. 1A). By contrast, neither mutation influenced the established ability of MED12 to function as a coactivator of β-catenin (10); diminished levels of β-catenin–dependent transactivation observed following MED12 knockdown were similarly rescued by WT and mutant MED12r derivatives (Fig. 1A). These results reveal a selective defect in the GLI3-specific suppressor as opposed to the β-catenin–specific coactivator function of MED12 arising from the FG/R961W and Lujan/N1007S mutations.
Fig. 1.
MED12 mutations R961W and N1007S disrupt a Mediator-imposed constraint on GLI3-dependent Shh signaling. (A and B, Upper) HeLa (A) or CH310T1/2 (B) cells were transfected or electroporated, respectively, with control (CNTL) or MED12-specific siRNA and subsequently transfected with an internal control pact–β-galactosidase expression plasmid along with a pG5-E1B-Luc reporter and either Gal4, Gal4-GLI3 MBD, or Gal4–β-catenin (A) or an 8 × 3′ Gli-BS-Luc reporter and GLI3 (B). Where indicated (MED12r), DNA transfections also included siRNA-resistant WT, FG/R961W, or Lujan/N1007S mutant MED12 expression plasmids. Where indicated in B. CH310T1/2 cells were treated with conditioned medium from control (−Shh) or Shh-expressing (+Shh) 293 cells. Normalized luciferase activities were calculated relative to the luciferase activity obtained in cells transfected with control siRNA and Gal4 (A) or cells transfected with control siRNA and treated with control-conditioned medium (B). For comparative purposes on the same plot in A, the relative luciferase activity for each activator in control siRNA-transfected cells was arbitrarily assigned a value of 1, yielding a relative transactivation level; their corresponding transactivation levels in MED12 siRNA-transfected cells are expressed relative to this value. The actual transactivation levels for Gal4–MBD and Gal4–β-catenin in these experiments averaged 531- and 211-fold, respectively. Data represent the mean ± SEM of at least three independent experiments performed in duplicate. Asterisks denote statistically significant differences compared with MED12r WT (Student t test, *P < 0.05, **P < 0.01). (A and B, Lower) Western blot (WB) analyses of extracts from representative knockdown/rescue assays using antibodies specific for either MED12, the FLAG epitope on MED12r derivatives, or transcription factor (TF) IIH TFIIH p89 as an internal loading control.
To investigate the impact of the FG and Lujan mutations in MED12 on GLI3-dependent transactivation induced by Shh, we comparatively examined WT MED12r and its corresponding R961W and N1007S mutant derivatives for their respective abilities to suppress enhanced Shh-induced activation of a GLI3-responsive reporter gene (8 × 3′ Gli-BS-Luc) in murine CH310T1/2 cells depleted of endogenous Med12 by RNAi. Previously, we showed that in CH310T1/2 cells, the 8 × 3′ Gli-BS reporter is induced by Shh in a manner dependent upon ectopically expressed GLI3 (12). Consistent with our previous observations (12), Med12 knockdown in CH310T1/2 cells enhanced GLI3-dependent reporter gene induction in response to Shh (Fig. 1B). Notably, both the FG/R961W and Lujan/N1007S mutations severely compromised the ability of MED12r to suppress enhanced GLI3-dependent transactivation induced by Shh (Fig. 1B). Taken together, these results reveal that the FG/R961W and Lujan/N1007S missense mutations in MED12 disrupt a Mediator-imposed constraint on GLI3-dependent Shh signaling.
CDK8 Is a Suppressor of GLI3 Transactivation Activity.
To investigate the mechanistic basis for this disruption, we initially considered the impact of the FG and Lujan mutations in MED12 on its biochemical properties critical for suppression of GLI3 transactivation activity. In this regard, we previously showed that the FG and Lujan mutations in MED12 do not deleteriously impact its intrinsic stability or its incorporation into Mediator (24). Furthermore, neither mutation disrupts the interaction of MED12 with GLI3. Thus, WT MED12 and each of its corresponding FG and Lujan mutant derivatives bound comparably to full-length GLI3 or the GLI3 MBD in coimmunoprecipation and GST pull-down assays, respectively (Fig. S1). Collectively, these findings suggest that the FG and Lujan mutations in MED12 disrupt its ability to suppress GLI3-dependent Shh signaling through a mechanism downstream of its role as a stable GLI3 interface in Mediator.
On the basis of these findings, we therefore sought to clarify the role of MED12 in Mediator-imposed suppression of GLI3 transactivation activity. Previously, we showed that MED12 anchors CDK8 in Mediator (10, 24), and others have implicated CDK8 directly in transcriptional repression (25, 26). Therefore, we asked whether CDK8 contributes to suppression of the GLI3 transactivation domain. To this end, we comparatively examined the influence of CDK8 versus MED12 knockdown on the transactivation activities of the GLI3 MBD (Gal4–MBD) as well as that of full-length GLI3 induced by Shh. As a control for these experiments, depletion of MED23, whose stable incorporation into Mediator is independent of either MED12 or CDK8 (24), was also evaluated for its influence on GLI3 transactivation activity. Knockdown of CDK8, but not MED23, triggered enhanced GLI3 MBD activity (Fig. 2 A and D) as well as Shh-induced GLI3-responsive reporter (Fig. 2 B and E) and endogenous Gli3 target gene (Fig. 2C) induction comparably to MED12 depletion, thus implicating CDK8 in suppression of GLI3 transactivation domain function. To determine whether CDK8 kinase activity is required for this suppression, we comparatively examined siRNA-resistant WT CDK8 (CDK8r WT) and a kinase-dead CDK8 derivative (CDK8r D173A) (25) for their respective abilities to suppress enhanced GLI3 MBD activity triggered by endogenous CDK8 knockdown. Compared with WT CDK8, kinase-dead CDK8 was unable to constrain the GLI3 MBD, confirming a role for enzymatically active CDK8 in suppression of GLI3 transactivation activity (Fig. 2F).
Fig. 2.
CDK8 kinase activity is required to suppress the Gli3 transactivation domain. HeLa (A) or CH310T1/2 (B and C) cells were transfected or electroporated, respectively, with control (CNTL), MED12-, CDK8-, or MED23-specific siRNAs and subsequently with an internal control pact–β-galactosidase expression plasmid along with a pG5-E1B-Luc reporter and either Gal4 or Gal4–GLI3 MBD (A) an 8 × 3′ Gli-BS-Luc reporter and GLi3 (B) or no plasmid DNA (C). Where indicated (B and C) CH310T1/2 cells were treated with conditioned medium from control (−Shh) or Shh-expressing (+Shh) 293 cells. Normalized luciferase activity is expressed relative to the luciferase activity obtained in cells transfected with control siRNA and Gal4 (A) or cells electroporated with control siRNA and treated with control-conditioned medium (B). Endogenous Gli1 mRNA levels in C were determined by RT-qPCR, normalized to β-actin levels, and expressed relative to the level of Gli1 RNA in cells treated with CNTL siRNA and control-conditioned medium. Data for all transient reporter assays including F represent the mean ± SEM of at least three independent experiments performed in duplicate. Asterisks denote statistically significant differences compared with CNTL siRNA (Student t test, *P < 0.05, **P < 0.01). (D and E) WB analyses of extracts from representative assays in A and B, using antibodies specific for MED12, CDK8, MED23, or TFIIH p89 as an internal loading control. (F, Upper) HeLa cells infected with lentiviruses expressing control (CNTL) or CDK8-specific shRNAs were subsequently transfected with internal control, reporter and activator plasmids as in A. Where indicated (CDK8r), DNA transfections also included siRNA-resistant WT or kinase dead (D173A) CDK8 derivatives. Relative luciferase activities were calculated as described in (A). (F, Lower) WB analysis from a knockdown/rescue assay using antibodies specific for CDK8 or transcription factor (TF) IIEβ as an internal loading control. Endogenous (end) CDK8 and ectopically (ect) expressed FLAG-tagged CDK8r derivatives are indicated.
FG and Lujan Mutant MED12/Mediator Complexes Exhibit Impaired Recruitment of CDK8 onto GLI3-Target Gene Promoters Leading to Hyperactivated GLI3-Dependent Shh Signaling.
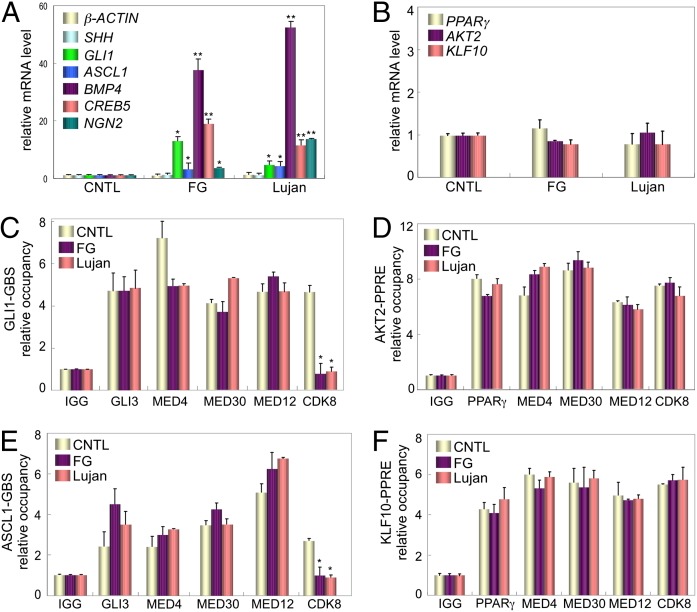
Because MED12 is an anchor in Mediator for CDK8, which in turn, suppresses GLI3 transactivation function, reduced incorporation of CDK8 into mutant MED12/Mediator complexes could explain how the FG and Lujan mutations in MED12 disrupt a Mediator-imposed constraint on GLI3-dependent SHH signaling. To explore this hypothesis, we examined both the levels of GLI3-dependent SHH signaling and the physical composition of Mediator in lymphoblastoid cell lines derived from FG and Lujan patients harboring, respectively, the R961W or N1007S MED12 missense mutations. First we monitored by RT-qPCR the expression levels of SHH pathway genes including SHH ligand, its downstream effector, and GLI3-target gene GLI1, and SHH/GLI3 target genes ASCL1, BMP4, CREB5, and NGN2 (12, 27, 28). SHH was detectably expressed at comparable levels in control (CNTL) as well as FG and Lujan patient-derived cell lines, indicative of constitutive pathway activation (Fig. 3A). Strikingly, however, expression levels of GLI1, ASCL1, BMP4, CREB5, and NGN2 were all significantly increased in FG and Lujan patient-derived cells, suggesting dysregulated (hyperactivated) signaling downstream of SHH in MED12 mutant cells (Fig. 3A). Importantly, enhanced expression of GLI1, ASCL1, BMP4, CREB5, and NGN2 in FG and Lujan patient-derived cells was inhibited by cyclopamine (Fig. S2) as well as RNAi-mediated GLI3 depletion (Fig. S3), confirming that their induction was indeed both SHH and GLI3 dependent.
Fig. 3.
SHH-responsive GLI3-target genes show enhanced expression and impaired CDK8 promoter recruitment in FG and Lujan patient-derived cells. (A and B) RNAs from control (CNTL), FG, and Lujan patient cells treated without cyclopamine (see Fig. S2 for cyclopamine treatment) were used for RT-qPCR. mRNA levels for each gene were normalized to β-actin mRNA and expressed relative to their corresponding mRNA levels in CNTL patient cells. Data represent the mean ± SEM of at least three independent experiments performed in duplicate. Asterisks denote statistically significant differences in the relative mRNA levels for each gene compared with their corresponding levels in CNTL cells (Student t test, *P < 0.05, **P < 0.01). (C–F) Soluble chromatin prepared from CNTL, FG, and Lujan patient cells was subjected to immunoprecipitation (IP) using the indicated antibodies. Immunoprecipitated chromatin was analyzed by qPCR using primers flanking GLI binding sites (GBSs) within the GLI1 and ASCL1 genes (C and E) or PPARγ response elements (PPREs) within the AKT2 and KLF10 genes (D and F). DNA occupancy for each protein is expressed relative to control IgG. Data represent the mean ± SEM of at least three independent experiments performed in triplicate. Asterisks denote statistically significant differences in relative DNA occupancy for a given protein compared with that in control (CNTL) patient cells (Student t test, *P < 0.05).
Next, we used chromatin immunoprecipitation (ChIP) to monitor the presence and subunit composition of GLI3 and Mediator on both the GLI1 and ASCL1 gene promoters in control as well as in FG and Lujan patient-derived lymphoblasts. We detected specific occupancy of GLI binding sites, but not upstream gene sequences, in the GLI1 and ASCL1 promoters by GLI3 and MED12-containing Mediator in control as well as in FG and Lujan patient-derived lymphoblasts (Fig. 3 C and E and Fig. S4). Strikingly, however, whereas CDK8 was readily detected on GLI binding sites within the GLI1 and ASCL1 promoters in control lymphoblasts, its respective presence on these promoter elements in FG and Lujan patient-derived lymphoblasts was significantly reduced, suggesting impaired recruitment of CDK8 by mutant MED12/Mediator complexes (Fig. 3 C and E).
Impaired CDK8 Recruitment by FG and Lujan Mutant MED12/Mediator Complexes Is Manifest in a Gene-Specific Manner.
To determine whether impaired CDK8 recruitment into FG and Lujan mutant MED12/Mediator complexes is restricted to GLI3-target genes, we extended ChIP analyses to other active genes in control, FG, and Lujan patient-derived lymphoblasts. For this purpose we monitored the composition of Mediator on validated PPARγ-target genes AKT2 and KLF10 (Fig. S5), because peroxisome proliferator-activated receptor (PPAR)γ is known to be expressed and active in lymphoid cells (29). AKT2 and KLF10 were comparably expressed in control, FG, and Lujan patient-derived cells, indicating no apparent dysregulation of PPARγ signaling in MED12 mutant cells (Fig. 3B). ChIP analysis further revealed specific occupancy of PPARγ response elements (PPREs), but not upstream gene sequences, in the AKT2 and KLF10 promoters by PPARγ and MED12-containing Mediator in control as well as in FG and Lujan patient-derived cells (Fig. 3 D and F and Fig. S4). Importantly, CDK8 was comparably detected on PPARγ response elements within the AKT2 and KLF10 promoters in control as well as FG and Lujan patient-derived cells, indicating that mutant MED12/Mediator complexes are not defective for CDK8 recruitment onto PPARγ-target genes (Fig. 3 D and F). Taken together, these findings reveal that impaired CDK8 recruitment into mutant MED12/Mediator complexes is manifest in a gene-specific manner, occurring on GLI3-target genes but not on PPARγ-target genes.
To extend this analysis further, we comparatively examined the composition of soluble (nonchromatin associated) Mediator immunoprecipitated from control, FG, and Lujan patient-derived cells using antibodies specific for MED30 or CDK8 within the Mediator core or kinase module, respectively. Resultant immunoprecipitates were then monitored for the presence of additional Mediator subunits, including components of the head (MED6 and MED18), middle (MED1 and MED4), tail (MED23), and kinase (CDK8 and MED12) modules. Analysis of MED30-specific immunoprecipitates from control and patient-derived cells revealed comparable levels of all Mediator subunits examined, including CDK8 (Fig. 4A). Concordantly, analysis of CDK8-specific immunoprecipitates from control and patient-derived cells revealed comparable levels of all Mediator subunits corresponding to the four Mediator modules (Fig. 4A). These data indicate that the FG and Lujan mutations in MED12 do not disrupt the recruitment of CDK8 into soluble Mediator. Furthermore, analysis of immunopurified WT and mutant MED12-containing kinase modules reconstituted from recombinant baculovirus-expressed proteins revealed comparable levels of all four kinase module subunits and MED12-dependent kinase activities, indicating that the FG and Lujan mutations in MED12 do not disrupt the compositional or functional integrity of a soluble kinase module (Fig. 4 B and C). Together, these findings indicate that mutant MED12/Mediator complexes are not compromised for CDK8 incorporation before their conscription onto GLI3-target genes, providing further support for the concept that impaired CDK8 recruitment by mutant MED12/Mediator complexes is manifest in a gene-specific context. Collectively, our findings suggest that a gene-specific CDK8 recruitment defect into Mediator as a consequence of pathogenic mutations in MED12 removes a constraint on GLI3 transactivation resulting in enhanced SHH signal output in individuals with FG and Lujan syndromes.
Fig. 4.
Soluble mutant MED12-containing Mediator and reconstituted kinase modules show no defect in CDK8 incorporation. (A) Nuclear extracts from control CNTL, FG, or Lujan patient cells were subjected to IP with goat IgG (IgG*), rabbit IgG, or antibodies specific for CDK8 (goat) or MED30 (rabbit) as indicated. Immunoprecipitates were processed by WB analysis using antibodies specific for the indicated Mediator subunits. Input corresponds to 20% of the nuclear extracts used for IP. (B and C) Lysates from High Five insect cells coexpressing the indicated combinations of epitope-tagged kinase module subunits CBP-MED13, MED12-HA (WT/R961W/N1007S), CDK8-FLAG, and 6HIS-CyclinC were subjected to IP with FLAG-specific antibodies. Note that because CDK8 expression is limiting relative to other kinase module subunits during viral infection, WT and mutant MED12 derivatives are thus present in excess levels at the input concentrations used in IPs. (B) Immunoprecipitates were processed by WB analysis using the indicated antibodies. (C) Immunoprecipitates were incubated with a purified GST-3xCTD substrate (corresponding to three heptapeptide repeats from the RNA polymerase II large subunit) in the presence of [γ-32P]ATP before resolution by SDS/PAGE and visualization of 32P-labeled GST-3xCTD by phosphorimager analysis (Upper) and total input GST-3xCTD by Coomassie blue staining (Lower). The amount of 32P-labeled GST-3xCTD catalyzed by each kinase module derivative was quantified and expressed relative to the amount catalyzed by WT MED12-containing module (100%). Values represent the average ± SEM of three independent experiments.
Discussion
Our findings provide a possible molecular explanation for the phenotypic overlap between congenital anomaly syndromes arising from mutations in MED12 and its direct interaction target GLI3. In this regard, we previously reported that the FG and Lujan mutations in MED12 disrupt epigenetic suppression of neuronal gene expression imposed by the RE1 silencing transcription factor REST, suggesting a possible basis to attribute the cognitive deficits associated with these XLID disorders with altered neuronal development (24). Our findings herein that the FG and Lujan mutations in MED12 also elicit aberrant GLI3-dependent SHH signaling not only suggests an additional basis for cognitive dysfunction through altered brain development (30, 31), but may further explain a broad range of clinically diverse non-CNS phenotypes associated with these syndromal disorders. In this regard, many of the digit, craniofacial, corpus callosal, and anorectal malformations that typify FG and/or Lujan syndromes are similarly observed, to varying extents, in congenital anomaly syndromes (GCPS and PHS) arising from mutations in GLI3 (14, 32, 33). Thus, mutagenic impairment of two interacting components within a common signaling pathway could serve to explain the phenotypic overlap observed in these monogenic syndromes. Phenotypes restricted to FG and/or Lujan syndromes are likely explained by defects in REST-dependent and possibly other SHH-independent signaling pathways that converge on MED12.
Our studies further illuminate the mechanistic basis by which Mediator modulates GLI3-dependent SHH signaling and how pathogenic mutations in MED12 disrupt this regulation. Previously, we reported that MED12 suppresses GLI3 transactivation activity in response to Shh, and we proposed a model whereby a Mediator-imposed constraint on GLI3 transactivation is reversed by direct interaction of the GLI3 MBD with the MED12 interface in Mediator (12) (Fig. 5A). In this context, we envision Mediator to function much like a rheostat to modulate the magnitude of GLI3-dependent SHH signal output. However, neither the mechanistic basis of Mediator-imposed GLI3 repression nor the specific contribution of MED12 to this process has heretofore been clarified. Here, we show that enzymatically active CDK8 is responsible for suppression of GLI3 transactivation activity, and further, that MED12 most likely contributes to this suppression through its role as an anchor for CDK8 in Mediator. A repressive requirement for CDK8 is further substantiated by our findings that the FG and Lujan mutations in MED12 disrupt Mediator-dependent CDK8 recruitment onto GLI3-target gene promoters and concordantly trigger their enhanced GLI3-dependent transactivation in response to SHH signaling. Based on these findings, we propose a model in which the FG and Lujan mutations in MED12 disrupt its ability to recruit CDK8 onto GLI3-target gene promoters, leading to unrestrained GLI3-dependent SHH signaling (Fig. 5B).
Fig. 5.
Schematic model for how XLID mutations in MED12 disrupt a Mediator-imposed constraint GLI3-dependent SHH signaling. (A) In a WT MED12 background, SHH-activated GLI3, through its MBD, physically binds the MED12 interface in Mediator to functionally reverse CDK8-mediated inhibition of GLI3 transactivation activity. The balance achieved between these antagonistic interactions contributes to the level of GLI3-dependent gene expression output in response to SHH. (B) FG/R961W and Lujan/N1007S mutations in MED12 disrupt its ability to recruit CDK8 onto GLI3-target gene promoters, effectively removing a constraint on Gli3 transactivation activity, leading to hyperactivated GLI3-dependent SHH signaling. The unknown fate of cyclin C and MED13 is indicated by a question mark.
Our studies reveal an unanticipated level of gene specificity in this model. In this regard, we found that the impaired ability of mutant MED12/Mediator complexes to recruit CDK8 is not a constitutive defect associated with these complexes, because CDK8 was efficiently recruited into soluble mutant MED12/Mediator complexes as well as those bound to PPARγ-target genes. Thus, impaired CDK8 recruitment by mutant MED12/Mediator complexes occurs selectively on GLI3-target genes. This may help to explain why the developmental phenotypes associated with FG and Lujan syndromes, although profound, are less catastrophic than might otherwise be expected from mutational disruption of an essential protein linked to multiple developmental pathways.
The identity of the CDK8 substrate involved in suppression of GLI3 transactivation activity is presently unknown, but could correspond to a component of the general transcription machinery or GLI3 itself, because CDK8 has previously been shown to inhibit transcription by phosphorylation of activators and general transcription factors (25, 26). Our initial efforts to distinguish among these possibilities indicate that CDK8 does not phosphorylate GLI3, pointing to the ostensible involvement of other functionally relevant CDK8 substrates. In this regard, our model for CDK8-mediated suppression of GLI3 is similar in some respects to that proposed for Mediator-dependent inhibition of the Rcs/Aft1 transcriptional activator in yeast, which occurs through CDK8-mediated phosphorylation of Saccharomyces cerevisiae Med2 (34). It is thus possible that CDK8-mediated phosphorylation of another subunit(s) in Mediator contributes to suppression of GLI3 transactivation activity.
Finally, regarding the basis by which the FG and Lujan mutations in MED12 disrupt CDK8 recruitment into Mediator, our findings imply a principal contribution from DNA-bound GLI3, because no CDK8 recruitment defect is associated with soluble mutant MED12/Mediator complexes or those bound to PPARγ-target genes. We speculate that a distinct GLI3-induced conformational change in mutant versus WT MED12 triggers the gene-specific dissociation of CDK8 from mutant MED12/Mediaor complexes. In this regard, prior work has shown that activator-induced structural shifts can trigger selective Mediator–cofactor interactions sufficient to dictate gene-specific transcription control (35). Our findings herein suggest that activator-induced structural shifts may also reveal the disruptive potential of otherwise latent mutations in Mediator, leading to gene-specific transcriptional defects. In summary, our findings uncover evidence for the gene-specific manifestation of pathogenic mutations in MED12, leading to dysregulated GLI3-dependent SHH signaling in individuals with FG and Lujan syndromes. These findings clarify the mechanistic basis of Mediator in the modulation of GLI3 transactivation activity and shed light on the pathology of MED12-associated XLID syndromes.
Methods
Expression and Reporter Plasmids.
A complete description of expression and reporter plasmids used in this study is provided in SI Methods.
Transfection, Reporter Assays, and RNAi.
A complete description of transfection-based transient reporter assays and RNAi is provided in SI Methods.
Cell Culture.
HeLa, 293, and CH310T1/2 cell lines were cultured as described previously (12, 24). Control, FG, and Lujan patient-derived lymphoblastoid cell lines were established by EBV transformation according to standard protocols after obtaining informed consent (Self Regional Healthcare Institutional Review Board, Greenwood, SC) and were cultured in RPMI 1640 supplied with 0.3 g/L l-glutamine, 2 g/L NaHCO3 (Sigma; R8758), 15% (vol/vol) FBS (Sigma), 100× antibiotic (Sigma; A5955), and (100 units penicillin/100 μg/0.25 μg)/mL at 37 °C, 10% CO2.
Antibodies.
A complete list of antibodies used in this study is provided in SI Methods.
ChIP.
Control, Lujan, and FG patient cell lines were treated with 1% formaldehyde for 15 min at room temperature. Cross-linking reactions were quenched with 0.125 M glycine, and soluble chromatin was obtained by sonication of pelleted cells in cell lysis buffer (50 mM Pipes [piperazine-N,N-bis(2-ethanesulfonic acid)], pH 8.0, 85 mM KCl, 1% Nonidet P-40) before processing as described previously (24). DNA isolated with a QIAquick PCR purification kit was subsequently used in qPCR using primers specific for the GLI-binding sites and upstream gene regions within the GLI1 and ASCL1 genes as well as the PPARγ response elements and upstream gene regions within the AKT2 and STAT3 genes. Primer sequences used for ChIP are provided in SI Methods.
Additional materials and methods are described in SI Methods.
Supplementary Material
Acknowledgments
We thank Robert Tjian for support. This work was supported by Grant MH085320 from the National Institute of Mental Health (to T.G.B.), and in part by a grant from the South Carolina Department of Disabilities and Special Needs (to C.E.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121120109/-/DCSupplemental.
See Commentary on page 19519.
References
- 1.Rogers RC, Stevenson RE, Simensen RJ, Holden KR, Schwartz CE. Finding new etiologies of mental retardation and hypotonia: X marks the spot. Dev Med Child Neurol. 2008;50(2):104–111. doi: 10.1111/j.1469-8749.2007.02022.x. [DOI] [PubMed] [Google Scholar]
- 2.Chiurazzi P, Schwartz CE, Gecz J, Neri G. XLMR genes: Update 2007. Eur J Hum Genet. 2008;16(4):422–434. doi: 10.1038/sj.ejhg.5201994. [DOI] [PubMed] [Google Scholar]
- 3.Risheg H, et al. A recurrent mutation in MED12 leading to R961W causes Opitz-Kaveggia syndrome. Nat Genet. 2007;39(4):451–453. doi: 10.1038/ng1992. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz CE, et al. The original Lujan syndrome family has a novel missense mutation (p.N1007S) in the MED12 gene. J Med Genet. 2007;44(7):472–477. doi: 10.1136/jmg.2006.048637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spaeth JM, Kim NH, Boyer TG. Mediator and human disease. Semin Cell Dev Biol. 2011;22(7):776–787. doi: 10.1016/j.semcdb.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30(5):235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11(11):761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36(12):3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larivière L, Seizl M, Cramer P. A structural perspective on Mediator function. Curr Opin Cell Biol. 2012;24(3):305–313. doi: 10.1016/j.ceb.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Kim S, Xu X, Hecht A, Boyer TG. Mediator is a transducer of Wnt/beta-catenin signaling. J Biol Chem. 2006;281(20):14066–14075. doi: 10.1074/jbc.M602696200. [DOI] [PubMed] [Google Scholar]
- 11.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet. 2006;38(8):896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 12.Zhou H, Kim S, Ishii S, Boyer TG. Mediator modulates Gli3-dependent Sonic hedgehog signaling. Mol Cell Biol. 2006;26(23):8667–8682. doi: 10.1128/MCB.00443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rocha PP, Scholze M, Bleiss W, Schrewe H. Med12 is essential for early mouse development and for canonical Wnt and Wnt/PCP signaling. Development. 2010;137(16):2723–2731. doi: 10.1242/dev.053660. [DOI] [PubMed] [Google Scholar]
- 14.Johnston JJ, et al. Molecular analysis expands the spectrum of phenotypes associated with GLI3 mutations. Hum Mutat. 2010;31(10):1142–1154. doi: 10.1002/humu.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lum L, Beachy PA. The Hedgehog response network: Sensors, switches, and routers. Science. 2004;304(5678):1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- 16.McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- 17.Hui CC, Angers S. Gli proteins in development and disease. Annu Rev Cell Dev Biol. 2011;27:513–537. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz i Altaba A, Mas C, Stecca B. The Gli code: An information nexus regulating cell fate, stemness and cancer. Trends Cell Biol. 2007;17(9):438–447. doi: 10.1016/j.tcb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rohatgi R, Scott MP. Patching the gaps in Hedgehog signalling. Nat Cell Biol. 2007;9(9):1005–1009. doi: 10.1038/ncb435. [DOI] [PubMed] [Google Scholar]
- 20.Bai CB, Stephen D, Joyner AL. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev Cell. 2004;6(1):103–115. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 21.Bok J, et al. Opposing gradients of Gli repressor and activators mediate Shh signaling along the dorsoventral axis of the inner ear. Development. 2007;134(9):1713–1722. doi: 10.1242/dev.000760. [DOI] [PubMed] [Google Scholar]
- 22.Buttitta L, Mo R, Hui CC, Fan CM. Interplays of Gli2 and Gli3 and their requirement in mediating Shh-dependent sclerotome induction. Development. 2003;130(25):6233–6243. doi: 10.1242/dev.00851. [DOI] [PubMed] [Google Scholar]
- 23.Wang C, Rüther U, Wang B. The Shh-independent activator function of the full-length Gli3 protein and its role in vertebrate limb digit patterning. Dev Biol. 2007;305(2):460–469. doi: 10.1016/j.ydbio.2007.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding N, et al. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol Cell. 2008;31(3):347–359. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akoulitchev S, Chuikov S, Reinberg D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature. 2000;407(6800):102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- 26.Nelson C, Goto S, Lund K, Hung W, Sadowski I. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature. 2003;421(6919):187–190. doi: 10.1038/nature01243. [DOI] [PubMed] [Google Scholar]
- 27.Ribes V, et al. Combinatorial signalling controls Neurogenin2 expression at the onset of spinal neurogenesis. Dev Biol. 2008;321(2):470–481. doi: 10.1016/j.ydbio.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Vokes SA, Ji H, Wong WH, McMahon AP. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 2008;22(19):2651–2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt MV, Brüne B, von Knethen A. The nuclear hormone receptor PPARγ as a therapeutic target in major diseases. ScientificWorldJournal. 2010;10:2181–2197. doi: 10.1100/tsw.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blaess S, Stephen D, Joyner AL. Gli3 coordinates three-dimensional patterning and growth of the tectum and cerebellum by integrating Shh and Fgf8 signaling. Development. 2008;135(12):2093–2103. doi: 10.1242/dev.015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rash BG, Grove EA. Shh and Gli3 regulate formation of the telencephalic-diencephalic junction and suppress an isthmus-like signaling source in the forebrain. Dev Biol. 2011;359(2):242–250. doi: 10.1016/j.ydbio.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiang C, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383(6599):407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 33.Mo R, et al. Anorectal malformations caused by defects in sonic hedgehog signaling. Am J Pathol. 2001;159(2):765–774. doi: 10.1016/S0002-9440(10)61747-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van de Peppel J, et al. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol Cell. 2005;19(4):511–522. doi: 10.1016/j.molcel.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 35.Ebmeier CC, Taatjes DJ. Activator-Mediator binding regulates Mediator-cofactor interactions. Proc Natl Acad Sci USA. 2010;107(25):11283–11288. doi: 10.1073/pnas.0914215107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.