Abstract
Chlamydia trachomatis is among the most clinically significant human pathogens, yet their obligate intracellular nature places severe restrictions upon research. Chlamydiae undergo a biphasic developmental cycle characterized by an infectious cell type known as an elementary body (EB) and an intracellular replicative form called a reticulate body (RB). EBs have historically been described as metabolically dormant. A cell-free (axenic) culture system was developed, which showed high levels of metabolic and biosynthetic activity from both EBs and RBs, although the requirements differed for each. EBs preferentially used glucose-6-phosphate as an energy source, whereas RBs required ATP. Both developmental forms showed increased activity when incubated under microaerobic conditions. Incorporation of isotopically labeled amino acids into proteins from both developmental forms indicated unique expression profiles, which were confirmed by genome-wide transcriptional analysis. The described axenic culture system will greatly enhance biochemical and physiological analyses of chlamydiae.
Keywords: metabolism, transcription, intracellular parasite
The bacterial obligate intracellular pathogen, Chlamydia trachomatis, comprises several different serological variants or serovars that cause distinctly different diseases in humans. Trachoma, the leading cause of infectious blindness, remains a serious problem in developing countries (1). C. trachomatis is also among the most common sexually transmitted pathogens (2). A rise in the number of infections in recent years and emergence of new clinical variants (3) illustrates the need for improved understanding of C. trachomatis physiology and virulence.
C. trachomatis undergoes an intracellular developmental cycle that involves a transition from the infectious elementary body (EB) to a vegetative reticulate body (RB) (4). Following replication, reversion to progeny EBs occurs asynchronously before release and reinfection. Although axenic metabolic activity and protein synthesis have been established for RBs (5–7), only limited activity has been described for EBs. Thus, EBs are frequently characterized as metabolically dormant (8–10).
With a genome of only 1.04 Mb (8), C. trachomatis metabolic activity is dependent on scavenging of many metabolic intermediates from the host cell. Indeed, analysis of the C. trachomatis genomic sequence has revealed that the organism is devoid of several enzymes and even entire metabolic pathways (8), underscoring the adaptations of C. trachomatis to intracellular parasitism. During infection of cultured host cells, C. trachomatis scavenges nutrients, including nucleotides (11), amino acids (12), and lipids (13–19). Here, we describe a host cell-free (axenic) medium for C. trachomatis and compare directly the energy source requirements and biosynthetic capacity of EBs and RBs.
Results
Characterization of C. trachomatis EB and RB Populations.
C. trachomatis EBs and RBs were purified from infected HeLa cells by density gradient centrifugation (20). Bacterial stocks were screened by comparison of inclusion-forming unit (IFU) titer and direct particle count (Table 1). Only preparations with an IFU/particle ratio of 0.9 or higher, indicating >90% viability, were used in experiments of EB metabolic activity. Transmission electron microscopy (TEM) was used to verify the ultrastructural characteristics of enriched EBs and RBs (Fig. S1A). TEM showed a uniform population of typical EBs (21), characterized by small size (∼0.3-μm diameter) and condensed chromatin. Conversely, the enriched RB population comprised larger cells (∼1.0-μm diameter) with relaxed chromatin (21). Consistent with the ultrastructural analysis, silver staining of bacterial lysates separated by SDS/PAGE revealed distinct protein profiles (Fig. S1B).
Table 1.
Infectivity per particle of enriched C. trachomatis EB preparations
| Stock no. | DPC | IFU titer | Infectivity, % |
| 1 | 3.78 × 1010 ± 1.81 × 109 | 3.47 × 1010 ± 6.98 × 108 | 91.8 |
| 2 | 2.53 × 1010 ± 1.09 × 109 | 1.63 × 1010 ± 6.21 × 108 | 64.4 |
| 3 | 3.35 × 1010 ± 1.16 × 109 | 2.76 × 1010 ± 1.82 × 109 | 82.4 |
| 4 | 1.82 × 1010 ± 7.45 × 108 | 1.74 × 1010 ± 1.39 × 109 | 95.6 |
Numbers represent the mean ± SEM of at least two independent experiments performed in duplicate. Percentage infectivity was calculated from the mean. DPC, direct particle count; IFU, inclusion-forming unit.
Because EBs and RBs differ in their ability to withstand osmotic stress (21), their structural integrity was compared by exposing enriched EBs and RBs to brief incubation in isotonic buffer or deionized water (dH2O) (Fig. S1C). Incubation of EBs and RBs in dH2O showed significantly more lysis of RBs than EBs.
Correlations between direct particle counts, optical densities, and protein content were established (Fig. S2). EB and RB suspensions normalized to an equivalent OD600 contained fourfold to fivefold more EBs than RBs, consistent with the smaller size of the EB (Fig. S2A). Populations of EBs and RBs were also normalized according to total protein. The RB protein density (total protein/particle) was found to be ∼10-fold higher than that of EBs (Fig. S2B).
Design of a Metabolically Permissive Nutrient Medium.
The chlamydial inclusion membrane is permeable to ions but not reporters >500 Da (22). Accordingly, the concentrations of potassium (K+) and sodium (Na+) in the inclusion lumen mimic that of the host cytosol (23). Therefore, a buffer referred to as intracellular phosphate buffer (IPB) (5 mM KH2PO4, 10 mM Na2HPO4⋅7H2O, 109.6 mM K-gluconate, 8 mM KCl, 1 mM MgCl2⋅6H2O) was designed that contains K+, Na+, and chloride (Cl−) concentrations reflecting those found in the eukaryotic cytoplasm.
[35S]Cys-Met incorporation by scintillation counting and autoradiography during incubation of enriched EBs in IPB supplemented with different nutrients was used as the basis for medium development. Dose–response analyses were carried out for individual nutrients. The additive effects of medium components with or without a carbon or energy source are shown in Fig. S3. Incubation of bacteria in IPB alone or supplemented with 1% FBS and 25 µM amino acids (except cysteine and methionine, which were present at 1 µM) resulted in minimal incorporation. However, addition of 0.5 mM glucose 6-phosphate (G6P), a predicted chlamydial primary carbon and energy source (24), produced a 16-fold increase in protein synthesis.
The structural integrity of EBs is conferred by a disulfide cross-linked outer membrane protein complex that can be relaxed by reducing agents such as DTT (21). Reduction of EB disulfide bonds promotes axenic metabolic activity (21, 25). EBs were incubated in IPB containing amino acids, FBS, and G6P, and supplemented with increasing concentrations of DTT (Fig. S3 C and D). A dose-dependent increase in incorporation was observed with a maximal 5.6-fold increase over controls.
Carbon and Energy Source Utilization by C. trachomatis in Axenic Medium.
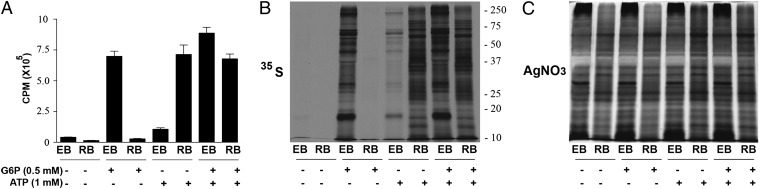
C. trachomatis EBs were incubated in medium containing 0, 0.1, 0.5, and 1 mM G6P, and ATP pools were measured (Fig. S4). Dose-dependent increases in bacterial [35S]Cys-Met incorporation and ATP pools indicate that EBs use G6P for production of ATP to drive bacterial de novo protein synthesis. However, RBs did not incorporate [35S]Cys-Met in the presence of G6P. Based on the acquisition of ATP by Chlamydia psittaci RBs, but not EBs (5), the possibility that EBs and RBs rely on different energy sources was assessed. Indeed, incubation of RBs in medium containing a concentration gradient of ATP showed dose-dependent [35S]Cys-Met incorporation (Fig. S4). Direct comparisons of EB and RB protein synthesis in media supplemented with optimal concentrations of G6P, ATP, or both, revealed differential energy source utilization between the cell forms (Fig. 1). EBs responded principally to G6P, but RBs responded exclusively to ATP and showed no additive effect with both substrates present.
Fig. 1.
Differential energy source utilization by EBs and RBs. Purified EBs and RBs were incubated in medium containing G6P, ATP, or both. (A) Scintillation counting and (B) autoradiography of bacterial lysates show differential energy source utilization by the two cell forms. (C) Silver staining shows equal loading of samples with distinct protein profiles. Data are expressed as the mean (n = 4) ± SEM. Background signal from heat-inactivated organisms was subtracted from all samples. An autoradiograph from a representative experiment is shown. Molecular weight markers are in kilodaltons.
Comparison of C. trachomatis EB and RB Protein Synthesis in Axenic Media.
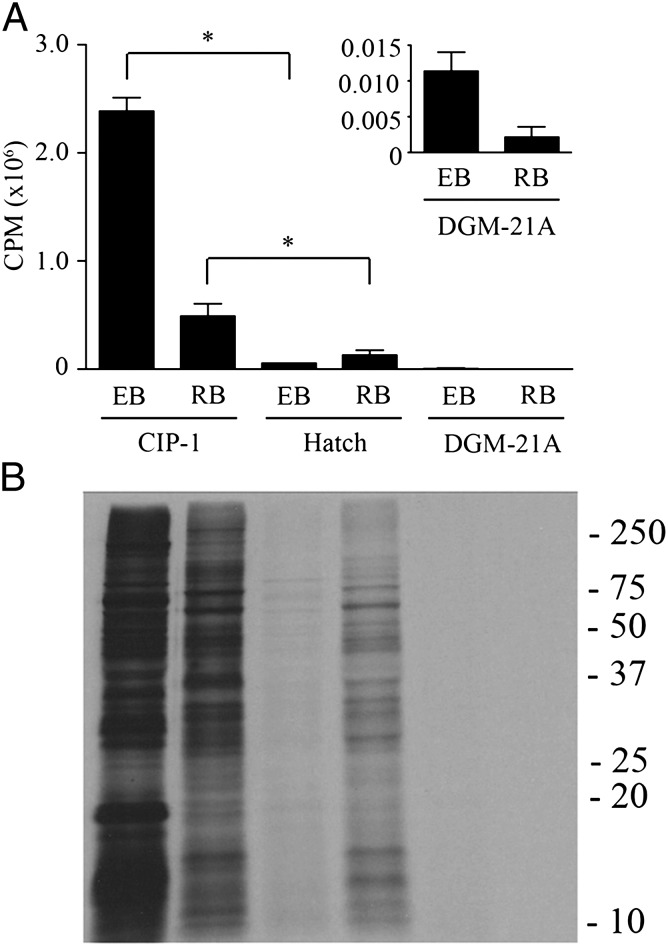
IPB supplemented with 1% FBS, 25 µM amino acids, 0.5 mM G6P, 1.0 mM ATP, 0.5 mM DTT, and 50 µM GTP, UTP, and CTP is referred to as Chlamydia intracellular phosphate-1 (CIP-1) medium (Table S1). Hatch et al. (6) demonstrated chlamydial axenic metabolic activity in a Tris⋅HCl-based medium containing amino acids, molecular ATP, as well as an ATP-regenerating system. Recently, Haider et al. (10) reported metabolic activity by C. trachomatis in DGM-21A medium. The ability of CIP-1 to support C. trachomatis protein synthesis was compared with that of Hatch medium and DGM-21A (Fig. 2). As reported (6), Hatch medium supported RB, but negligible EB protein synthesis. DGM-21A supported marginal incorporation by either RBs or EBs. Overall, C. trachomatis EB and RB protein synthesis was higher in CIP-1 compared with alternative media used previously. This protein synthetic activity in CIP-1 plateaued after 6 h for RBs but remained active for up to 12 h for EBs (Fig. S5).
Fig. 2.
Comparison of EB and RB protein synthesis in axenic media. Protein synthesis by EBs and RBs normalized to total protein content was assessed in CIP-1 and two previously described media by measuring incorporation of [35S]Cys-Met into bacterial proteins after 6 h of incubation. (A) Scintillation count analysis showed strong incorporation of radiolabel in both EBs and RBs when incubated in CIP-1 medium, and moderate radiolabel incorporation by RBs, but not EBs when incubated in Hatch medium. Medium DGM-21A supported negligible levels of radiolabel incorporation by either EBs and RBs. (Inset) Counts per minute detected in EB and RB lysates after incubation in DGM-21A on adjusted scale. Data are expressed as the mean (n = 4) ± SEM. The asterisk indicates statistical significance (P < 0.05). Background signal measured from incubation with heat-inactivated organisms was subtracted from all samples. (B) SDS/PAGE and autoradiography of bacterial lysates confirmed incorporation into bacterial proteins. An autoradiograph from a representative experiment is shown. Molecular weight markers are in kilodaltons.
C. trachomatis Protein Synthesis in CIP-1 Is Sensitive to Translational and Transcriptional Inhibitors.
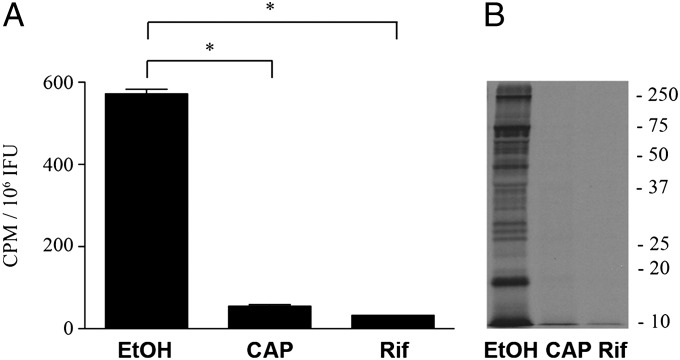
C. trachomatis EBs are known to contain mRNA transcripts, or carry over RNA, entrapped during differentiation of RBs back to EBs (4, 26). To assess the requirements for protein synthesis, EBs were incubated in the presence or absence of chloramphenicol or rifamycin, and [35S]Cys-Met incorporation was determined (Fig. 3). Only background levels of incorporation were observed in the presence of either antibiotic, indicating that axenic protein synthesis by C. trachomatis EBs is dependent on de novo transcription. Because transcriptional activity is necessary for chlamydial protein synthesis, we examined the effects of supplementation with the ribonucleotides GTP, UTP, and CTP (Fig. S6). EBs and RBs responded to NTP supplementation in a dose-dependent manner.
Fig. 3.
Pharmacologic inhibition of chlamydial axenic protein synthesis. C. trachomatis de novo translation and transcription was assessed by analyzing incorporation of [35S]Cys-Met after incubation of EBs in medium containing ethanol solvent, or medium supplemented with 10 µg/mL chloramphenicol or rifampicin. (A and B) Data are expressed as the mean (n = 6) ± SEM. The asterisk indicates statistical significance (P < 0.05). Background signal measured from heat-inactivated organisms was subtracted from all samples. A representative autoradiograph is shown. Molecular weight markers are in kilodaltons.
Microaerobic Conditions Enhance C. trachomatis Metabolic Activity in Axenic Medium.
Genome sequence analysis indicates that cytochrome bd, a respiratory chain terminal oxidase with high affinity for oxygen, is encoded by C. trachomatis. Because cytochrome bd is associated with bacterial microaerobic metabolism, the effect of reduced oxygen tension on C. trachomatis metabolic activity was tested. EBs were preincubated for 24 h in CIP-1 under 20, 10, 2.5% oxygen tensions or under anaerobic conditions. Anaerobic conditions for 24 h resulted in loss of EB metabolic activity. However, increases of 1.6- and 2.3-fold in [35S]Cys-Met incorporation were observed for EBs incubated in 10 or 2.5% oxygen compared with organisms incubated in 20% oxygen, respectively (Fig. S7). RB metabolic activity was similarly optimal under microaerobic conditions (Fig. S7).
Cell Type-Specific C. trachomatis Transcriptional Activity During Axenic Incubation.
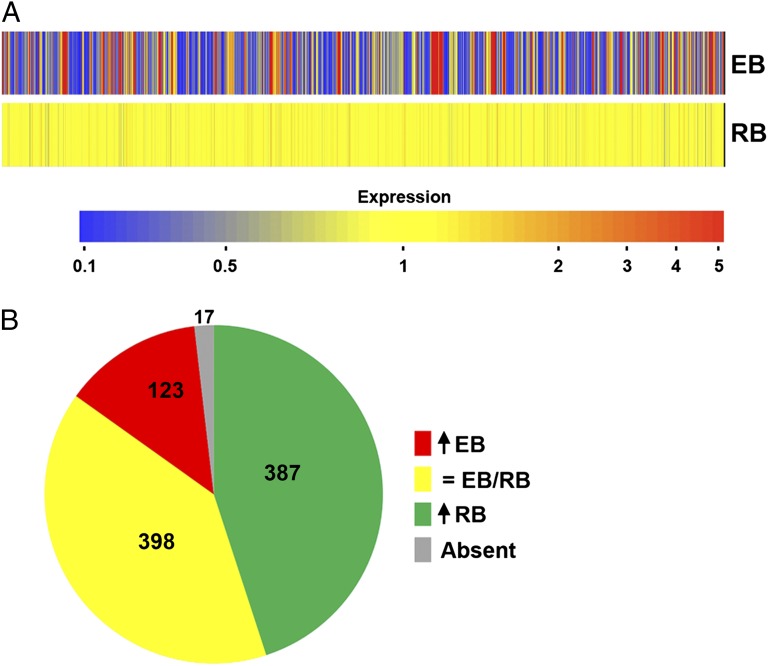
C. trachomatis undergoes a programmed sequence of temporal gene expression as they progress through their developmental cycle (4, 26). To directly test the extent of C. trachomatis developmental gene regulation during axenic metabolism, genome-wide transcription microarray analysis was conducted on EBs and RBs incubated in CIP-1 for 3 h in a 2.5% oxygen environment. Incubation in the presence of rifampicin was used to confirm de novo transcription during incubation in CIP-1. Of 925 probe sets corresponding to the C. trachomatis chromosome, averages of 655 and 893 transcripts were detected for EBs and RBs, respectively. Of these, 123 were specific (greater than threefold up-regulation) to EBs and 387 specific to RBs (Fig. 4). Between the two cell types, >90% of the entire transcriptome was detected. As shown in Table S2, EBs and RBs maintain numerous developmental stage-specific transcriptional signatures during axenic metabolism. The up-regulation of chlamydial transcription further confirms that the activity observed is attributable to chlamydiae and not contaminating host organelles in these highly purified preparations. The complete data set is available at www.ncbi.nlm.nih.gov/geo (accession no. GSE39530).
Fig. 4.
Developmental stage-specific transcription of purified EBs and RBs. EBs and RBs were incubated in CIP-1 for 3 h under 2.5% oxygen and RNA extracted for microarray analysis. (A) Heat map showing axenic EB gene expression relative to axenic RBs. (B) Number of genes up-regulated more than threefold in EBs, up-regulated more than threefold in RBs, or expressed at relatively equivalent levels in both. Seventeen transcripts were not detected.
Discussion
Here, we describe the development of a medium that supports axenic metabolic activity of chlamydial EBs and RBs as measured by de novo protein and ATP synthesis. EBs and RBs showed unique preferences for energy sources with EBs requiring G6P and RBs showing enhanced activity in the presence of ATP. Isotopic labeling of de novo-synthesized proteins in CIP-1 demonstrated EB- and RB-specific profiles that were dependent on chlamydial transcriptional activity. Developmental form-specific transcription in CIP-1 was confirmed by microarray analysis. Overall, C. trachomatis de novo protein synthesis in CIP-1 shows a substantial improvement over previously described media (6, 10, 27) and will facilitate analysis of chlamydial physiology and metabolism in the absence of the background of a host cell.
Access to a suitable carbon and/or energy source is critically important to generate metabolic intermediates and fuel cellular reactions. Both experimental (7, 9) and bioinformatic (8) analyses suggest C. trachomatis can use G6P. With the exception of the first enzyme of glycolysis, hexokinase, C. trachomatis has a full complement of glycolytic enzymes. Additionally, C. trachomatis appears to have a functional pentose phosphate pathway (8). Although G6P can be oxidized via either pathway, only oxidation via the glycolytic pathway would result in ATP production. Notably, in CIP-1, EBs and RBs show differential ability to use extracellular G6P and ATP as energy sources for de novo protein synthesis. Overall, this physiological profile indicates that EBs primarily rely on substrate level and oxidative phosphorylation to meet cellular energy demands, whereas RBs primarily scavenge ATP.
Reduced oxygen tension is critical for axenic replication of the obligate intracellular parasite Coxiella burnetii (28). Similarly, C. trachomatis encodes cytochrome bd, a terminal oxidase with increased affinity for oxygen and thus associated with metabolic activity under reduced oxygen conditions. Maintenance of EB and RB metabolic activity was significantly improved when C. trachomatis was incubated in a reduced oxygen environment.
Chlamydial EBs are adapted for extracellular survival and primed for infection of susceptible host cells. The dogma has been that EBs are relatively metabolically dormant. Although the data herein do not speak to the activity of EBs in their natural environment, they demonstrate that EBs have the capacity for considerable metabolic and biosynthetic activity under the appropriate conditions. It is unclear what specific signal(s) trigger EB germination but the availability of glucose-6-phosphate in CIP-1 stimulates both ATP production and protein synthesis and is accompanied by a transcriptional up-regulation of many previously identified early transcripts. Whether EBs can metabolize to maintain ATP stores and viability in more natural extracellular environments, such as mucosal surfaces, remains to be determined.
Chlamydiae have been proposed to undergo a temporal program of gene expression consisting of early, midcycle, and late genes (4). Among the early genes are many of those associated with inclusion development to create an appropriate intracellular niche. Late genes include those involved in the secondary differentiation of RBs back to EBs such as the histone-like proteins involved in DNA condensation and the cysteine-rich proteins that stabilize the cell wall outer membrane complex. Host-free RNA synthesis has been observed from freshly purified chlamydial developmental forms isolated at 1 or 24 h postinfection (29). The transcripts from 1- and 24-h specimens hybridized to unique genomic profiles, although with the exception of the gene encoding MOMP, the genes transcribed were not identified. Genes expressed by EBs in CIP-1 include many expected during early transcription, such as those encoding inclusion membrane proteins (Incs). A few classical late genes such as those encoding the cysteine-rich outer membrane complex proteins, OmcA and B and the histone-like protein, Hc2, were also up-regulated in EBs. Another prominent late gene, encoding Hc1, was not up-regulated. Thus, although the transcriptional profiles of EBs and RBs were unique, they did not entirely reflect the in vivo situation. The potential for recognizing additional developmental regulatory signals is enhanced by the availability of an axenic medium supporting transcription and translation. Whether shifts in developmental stage-specific gene expression profiles occur or can be induced during axenic culture will be a prominent goal of future studies.
The findings presented herein will facilitate the study of chlamydial metabolism and physiology under controlled nutritional and physiochemical conditions. Moreover, robust axenic metabolic activity will complement current research tools used to study chlamydiae, including the recent development of basic genetic tools (30, 31). The identification of metabolic activity by C. trachomatis EBs challenges the categorization of the chlamydial EB as metabolically inert/dormant. As such, EBs may be best characterized as conditionally active. CIP-1 can be used as a foundation for further studies on C. trachomatis axenic metabolic activity and serves as a basis for exploration of additional factors that may stimulate differentiation and eventually replication.
Materials and Methods
Propagation and Purification of C. trachomatis.
C. trachomatis serovar L2 (LGV 434) was propagated in HeLa 229 cells and purified by Renografin density gradient centrifugation as described (20). RBs were isolated 14–16 h postinfection, whereas EBs were isolated 43–45 h postinfection. IFUs were determined as previously described (32) using indirect immunofluorescence with a rabbit polyclonal anti-C. trachomatis L2 EB antibody followed by an anti-rabbit IgG Alexa Fluor 488-conjugated secondary antibody (Invitrogen).
TEM.
Ultrastructural analysis of C. trachomatis EBs and RBs were analyzed by TEM as described (33). Micrographs were captured using a Hitachi H-7500 electron microscope (Hitachi) equipped with a Hamamatsu XR-100 digital camera system (AMT).
Normalization of EB and RB Populations.
EBs and RBs were fixed [2.5% (vol/vol) Glut/0.05 M sucrose in 0.1 M NaCl, pH 6.8], and cellular densities were adjusted to OD600 of 0.5, 0.25, and 0.1. Direct particle counts were determined as previously described (34). For measurement of bacterial total protein content, bacterial suspensions were lysed in 1% (wt/vol) SDS for 5 min at 100 °C, and total protein was determined using a Bio-Rad Dc Protein Assay kit (Bio-Rad).
C. trachomatis Osmotic Stability.
Differential sensitivity of enriched C. trachomatis EBs and RBs to hypoosmotic stress was determined as previously described (21) by suspension in K-36 buffer (0.05 M K2HPO4/KH2PO4, 0.1 M KCl, 0.15 M NaCl, pH 7.0) or dH2O for 5 min. The OD600 was then determined.
Protein Synthesis by [35S]Cysteine–Methionine Incorporation.
C. trachomatis protein synthesis was measured by incorporation of [35S]Cys-Met (MP Biomedicals). Chlamydiae were incubated for 6 h under the indicated conditions in microcentrifuge tubes containing 500 µL of medium supplemented with 50–100 µCi of [35S]Cys-Met. Organisms were then pelleted, washed with K-36, and lysed in Laemmli sample buffer at 100 °C for 5 min. Equal volumes of lysate were used for quantification of [35S]Cys-Met incorporation by scintillation counting. For qualitative analysis of radiolabel incorporation into bacterial proteins, equal volumes of sample lysate were separated by SDS/PAGE and proteins were visualized by autoradiography by standard methods as described (35). Silver staining of SDS/PAGE gels was done using a SilverQuest kit (Invitrogen) according to the manufacturer’s recommendations. For direct comparison of EB and RB metabolic activity, bacterial inocula were normalized according to total protein content. Incubations of bacteria in microaerobic or anaerobic environments were performed using Innova CO-48 (New Brunswick Scientific) and MCO-5M (Sanyo) incubators, or a GasPak jar system (BD Biosciences), respectively.
Measurement of Chlamydial ATP Pool.
ATP pools were measured using an ATP Determination Kit (Invitrogen) according to the manufacturer's instructions. Luminescence was measured using a Synergy-4 Multi-Mode Microplate Reader (BioTek Instruments). ATP content was determined by comparison with a standard curve.
Transcription Microarray Analysis.
Density gradient-purified EBs and RBs were incubated in quadruplicate in four-well plates containing 500 μL of CIP-1 for 3 h under 2.5% oxygen in the presence or absence of rifamycin (10 µg/mL). Following incubation, bacteria were collected by centrifugation. Bacterial RNA was stabilized immediately by addition of 500 µL of RLT lysis buffer (Qiagen) and samples were frozen at −80 °C. RNA was extracted using a RNeasy mini kit according to manufacturer’s recommendations (Qiagen). DNase treatment of bacterial RNA was performed in 100-μL reactions using 20 U of RNAase-free DNase I (New England Biolabs) per reaction. cDNA synthesis and fragmentation, and 3′-terminal labeling were performed according to the Affymetrix instructions for “Prokaryotic Target Preparation” (Affymetrix). The quality of the RNA starting material, cDNA, and the fragmentation efficiency were monitored using a 2100 Bioanalyzer (Agilent Technologies). Labeled cDNA target was hybridized to custom Affymetrix GeneChip microarrays containing 926 C. trachomatis probe sets according to standard procedures essentially as described (36). Chips were scanned using a 7Gplus GeneChip scanner (Affymetrix), and data were analyzed using GeneSpring GX 7.3 (Agilent Technologies) and Partek Genomics Suite software (Partek). Additional experimental details are provided at www.ncbi.nlm.nih.gov/geo (accession no. GSE39530).
Supplementary Material
Acknowledgments
We thank A. Whitney and F. DeLeo for advice and assistance with RNA processing and K. Virtaneva, K. Kanakabandi, and S. F. Porcella for technical help with optimization of sample preparation for the transcription microarray analysis. Additional electron microscopy by E. R. Fischer and B. Hansen is also appreciated. This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE39530).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212831109/-/DCSupplemental.
References
- 1.Burton MJ, Mabey DC. The global burden of trachoma: A review. PLoS Negl Trop Dis. 2009;3:e460. doi: 10.1371/journal.pntd.0000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: Implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol. 2005;5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- 3.Unemo M, Clarke IN. The Swedish new variant of Chlamydia trachomatis. Curr Opin Infect Dis. 2011;24:62–69. doi: 10.1097/QCO.0b013e32834204d5. [DOI] [PubMed] [Google Scholar]
- 4.Shaw EI, et al. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol. 2000;37(4):913–925. doi: 10.1046/j.1365-2958.2000.02057.x. [DOI] [PubMed] [Google Scholar]
- 5.Hatch TP, Al-Hossainy E, Silverman JA. Adenine nucleotide and lysine transport in Chlamydia psittaci. J Bacteriol. 1982;150:662–670. doi: 10.1128/jb.150.2.662-670.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatch TP, Miceli M, Silverman JA. Synthesis of protein in host-free reticulate bodies of Chlamydia psittaci and Chlamydia trachomatis. J Bacteriol. 1985;162:938–942. doi: 10.1128/jb.162.3.938-942.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss E, Wilson NN. Role of exogenous adenosine triphosphate in catabolic and synthetic activities of Chlamydia psittaci. J Bacteriol. 1969;97:719–724. doi: 10.1128/jb.97.2.719-724.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stephens RS, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 9.Iliffe-Lee ER, McClarty G. Glucose metabolism in Chlamydia trachomatis: The “energy parasite” hypothesis revisited. Mol Microbiol. 1999;33:177–187. doi: 10.1046/j.1365-2958.1999.01464.x. [DOI] [PubMed] [Google Scholar]
- 10.Haider S, et al. Raman microspectroscopy reveals long-term extracellular activity of Chlamydiae. Mol Microbiol. 2010;77:687–700. doi: 10.1111/j.1365-2958.2010.07241.x. [DOI] [PubMed] [Google Scholar]
- 11.Tipples G, McClarty G. The obligate intracellular bacterium Chlamydia trachomatis is auxotrophic for three of the four ribonucleoside triphosphates. Mol Microbiol. 1993;8:1105–1114. doi: 10.1111/j.1365-2958.1993.tb01655.x. [DOI] [PubMed] [Google Scholar]
- 12.Hatch TP. Competition between Chlamydia psittaci and L cells for host isoleucine pools: A limiting factor in chlamydial multiplication. Infect Immun. 1975;12:211–220. doi: 10.1128/iai.12.1.211-220.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derre I, Swiss R, Agaisse H. The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Pathog. 2011;7:e1002092. doi: 10.1371/journal.ppat.1002092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wylie JL, Hatch GM, McClarty G. Host cell phospholipids are trafficked to and then modified by Chlamydia trachomatis. J Bacteriol. 1997;179:7233–7242. doi: 10.1128/jb.179.23.7233-7242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cocchiaro J, Kumar Y, Fischer ER, Hackstadt T, Valdivia RH. Cytoplasmic lipid droplets are translocatedd into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc Natl Acad Sci USA. 2008;105(27):9379–9384. doi: 10.1073/pnas.0712241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beatty WL. Trafficking from CD63-positive late endocytic multivesicular bodies is essential for intracellular development of Chlamydia trachomatis. J Cell Sci. 2006;119:350–359. doi: 10.1242/jcs.02733. [DOI] [PubMed] [Google Scholar]
- 17.Hackstadt T, Scidmore MA, Rockey DD. Lipid metabolism in Chlamydia trachomatis-infected cells:directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc Natl Acad Sci USA. 1995;92:4877–4881. doi: 10.1073/pnas.92.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carabeo RA, Mead DJ, Hackstadt T. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc Natl Acad Sci USA. 2003;100:6771–6776. doi: 10.1073/pnas.1131289100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elwell CA, et al. Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development. PLoS Pathog. 2011;7:e1002198. doi: 10.1371/journal.ppat.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caldwell HD, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackstadt T, Todd WJ, Caldwell HD. Disulfide-mediated interactions of the chlamydial major outer membrane protein: Role in the differentiation of chlamydiae? J Bacteriol. 1985;161:25–31. doi: 10.1128/jb.161.1.25-31.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinzen RA, Hackstadt T. The Chlamydia trachomatis parasitophorous vacuolar membrane is not passively permeable to low-molecular-weight compounds. Infect Immun. 1997;65:1088–1094. doi: 10.1128/iai.65.3.1088-1094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grieshaber S, Swanson JA, Hackstadt T. Determination of the physical environment within the Chlamydia trachomatis inclusion using ion-selective ratiometric probes. Cell Microbiol. 2002;4(5):273–283. doi: 10.1046/j.1462-5822.2002.00191.x. [DOI] [PubMed] [Google Scholar]
- 24.Weiss E. Adenosine triphosphate and other requirements for the utilization of glucose by agents of the psittacosis-trachoma group. J Bacteriol. 1965;90:243–253. doi: 10.1128/jb.90.1.243-253.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bavoil P, Ohlin A, Schachter J. Role of disulfide bonding in outer membrane structure and permeability in Chlamydia trachomatis. Infect Immun. 1984;44:479–485. doi: 10.1128/iai.44.2.479-485.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belland RJ, et al. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci USA. 2003;100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allan I, Hatch TP, Pearce JH. Influence of cysteine deprivation on chlamydial differentiation from reproductive to infective life-cycle forms. J Gen Microbiol. 1985;131:3171–3177. doi: 10.1099/00221287-131-12-3171. [DOI] [PubMed] [Google Scholar]
- 28.Omsland A, et al. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci USA. 2009;106:4430–4434. doi: 10.1073/pnas.0812074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crenshaw RW, Fahr MJ, Wichlan DG, Hatch TP. Developmental cycle-specific host-free RNA synthesis in Chlamydia spp. Infect Immun. 1990;58:3194–3201. doi: 10.1128/iai.58.10.3194-3201.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, et al. Development of a transformation system for Chlamydia trachomatis: Restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 2011;7:e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kari L, et al. Generation of targeted Chlamydia trachomatis null mutants. Proc Natl Acad Sci USA. 2011;108:7189–7193. doi: 10.1073/pnas.1102229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furness G, Graham DM, Reeve P. The titration of trachoma and inclusion blennorrhoea viruses in cell cultures. J Gen Microbiol. 1960;23:613–619. doi: 10.1099/00221287-23-3-613. [DOI] [PubMed] [Google Scholar]
- 33.Omsland A, et al. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl Environ Microbiol. 2011;77:3720–3725. doi: 10.1128/AEM.02826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moos A, Hackstadt T. Comparative virulence of intra- and interstrain lipopolysaccharide variants of Coxiella burnetii in the guinea pig model. Infect Immun. 1987;55:1144–1150. doi: 10.1128/iai.55.5.1144-1150.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Omsland A, Cockrell DC, Fischer ER, Heinzen RA. Sustained axenic metabolic activity by the obligate intracellular bacterium Coxiella burnetii. J Bacteriol. 2008;190:3203–3212. doi: 10.1128/JB.01911-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M, Villaruz AE, Cha DJ, Sturdevant DE, Otto M. Gram-positive three-component antimicrobial peptide-sensing system. Proc Natl Acad Sci USA. 2007;104:9469–9474. doi: 10.1073/pnas.0702159104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.