Abstract
Most nucleosomes that package eukaryotic DNA are assembled during DNA replication, but chromatin structure is routinely disrupted in active regions of the genome. Replication-independent nucleosome replacement using the H3.3 histone variant efficiently repackages these regions, but how histones are recruited to these sites is unknown. Here, we use an inducible system that produces nucleosome-depleted chromatin at the Hsp70 genes in Drosophila to define steps in the mechanism of nucleosome replacement. We find that the Xnp chromatin remodeler and the Hira histone chaperone independently bind nucleosome-depleted chromatin. Surprisingly, these two factors are only displaced when new nucleosomes are assembled. H3.3 deposition assays reveal that Xnp and Hira are required for efficient nucleosome replacement, and double-mutants are lethal. We propose that Xnp and Hira recognize exposed DNA and serve as a binding platform for the efficient recruitment of H3.3 predeposition complexes to chromatin gaps. These results uncover the mechanisms by which eukaryotic cells actively prevent the exposure of DNA in the nucleus.
Keywords: nucleosome assembly, transcription
DNA in the eukaryotic nucleus is associated with histone proteins to form nucleosomes, the fundamental units of chromatin. Most nucleosomes are assembled during DNA replication, but chromatin structure is routinely disrupted in active regions of the genome. These regions are repackaged by replication-independent (RI) nucleosome replacement using the H3.3 histone variant (1, 2). This process results in the enrichment of the H3.3 histone variant at all sites where nucleosomes are unstable or disrupted (3, 4).
How H3.3 is delivered to dynamic chromatin sites is unknown. However, biochemical isolation of predeposition complexes has identified shared and distinctive assembly factors that associate with the H3 and H3.3 histones and mediate the replication-coupled or RI assembly of nucleosomes, respectively (4–6). These factors include histone chaperones and chromatin remodelers that are important for new nucleosome assembly, and might potentially target histones to active chromatin regions. However, mutants in some of these factors have surprisingly limited phenotypes. The Hira chaperone promotes H3.3 deposition at genes (4) but is only essential for H3.3 deposition on sperm chromatin during fertilization (7, 8). In Drosophila the ATRX/XNP remodeler homolog Xnp colocalizes with H3.3 in somatic cells, but is not essential (9). In mammals, ATRX/XNP promotes H3.3 deposition only at telomeres and some heterochromatic sequences (4, 6, 10). These results have raised the possibilities that H3.3 assembly factors are redundant or that additional factors involved in the deposition of this histone variant exist. Loss of H3.3 itself can be compensated in somatic cells by the major H3 histone, suggesting that assembly of any nucleosome suffices (11).
In this work we use an inducible system that produces nucleosome-depleted chromatin at the Hsp70 genes in Drosophila to study the mechanism of nucleosome replacement. We provide evidence that H3.3 predeposition factors mediate two separable steps in RI nucleosome assembly. We show that the Xnp and Hira factors bind genomic sites when nucleosomes are disassembled, thereby marking sites for RI assembly. Strikingly, we also demonstrate that Hira and Xnp are redundant for RI nucleosome assembly in somatic nuclei. Our results further reveal that RI nucleosome replacement is essential for chromatin structure and viability, and uncover a cellular system that surveys chromatin for defects and promotes its repair.
Results and Discussion
Induced Genes Recruit RI Assembly Factors.
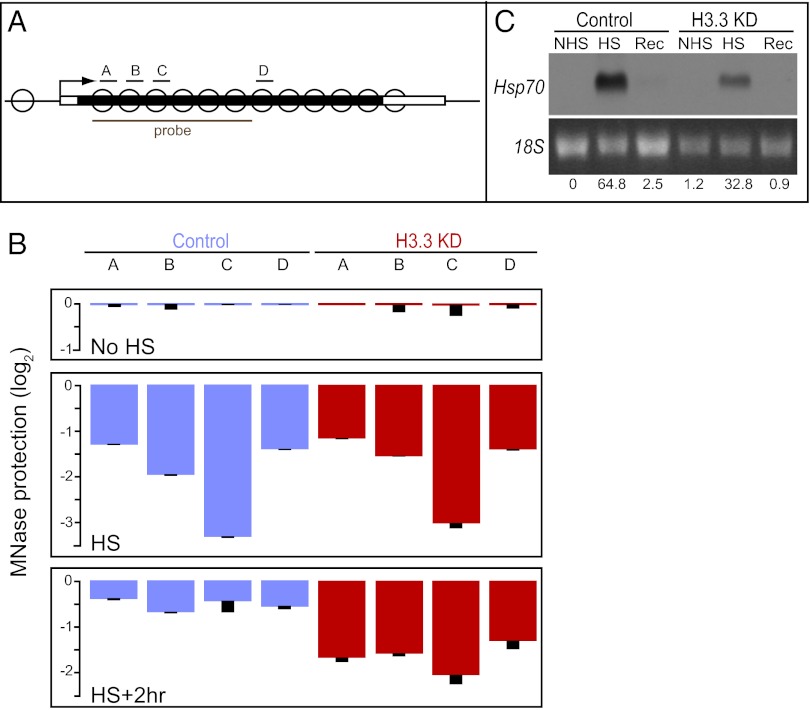
We used the inducible Hsp70 genes as a controlled in vivo system to deplete nucleosomes from chromatin. Heat-shock activates transcription of Hsp70, displacing nucleosomes and increasing the sensitivity of the locus to digestion by micrococcal nuclease (MNase) (12). After heat-shock, H3.3-containing nucleosomes repackage the locus (13). To generate persistently nucleosome-depleted chromatin, we induced the Hsp70 genes in H3.3-deficient cells. ChIP experiments using an anti-H3 antibody show similar amounts of histone H3 at the Hsp70 genes in wild-type and H3.3-deficient salivary glands (0.8% of input ±0.6 vs. 1.3% of input ±0.4), and the genes show similar protection from MNase digestion (Fig. 1A). These data demonstrate that Hsp70 sequences are fully protected by nucleosomes before induction in wild-type and H3.3-deficient salivary glands. In both genotypes induced Hsp70 genes become hypersensitive to MNase as nucleosomes are lost (Fig. 1 A and B). In wild-type cells after heat-shock nuclease protection is restored, but is not restored in H3.3-deficient glands (Fig. 1B). This finding demonstrates that nucleosomes are not replaced after Hsp70 induction in the absence of H3.3. This system allows us to tease apart cause-and-effect in analyzing the effects of nucleosome assembly factors.
Fig. 1.
Nucleosomes are not restored to repressed Hsp70 genes in H3.3-deficient cells. (A) Schematic depiction of the nucleosomal Hsp70 gene. The location of PCR amplicons and the Northern probe are shown. (B) DNA survival after MNase digestion. Chromatin was purified from salivary glands before (NoHS), during heat-shock (HS), or 2 h into recovery after heat-shock (HS+2hr). Blue, DNA survival in wild-type controls; red, DNA survival in H3.3 knock-down glands. PCR Cycle-to-threshold (Ct) values were normalized to a background intergenic amplicon and to NoHS values. DNA protection is presented in a log2 scale, and the SD for each measurement is indicated. (C) Northern detection of Hsp70 transcripts in control and H3.3 knock-down salivary glands. The amount of the Hsp70 signal was calculated relative to the 18S signal (stained with ethidium bromide) after subtraction of background.
We first used our inducible system for nucleosome depletion to characterize the targeting of histone chaperones and chromatin remodelers implicated in H3.3 deposition. We have previously described that the Xnp chromatin remodeler colocalizes with H3.3 in chromatin (9). Mammalian data has suggested that the Xnp homolog ATRX mediates H3.3 deposition at telomeres and at transcribed heterochromatic repeat sequences (4, 6), although its nuclear distribution is much broader (14). The Hira histone chaperone has been implicated in global nucleosome replacement after the removal of protamines from the sperm nucleus during fertilization (7) and near genes in mammalian cells (4). Purification of nuclear soluble H3.3-containing complexes showed that ATRX and Hira are in two separate complexes that mediate H3.3 deposition at distinct target sites (4–6). ASF1 is a general histone chaperone that complexes with new histone dimers in the cytoplasm and escorts them into the nucleus (15). We used antibodies to Drosophila Xnp, Hira, and ASF1 to track their localization during Hsp70 induction and subsequent RI nucleosome assembly in polytene chromosomes of larval salivary glands.
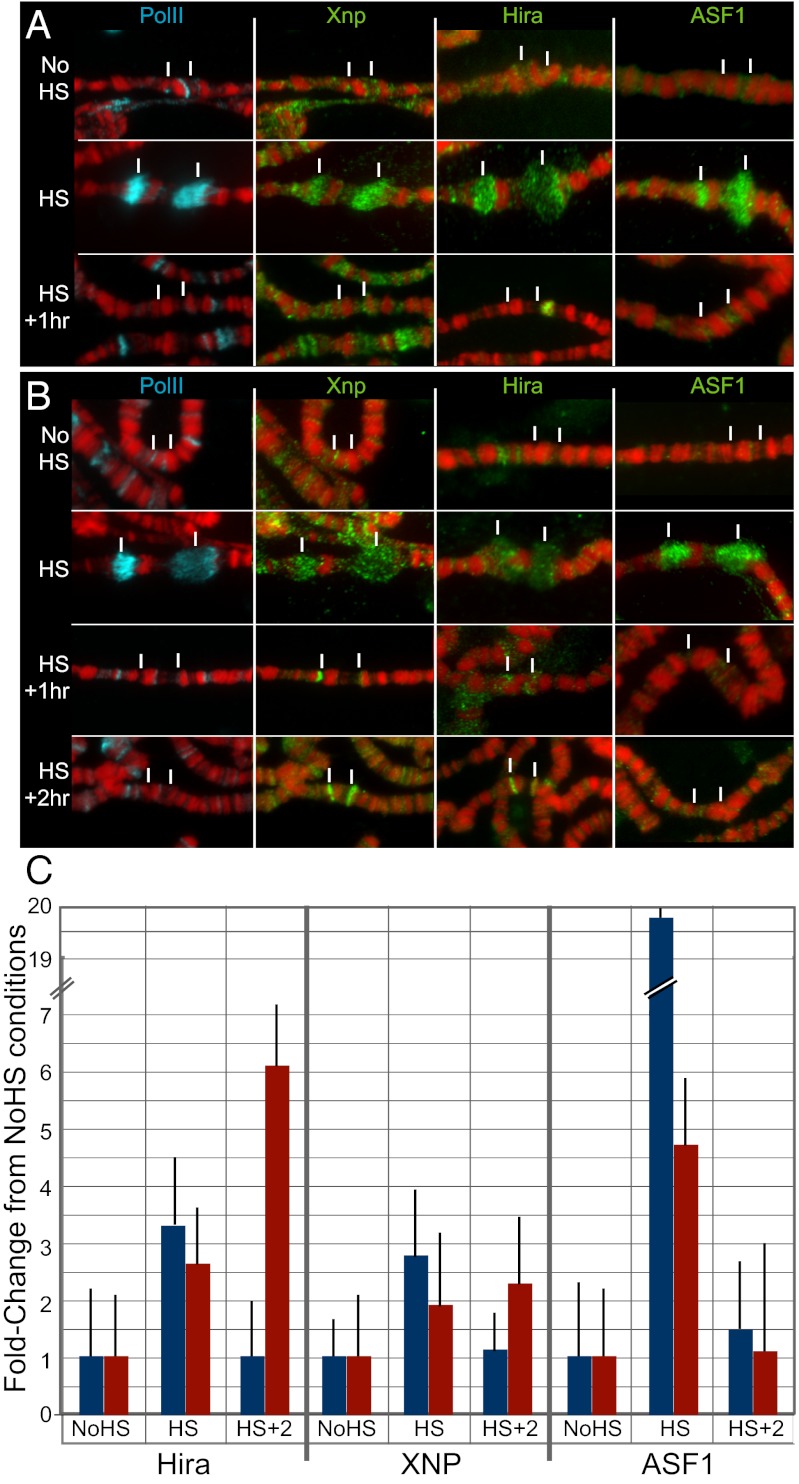
Heat-shock rapidly activates transcription of Hsp70, and elongating RNA polymerase II becomes strongly localized in puffs at the transcribing genes (Fig. 2A). After cessation of a heat-shock, the puffs regress and RNA polymerase II leaves the locus within 30 min. We found that neither Xnp, Hira, nor ASF1 are enriched at the Hsp70 loci before induction. However, upon induction all three proteins are rapidly recruited to the Hsp70 loci (Fig. 2 A and C). Xnp, Hira, and ASF1 remain associated with chromatin and the genes are transcribed, but then leave the Hsp70 genes after heat-shock. Their dynamic recruitment implicates these assembly factors in cotranscriptional nucleosome dynamics.
Fig. 2.
The Xnp and Hira assembly factors are recruited to nucleosome-depleted chromatin. Polytene chromosomes were prepared from larvae heat-shocked at 37° for 20 min with or without recovery at 25°, and imaged with a 100× objective. The location of the Hsp70 genes at cytological positions 87A and 87C are indicated (white lines). DAPI is shown in red. (A) Heat-shock of wild-type larvae causes the rapid recruitment of elongating RNA polymerase II (blue) to the Hsp70 loci. RNA polymerase leaves within 60 min of recovery as the genes are repressed. Xnp, Hira, and ASF1 are rapidly recruited to induced Hsp70 genes, and leave during recovery. (B) Polytene chromosomes from H3.3 knock-down salivary glands. Elongating RNA polymerase II (blue) and ASF1 label the induced Hsp70 loci, and leave within 60 min of recovery. Xnp and Hira are also recruited to induced Hsp70 genes, but remain bound 2 h after recovery. (C) ChIP of assembly factors from wild-type (blue) and H3.3-deficient (red) salivary glands. The association of factors at the “C” amplicon in Fig. 1 was assessed before, during and after heat-shock.
We then followed the induction of Hsp70 in H3.3-deficient salivary glands, where transcription produces nucleosome-depleted chromatin. Both Northern analysis and cytological observations of RNA polymerase II showed that Hsp70 was induced in wild-type and H3.3-deficient salivary glands, although H3.3-deficient glands produce ∼50% less mRNA (Figs. 1C and 2B). We observed two effects on the recruitment of RI assembly factors. First, Xnp, Hira, and ASF1 are all rapidly recruited to the induced Hsp70 genes at moderately reduced levels, indicating that the Hsp70 genes are less efficiently induced in this genotype. (Fig. 2 B and C). Second, both Xnp and Hira—but not ASF1—accumulate and persist at the Hsp70 genes after heat-shock (Fig. 2 B and C). The correspondence between persistent nucleosome depletion and the persistent binding of these factors is striking. This correspondence is a distinctive property of these two RI assembly factors, because the histone chaperone ASF1 is also cotranscriptionally recruited but dissociates both in wild-type and in H3.3-deficient cells (Fig. 2). Finally, retention of Xnp and Hira is not a result of ongoing transcription, because RNA polymerase II rapidly leaves the Hsp70 loci after heat-shock (Fig. 2B) and transcript production ceases (Fig. 1C). We conclude that once Xnp and Hira bind nucleosome-depleted chromatin, they are only displaced when new nucleosomes assemble.
How are Xnp and Hira recruited to nucleosome-depleted chromatin? These factors may be directly recruited by transcriptional machinery to active genes. Alternatively, Xnp and Hira may bind a structural feature common to chromatin gaps, or may simply bind exposed DNA. These factors might be complexed with DNA-binding factors or may bind DNA themselves. The homologous ATRX remodeler contains an ADD (ATRX-DNMT3-DNMT3L) domain that can bind DNA or histone tails (16–18). Indeed, ATRX is recruited to the genomes of DNA viruses as they enter the nucleus, suggesting that it may directly bind exposed DNA (19). A recent study has shown that the mammalian Hira chaperone may also directly bind exposed DNA at chromatin gaps (20). We find that Xnp and Hira bind independently at induced Hsp70 genes in null mutants of the other factor (Fig. S1), implying that there may be multiple ways that RI assembly factors recognize exposed DNA.
Xnp and Hira Are Essential for H3.3 Deposition.
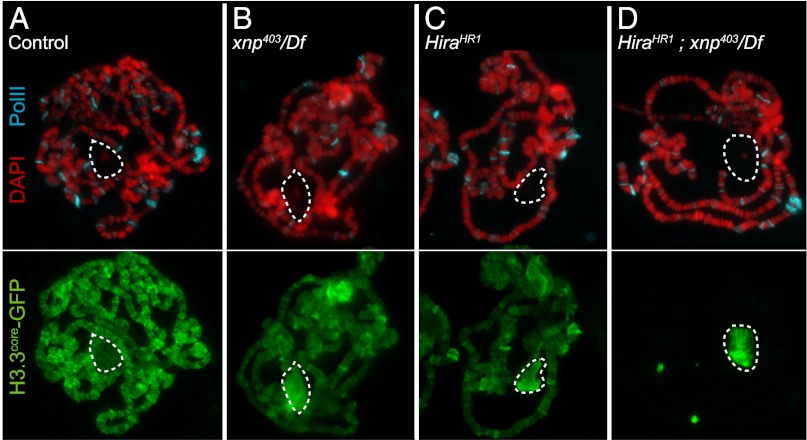
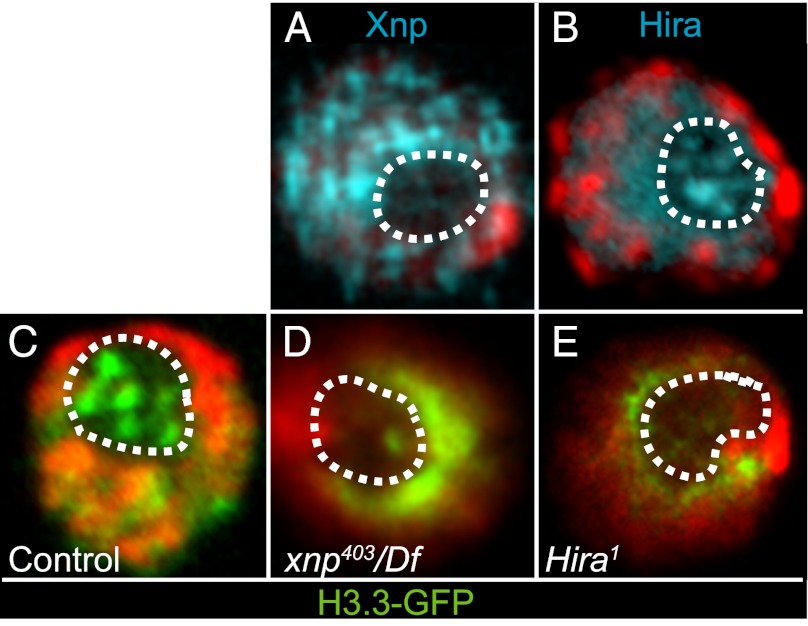
Although both Xnp and Hira have been implicated in RI nucleosome assembly, mutants in these factors have surprisingly limited phenotypes. These results have raised the possibility that H3.3 assembly factors are redundant. To test if these factors promote nucleosome replacement at these sites, we assayed deposition of GFP-tagged truncated H3.3 histone into chromatin in wild-type and mutant genotypes. H3.3core-GFP can only be incorporated by RI nucleosome assembly, and the histone labels active genes (1, 13). We therefore produced a pulse of H3.3core-GFP in salivary glands and prepared chromosome spreads 2 h later to assess the efficiency of RI nucleosome assembly (Fig. 3). In wild-type cells, H3.3core-GFP strongly labels chromosome arms and active genes (Fig. 3A). In contrast, the H3.3core-GFP protein is efficiently produced in xnp-null mutant cells, but only a fraction of the protein deposits onto chromosomes; instead, most of the protein accumulates within the nucleolus (Fig. 3B). This protein does not coincide with DNA in the nucleolus, and may be predeposition or aggregated histones. Hira mutants have a similar reduction in H3.3 deposition: H3.3core-GFP protein is produced, but most protein accumulates in the nucleolus (Fig. 3C). These results demonstrate that the rate of RI nucleosome assembly is reduced in both xnp and Hira mutants, although some assembly can still occur. Indeed, longer expression of tagged histones in xnp or Hira mutants does achieve apparently normal levels (Fig. S2) (8, 9).
Fig. 3.
Xnp and Hira are required for H3.3 nucleosome replacement. Deposition of H3.3core-GFP (green) in polytene chromosomes after recovery from heat-shock. Polytene spreads were imaged with a 100× objective. DAPI is in red, elongating RNA Polymerase II is in blue, and the nucleolus is outlined with a dotted line. (A) H3.3core-GFP is efficiently deposited along chromosome arms in wild-type, and the nucleolus is devoid of soluble tagged protein. (B) xnp403/Df(3R)Exel6202 and (C) HiraHR1 mutants show reduced H3.3core-GFP signals along chromosome arms, and increased accumulation of the tagged protein within nucleoli. (D) Deposition of H3.3core-GFP is completely blocked in HiraHR1;xnp403/Df(3R)Exel6202 double-mutants and the tagged protein accumulates within the nucleolus.
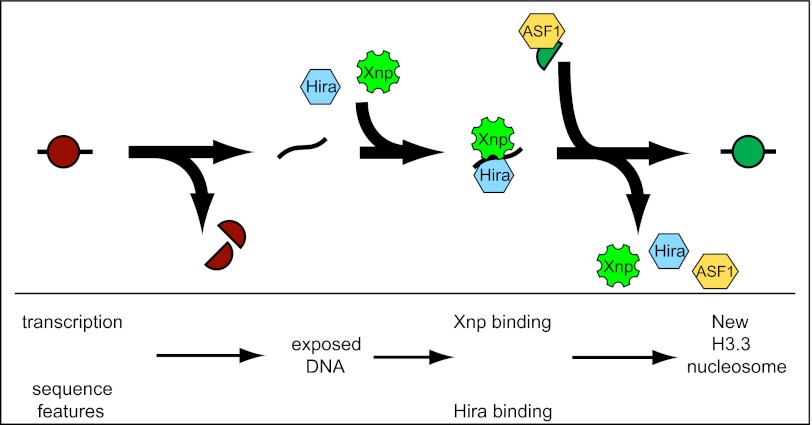
To test if Xnp and Hira have redundant roles in nucleosome replacement, we generated Hira;xnp double-mutant animals. Single-mutants are fully viable, but we found that double-mutant larvae grow slower than wild-type siblings and die during larval development. We therefore measured H3.3core-GFP deposition in these double-mutants. Strikingly, high levels of H3.3core-GFP were produced in Hira;xnp animals, but all of the protein accumulates in the nucleolus, with no detectable staining of chromosomes (Fig. 3D). We conclude that both Hira and Xnp contribute to the efficiency of H3.3 RI nucleosome assembly, but this fails when both factors are eliminated. Our results lead us to suggest that Xnp and Hira identify nucleosome-depleted chromatin and promote new nucleosome assembly through a stepwise process (Fig. 4). In the first step, Xnp and Hira bind exposed DNA at chromatin gaps, thereby marking sites where a nucleosome has been displaced. In the second step, delivery factors carrying new histones are recruited by binding to Xnp and Hira at chromatin gaps. In the final step, these factors assist in the transfer of histones from delivery chaperones to DNA, and Xnp and Hira are released when nucleosome assembly is complete. In this way, these factors may act to survey chromatin for gaps and promote their repair.
Fig. 4.
Step-wise model for recognition of chromatin gaps and delivery of predeposition H3.3•H4 histones. Old nucleosomes (red) are disrupted by diverse processes on chromatin, displacing histones. Xnp and Hira independently bind exposed DNA at chromatin gaps. A separate predeposition complex containing ASF1 and new H3.3•H4 heterodimers (green) is recruited to chromatin-bound Xnp and Hira. In the final step, Xnp and Hira pry histones off ASF1 and wrap them with DNA to rebuild nucleosomes.
Xnp and Hira Assembly Factors Have Some Distinct Target Sites.
We previously reported that the Xnp remodeler is found at all sites where H3.3 is enriched, including active genes (9). The Hira chaperone also localizes at active genes (Fig. S3B). However, there are additional sites in the genome where the two assembly factors do not coincide. First, the major site for Xnp binding is at a nucleosome-depleted satellite block, where Hira is not found (Fig. S3 A and B). Second, most Hira is localized to the repeated ribosomal DNA (rDNA) within the nucleolus, where Xnp is not found (Fig. S3B). This finding implies that the two factors are not redundant at all sites where H3.3 is deposited.
The rDNA genes are repressed in late-stage salivary glands. Therefore, we assayed deposition of H3.3 in the somatic follicle cells of ovaries, where rDNA is highly transcribed. In this cell type, Xnp localizes broadly in the nucleus but not within the nucleolus, but most Hira protein forms foci within the nucleolus (Fig. 5 A and B). A pulse of epitope-tagged H3.3 produced in follicle cells broadly labels the nucleus and foci within the nucleolus, corresponding to transcriptionally active sites in this cell type (Fig. 5C). Follicle cells from xnp mutant ovaries also show nucleolar labeling with H3.3-GFP (Fig. 5D), demonstrating that Xnp is not required for nucleolar RI assembly. In contrast, a pulse of epitope-tagged H3.3 in Hira mutant follicle cells does not deposit in the nucleolus (Fig. 5E). Thus, rDNA chromatin must rely on the Hira chaperone for H3.3 deposition. We conclude that Xnp and Hira are redundant at many sites within the genome, but some sites rely on individual factors for replacement nucleosome assembly. Although neither Hira nor Xnp are individually required for viability, there may be more subtle phenotypes that occur in repetitive sites in the genome.
Fig. 5.
Hira is required for H3.3 nucleosome replacement at active rDNA genes. Assembly factor localization and H3.3 deposition in stage 10 somatic follicular epithelia of adult ovaries, imaged with a 60× objective. DNA is in red. The nucleolus is a clear area within each nucleus. (A) Hira-GFP (green) stains chromatin throughout the nucleus and labels foci within the nucleolus. (B) Xnp (green) broadly stains chromatin but does not localize within the nucleolus. (C–E) Hira but not Xnp is required for H3.3 deposition (green) at active rDNA genes. (C) A pulse of H3.3-GFP rapidly accumulates in nucleolar foci in wild-type follicle nuclei. (D) A pulse of H3.3-GFP deposits within the nucleolus in xnp403/Df(3R)Exel6202 cells. (E) H3.3-GFP does not deposit within the nucleolus in Hira1 adults.
Xnp and Hira Recognize Chromatin Defects.
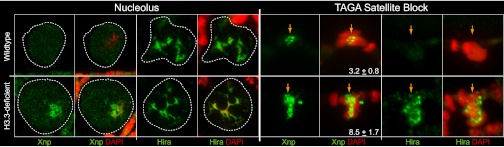
If Xnp and Hira are generally involved in recognizing chromatin gaps, the localization of these factors should be affected in H3.3-deficient cells. Indeed, new binding sites for Xnp appear in rDNA chromatin after H3.3 knock-down (Fig. 6 A and B, and Fig. S3). Hira is also recruited to the nucleosome-depleted satellite block after H3.3 knock-down (Fig. 6 G and H). Strikingly, the area of the nucleosome-depleted satellite block is ∼2.5-times larger after H3.3 knock-down, and the Xnp signal at this site is elevated (Fig. 6 E and F). This finding implies that RI assembly is normally required for compaction of the satellite. The redundancy of Hira and Xnp implies that their importance for H3.3 deposition is underestimated in single-mutants. Both the relocalization and increased binding of Xnp and Hira after H3.3 knock-down support the idea that these assembly factors are recruited to aberrant, nucleosome-depleted chromatin. As persistent exposure of DNA may disrupt transcriptional regulation or allow DNA damage, surveying chromatin for gaps with RI assembly factors may be critical for genome stability and function.
Fig. 6.
Xnp and Hira mark chromatin defects in H3.3-deficient cells. Magnifications of the nucleolus (Left) and the nucleosome-depleted TAGA satellite block (Right) from polytene chromosome spreads imaged with a 100× objective. Chromosomes were prepared from larvae carrying the In(1)wm4 inversion to easily visualize the TAGA satellite block away from the compacted chromocenter. Images for wild-type (Upper) and H3.3-deficient (Lower) cells are shown stained with anti-Xnp or anti-Hira antibodies. DAPI is in red. Xnp does not normally associate with nucleolar chromatin in wild-type spreads, but is strongly enriched on nucleolar chromatin in H3.3-deficient glands. Hira stains the nucleolus in both wild-type and H3.3-deficient spreads. Xnp associates with the nucleosome-depleted satellie block in both wild-type and H3.3-deficient cells, but the area of the block expands in H3.3-deficient cells. The area of the Xnp-stained block (± SD) is shown in arbitrary units. Hira does not normally associate with the nucleosome-depleted TAGA satellite block, but is strongly enriched at the TAGA satellite block in H3.3-deficient cells.
Experimental Procedures
Fly Stocks and Crosses.
All stocks and crosses were grown at 25 °C. Heat-shock induction was conducted at 37 °C water-bath, as previously described (13). Null alleles for xnp [xnp403 and Df(3R)Exel6202] were previously described in ref. 9, and the null HiraHR1 allele in ref. 8. The Hira1 null allele was derived from HiraHR1 by Cre recombinase-mediated excision of the miniw+ marker (21). The H3.3-null alleles His3.3B0 and His3.3A2X1 were described in ref. 11. Inducible P[H3.3-GFP]B6 and P[H3-GFP]C4 lines were described previously (13). The GAL4-inducible knockdown line 5825R-3 (22) was obtained from the National Institute of Genetics Fly Stock Center (Japan) and was expressed using the P[SGS3-GAL4]TP1 driver line (BloomingtonDrosophila Stock Center). This genotype efficiently knock-down new production of H3.3 histones (Fig. S4). For construction of the Hira-GFP fusion gene, the eGFP coding sequence was inserted between the Hira and FLAG tag sequences of PW8-Hira-3xFLAG (7), and the construct was transformed into flies by standard procedures (23).
Polytene Chromosome Cytology.
Salivary glands from male larvae were fixed and spread and slides were treated as described previously (9), except using 1% (wt/vol) BSA in PBST as a blocking solution. Antibodies used for Xnp (1:2,000), Hira (PG1, 1:100), and ASF1 have been previously described (8, 9, 24). Other antibodies used were against elongating RNA Polymerase II (Convance; H5, 1:100), GFP (Clontech, JL8; Abcam, ab6556, 1:100). Fluorescently labeled secondary antibodies (Jackson ImmunoResearch) were used at a 1:200 dilution. Quantitative comparisons of Xnp staining were made by cosquashing and staining glands from H3.3-knock-down strains with glands from animals carrying the In(1)wm4 inversion. Polytene spreads were imaged using a 60× or 100× objective on a Nikon 80i Microscope using UV-2E/C, FITC HQ and TRITC HQ filter sets. Images were collected with a Hamamatsu ORCA-R2 camera using the MetaMorph image software. Image processing was performed with IPLab imaging software, and assembled in Adobe Photoshop and Adobe Illustrator. The relative area of the Xnp-stained satellite block (n = 6) was measured as the ratio of the number of pixels in the block divided by the number in a nearby control band to normalize for the size of the chromosomes.
Chromatin Immunoprecipitation.
Wild-type or H3.3-knockdown larvae were heat-shocked for 30 min at 37 °C, followed or not by 2 h of aging at 25 °C, or not treated. For each group, 20 pairs of salivary glands were dissected in 80T buffer (80 mM NaCl, 2 mM MgCl2, 2 mM EGTA, 0.1% (vol/vol) Triton X-100, 0.5 mM PMSF, pH 7.4,) with 1% formaldehyde and fixed at room temperature for 1 min. Samples were next transferred to 80T buffer containing 125 mM glycine for 3 min before being stocked in 80T buffer containing 1 mM EDTA and 0.1% (wt/vol) SDS. Chromatin was sonicated five times (20-s ON/20-s OFF cycles), which yielded fragments of 100–500 bp. Chromatin samples were pelleted for 1 min at 8,000 × g and soluble chromatin was incubated with antibodies (1:150 dilution) overnight at 4 °C, and an aliquot was set aside as an input sample. Chromatin-antibody complexes were immunoprecipitated using 10 µL of ProteinG magnetic beads (Invitrogen; Dynabeads Protein G, 100.04D) for 2 h at 4 °C. Beads were next washed twice in 80T buffer, treated with 0.5 µg of RNAseA (Sigma; R6513) for 30 min at 37 °C, supplemented with 0.5% (wt/vol) SDS and 20 µg of Proteinase K (Promega; V3021), and incubated at 70 °C for at least 4 h to reverse cross-links. DNA was purified with phenol/chloroform and precipitated with 1/10 volume of 3 M NaAc (pH 5.0), 20 µg of glycogen (Calbiochem), and 2.5 volumes of ethanol. DNA was resuspended in 50 µL buffer TE and used directly in PCR (input samples were used at a 1/10 dilution). Real-time PCR was performed in triplicates in 96-well plates using an Applied Biosystems 7500 Fast cycler. Ct values for immuno-precipitated Hsp70 DNA was normalized to a background primer set and to the input samples.
Ovarian Follicle Cell Cytology.
Two- to 4-d-old adult females were heat shocked for 1 h at 37 °C in a water bath to induce expression from P[H3.3-GFP]B6 or P[H3.3core-GFP]G5A constructs, and then allowed to recover for 30 min at 25° before dissection in PBST. Ovaries were fixed in 4% (wt/vol) paraformaldehyde/PBT for 25 min, and washed three times for 10 min with PBST before blocking with 10% (wt/vol) milk in PBT or 1% (wt/vol) BSA in PBST. Antibodies to Xnp were applied overnight at 4°, and detected with secondary antibodies (Molecular Probes or Jackson ImmunoResearch). FITC filter sets were used to detect fluorescence from H3.3-GFP and Hira-GFP. Images were collected from stage 10 egg chambers by confocal or wide-field microscopy.
Nuclease Protection Assays.
Micrococcal nuclease digests were performed as previously described (25), with the following modifications: four to five gland pairs were isolated from each sample in cold M-buffer supplemented with 1 mM ZnCl2 to inhibit endogenous nucleases, and incubated in the same buffer on ice. Glands were permeabilized with 0.5% (vol/vol) Nonidet P-40 for 10 min on ice, centrifuged at 2,100 × g for 5 min at 4 °C, and resuspended in 200 µL of M-buffer supplemented with 2 mM CaCl2 and 400 U of MNase (USB, 70196Y). Digests were carried out at 25 °C for 40 min with occasional agitation. The reactions were then stopped by adjusting samples to final concentrations of 20 mM EDTA and 0.5% (wt/vol) SDS before Protease K treatment and DNA purification. This treatment produced predominantly mononucleosomal DNA fragments. Real-time PCR measurements were performed in triplicate using the 7500 Fast Real-Time PCR System (Applied Biosystems) in 96-well plates. Primers for Hsp70 were hsp70 +334F/+423R (“A”), hsp70 +645F/+718R (“B”), hsp70 +872F/+1019R (“C”), and hsp70 +1649F/+1754R (“D”) described in ref. 26. Ct values for each digest were normalized to background reads (“BackF” 5-TTGCACTCACCGTGATTGGAATG-3; “BackR” 5- GTCACAATGCTAACATCTCCTTAT-3) to give ΔCt values. Two biological replicates were performed for each experiment.
RNA Analysis.
RNA for Northern blotting was extracted from five pairs of salivary glands using TRIzol (Invitrogen) and precipitated with glycogen, yielding ∼1.25 µg RNA per sample. RNA was resuspended in 1XMOPS/formamide/formaldehyde buffer containing 20 ng ethidium bromide according to standard protocols, and run on a 1.2% (vol/vol) formaldehyde gel. The gel was imaged with UV and capillary-transferred overnight to a positively charged nylon membrane (Roche). DIG-labeled probe for Hsp70 mRNA was made using a DNA template generated with primers hsp70 +334F/+1490 (26) and detected using the DIG DNA Labeling and Detection Kit (Roche) and CSPD ready-to-use substrate (Roche). Membrane was exposed to autoradiography film, and integrated band intensities were calculated using the ImageJ software.
Supplementary Material
Acknowledgments
We thank Joe Geisberg for assistance with real-time PCR. Microscopy data for this study were acquired in the Nikon Imaging Center at Harvard Medical School and fly lines were provided by the Bloomington Stock Center (University of Indiana, Bloomington). Work in the B.L. laboratory was supported by the Centre National de la Recherche Scientifique, the French Ministry of Research, the Agence Nationale de Recherche (ANR-08-BLAN-0139-01), and the Association pour la Recherche sur le Cancer (DOC20100601010).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206629109/-/DCSupplemental.
References
- 1.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9(6):1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 2.Elsaesser SJ, Goldberg AD, Allis CD. New functions for an old variant: No substitute for histone H3.3. Curr Opin Genet Dev. 2010;20(2):110–117. doi: 10.1016/j.gde.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37(10):1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg AD, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140(5):678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116(1):51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 6.Drané P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24(12):1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loppin B, et al. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437(7063):1386–1390. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- 8.Bonnefoy E, Orsi GA, Couble P, Loppin B. The essential role of Drosophila HIRA for de novo assembly of paternal chromatin at fertilization. PLoS Genet. 2007;3(10):1991–2006. doi: 10.1371/journal.pgen.0030182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneiderman JI, Sakai A, Goldstein S, Ahmad K. The XNP remodeler targets dynamic chromatin in Drosophila. Proc Natl Acad Sci USA. 2009;106(34):14472–14477. doi: 10.1073/pnas.0905816106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong LH, et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20(3):351–360. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakai A, Schwartz BE, Goldstein S, Ahmad K. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr Biol. 2009;19(21):1816–1820. doi: 10.1016/j.cub.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134(1):74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz BE, Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 2005;19(7):804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDowell TL, et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc Natl Acad Sci USA. 1999;96(24):13983–13988. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campos EI, et al. The program for processing newly synthesized histones H3.1 and H4. Nat Struct Mol Biol. 2010;17(11):1343–1351. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardoso C, et al. ATR-X mutations cause impaired nuclear location and altered DNA binding properties of the XNP/ATR-X protein. J Med Genet. 2000;37(10):746–751. doi: 10.1136/jmg.37.10.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Argentaro A, et al. Structural consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc Natl Acad Sci USA. 2007;104(29):11939–11944. doi: 10.1073/pnas.0704057104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otani J, et al. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10(11):1235–1241. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukashchuk V, McFarlane S, Everett RD, Preston CM. Human cytomegalovirus protein pp71 displaces the chromatin-associated factor ATRX from nuclear domain 10 at early stages of infection. J Virol. 2008;82(24):12543–12554. doi: 10.1128/JVI.01215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ray-Gallet D, et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell. 2011;44(6):928–941. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Gong WJ, Golic KG. Genomic deletions of the Drosophila melanogaster Hsp70 genes. Genetics. 2004;168(3):1467–1476. doi: 10.1534/genetics.104.030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Umemori M, et al. RNAi-mediated knockdown showing impaired cell survival in Drosophila wing imaginal disc. Gene Regul Syst Bio. 2009;3:11–20. doi: 10.4137/grsb.s2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218(4570):348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 24.Tyler JK, et al. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol Cell Biol. 2001;21(19):6574–6584. doi: 10.1128/MCB.21.19.6574-6584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corona DF, et al. ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS Biol. 2007;5(9):e232. doi: 10.1371/journal.pbio.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23(21):7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.