Abstract
Hydra’s unlimited life span has long attracted attention from natural scientists. The reason for that phenomenon is the indefinite self-renewal capacity of its stem cells. The underlying molecular mechanisms have yet to be explored. Here, by comparing the transcriptomes of Hydra’s stem cells followed by functional analysis using transgenic polyps, we identified the transcription factor forkhead box O (FoxO) as one of the critical drivers of this continuous self-renewal. foxO overexpression increased interstitial stem cell and progenitor cell proliferation and activated stem cell genes in terminally differentiated somatic cells. foxO down-regulation led to an increase in the number of terminally differentiated cells, resulting in a drastically reduced population growth rate. In addition, it caused down-regulation of stem cell genes and antimicrobial peptide (AMP) expression. These findings contribute to a molecular understanding of Hydra’s immortality, indicate an evolutionarily conserved role of FoxO in controlling longevity from Hydra to humans, and have implications for understanding cellular aging.
Keywords: adult stem cell, differentiation
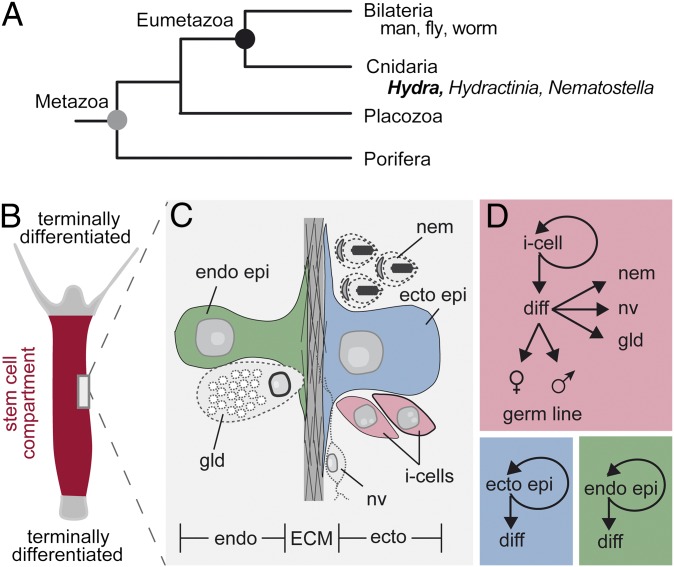
The unlimited life span of Hydra, due to the indefinite self-renewal capacity of its stem cells, has long attracted attention from biologists, as it promises insights into the mechanisms controlling longevity in more advanced animals including humans. Analysis of the mortality patterns and reproductive rates of Hydra vulgaris could not detect any evidence for aging and no apparent signs of decline in reproductive rates (1).Thus, Hydra, a member of the phylogenetically old animal phyla Cnidaria (Fig. 1A), has been suggested to not undergo senescence and to be biologically immortal. In vivo tracking of individual enhanced green fluorescent protein (eGFP)-expressing cells provides experimental proof that both epithelial and interstitial stem cells in Hydra are continuously self-renewing and differentiating cells (2–4). Offspring of individual cells were tracked for several years without obtaining evidence for apparent signs of decline in proliferation rates. The apparent lack of cellular senescence results in an adult polyp with indefinite proliferative life span and adds support to the study by Martinez (1). The biological reason behind that seemingly unique potential to escape mortality and senescence is simple: the animals have adopted a life cycle in which proliferation and population growth occurs exclusively asexually by budding (2). This asexual mode of reproduction not only demands that each individual polyp maintains continuously self-renewing stem cells but also points to a strong selective constraint that equips the adult polyp’s tissue with cells that are capable of continuous self-renewal and differentiation. In Hydra, three stem cell lineages—ectodermal and endodermal epitheliomuscular stem cells and interstitial stem cells—contribute to the continuously self-renewing epithelium (Fig. 1 B–D) (2, 5). The two epitheliomuscular stem cell lineages shape the diploblastic body of the polyps and are responsible for all its morphogenetic properties (6). The multipotent interstitial stem cells located in the ectoderm of the gastric region have a developmental potency much wider than that of epithelial cells by being capable of giving rise not only to a number of somatic cell types but also to gametes (2, 5) (Fig. 1D). Although the mechanisms controlling self-renewal and differentiation of Hydra stem cells could reveal long-sought secrets about longevity, until now such mechanisms have not been identified due to the lack of unique markers and the absence of functional approaches (7).
Fig. 1.
Stem cells in Hydra are continuously self-renewing. (A) Schematic phylogenetic tree showing the main branches in metazoan evolution. (B) Scheme of the stem cell compartment in Hydra with sharp boundaries towards terminally differentiated head and foot tissue. (C) The major cell types in Hydra. Stem cell lineages are colored, with derivatives of the interstitial cell lineage in gray. (D) The three independent stem cell systems in Hydra. Both epithelial cell lineages represent unipotent stem cells whereas interstitial stem cells exhibit multipotent features as they are able to differentiate into various derivatives. diff, differentiation ecto, ectoderm; ECM, extracellular matrix; ecto epi, ectodermal epithelial cell; endo, endoderm; endo epi, endodermal epithelial cell; gld, gland cell; i-cell, interstitial stem cell; nv, nerve cell; nem, nematocyte.
To uncover the regulatory events and signaling pathways that control stem cells in an evolutionarily old animal that branched off as the bilaterian ancestor (Fig. 1A) almost 600 million years ago, we recently separated eGFP-labeled stem cells of all three stem cell lineages by fluorescence-activated cell sorting and addressed the commonalities and differences among the three stem cell lineages by deep RNA sequencing and gene expression studies (8). This first-time characterization of the stem cell transcriptomes in an animal at the base of evolution revealed not only that stem cells in the metazoan ancestors were multifunctional, but also that they bear a defined molecular signature composed of distinct sets of transcription factors, signal transducers, and effector genes (8). In search of transcription factors that are strongly expressed and shared by all three stem cell lineages and therefore that may play a role in the regulation of self-renewal and differentiation in all three stem cell lineages, we discovered FoxO, a member of the forkhead box (Fox) family of proteins (Fig. 2A) (8). Recent work has implicated FoxO in conferring increased life span and stress resistance in flies and worms and identified foxO3a as an important component of the genetic signatures of human exceptional longevity (9–13). In addition, FoxO3a was shown to play a role in maintenance of adult hematopoietic stem cells (14, 15) as well as neural stem cells (16, 17), whereas FoxO1 is important for embryonic stem cell maintenance (18). Thus, we hypothesized that the role of FoxO in controlling stem cell behavior and longevity might be evolutionarily highly conserved. Here we show by gain-of-function and loss-of-function analysis that transcription factor FoxO indeed is a critical component of the mechanisms controlling stem cell behavior in Hydra.
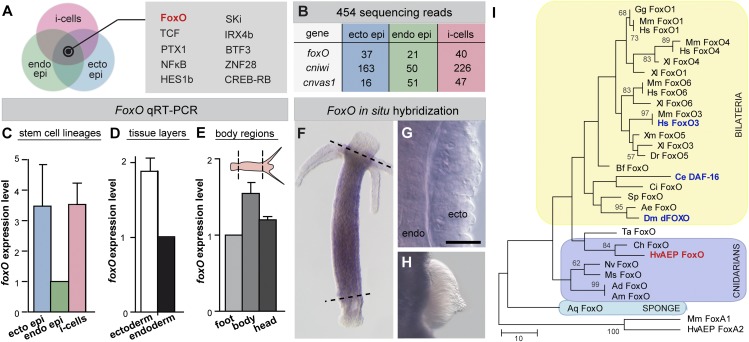
Fig. 2.
foxO is expressed in all three stem cell types in Hydra. (A) Venn diagram of conserved transcription factors being expressed in all three stem cell types according to 454 transcriptome sequencing. (B) Reads from 454 sequencing for foxO, cnvas1, and cniwi. (C–E) qRT-PCR reveals foxO expression in (C) all three stem cell lineages, (D) both tissue layers, and (E) in the body column (n = 2 replicates). Expression in endodermal epithelial cells, endodermal tissue layer, and foot tissue was used as calibrator. (F–H) In situ hybridization shows that foxO is expressed in both tissue layers along the body axis and absent from terminally differentiated cells in head, foot, and (H) gonads. (I) Maximum-parsimony phylogram of the forkhead domain from selected FoxO proteins rooted using Mus musculus FoxA1 and Hydra FoxA2. Numbers at nodes are bootstrap support values calculated by 1,000 replicates of maximum parsimony. Bootstrap values under 50 are not shown. Aa: Aedes aegypti; Ad: Acropora digitifera; Am: Acropora millepora, Aq: Amphimedon queenslandica; Bf: Branchiostoma floridae; Ce: C. elegans; Ch: Clytia hemisphaerica; Ci: Ciona intestinalis; Dm: Drosophila melanogaster; Dr: Danio rerio; Gg: Gallus gallus; Hs: Homo sapiens; HvAEP: H. vulgaris strain AEP; Mm: M. musculus; Ms: Metridium senile; Nv: Nematostella vectensis; Sp: Strongylocentrotus purpuratus; Ta: Trichoplax adhaerens; Xl: Xenopus laevis; and Xm: Xiphophorus maculates. (Scale bar in G: 20 µm.)
Results
foxO Expression Correlates with the Undifferentiated Stem Cell State.
When analyzing the stem cell signatures of the three stem cell lineages in Hydra, foxO emerged as an obvious candidate for controlling stem cell self-renewal as it is highly expressed in all three stem cell lineages (Fig. 2 A and B). To independently confirm foxO expression in all three stem cell lineages, we performed an additional cell-sorting experiment followed by quantitative real-time PCR (qRT-PCR) (Fig. 2C). Further independent support for this conclusion comes from mechanical separation of ectodermal and endodermal tissue followed by qRT-PCR (Fig. 2D) as well as by in situ hybridization (Fig. 2 F and G). Interestingly, foxO is down-regulated upon terminal differentiation in both head and foot tissue (Fig. 2 E and F) as well as in differentiating gametes within gonads (Fig. 2H), suggesting potential FoxO functions in stem cell regulation. Similar to all other invertebrate species, the Hydra magnipapillata genome contains only a single FoxO gene that, according to phylogenetic analyses (Fig. 2I and Fig. S1), groups in a basal position to all bilaterian FoxO genes, including human foxO3a, Caenorhabditis elegans dauer-formation 16 (DAF-16)/foxO, and Drosophila dFOXO.
Overexpression of foxO in Interstitial Stem Cell Lineage Increases Stem Cell and Progenitor Cell Proliferation and Induces Expression of Stemness Genes in Terminally Differentiated Nematocytes.
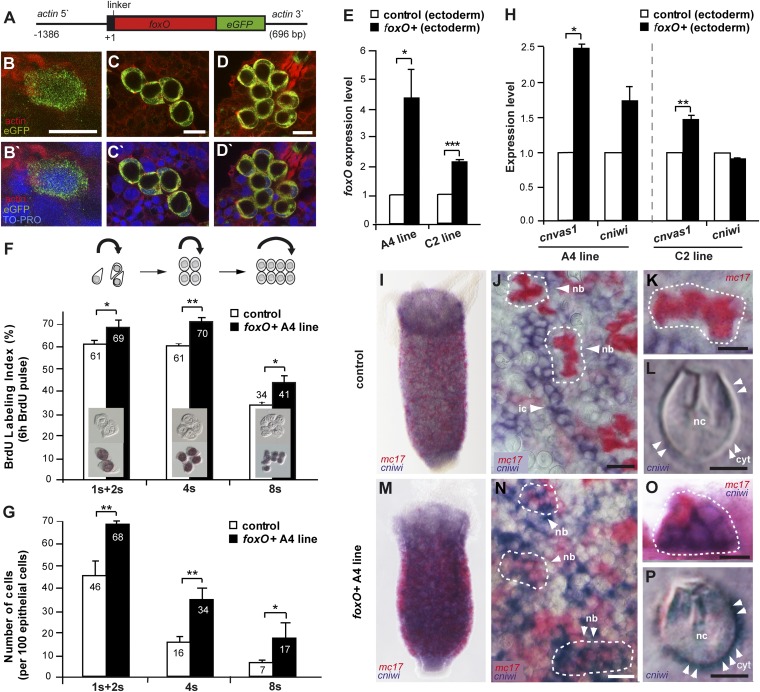
To examine whether FoxO plays essential regulatory roles in the processes that influence interstitial stem cell number and function, we generated two independent lines of polyps containing an actin-driven foxO-eGFP transgene (Fig. 3A) expressed in interstitial stem cells and nematoblast precursors (Fig. 3 B–D). qRT-PCR results confirm that levels of foxO transcripts are significantly elevated in polyps of both foxO-eGFP transgenic lines, compared with eGFP-expressing control polyps (Fig. 3E). Confocal microscopy of polyps overexpressing foxO in the interstitial cell lineage (Fig. 3 B–D) and in ectodermal epithelial cells (Fig. S2) shows that FoxO-eGFP fusion protein is loacalized in both nucleus and cytoplasm of stem cells (Fig. 3B and Fig. S2A). In terminally differentiated cells in tentacles and hypostome, FoxO-eGFP localization is mostly cytoplasmic (Fig. S2 B and C). To assess the role of FoxO in controlling interstitial cell proliferation, we determined the 5-bromo-2′-deoxyuridine (BrdU)-labeling index of interstitial stem cells and nematoblast precursors. Fig. 3F indicates that foxO overexpression in the interstitial cell lineage is accompanied by a significant increase in proliferation of interstitial stem cells and nematoblast precursors, which results in an increased number of interstitial stem cells and nematoblast precursors per polyp (Fig. 3G).
Fig. 3.
foxO overexpression. (A) Construct used for foxO overexpression. (B–D) foxO overexpressing (B) interstitial stem cells and (C and D) groups of nematoblasts. (E) foxO expression levels in ectodermal tissue layer of foxO+ polyps (A4 and C2 line), analyzed by qRT-PCR (n = 3 replicates). Asterisks indicate significant changes in expression levels (t test); P values: A4 line = 0.014, C2 line = 0.0005. (F) BrdU-labeling index of interstitial stem cells (1s + 2s) and nematoblast progenitor cells (4s, 8s) in foxO+ (A4 line) and control polyps after 6 h exposure to BrdU (n1s+2s = ∼1,000, n4s = ∼200, n8s = ∼100 per replicate; n = 3 replicates). Asterisks indicate significant changes in cell numbers (t test); P values: 1s + 2s = 0.0265, 4s = 0.0034, 8s = 0.0136. (G) Numbers of interstitial stem cells (1s + 2s) and nematoblasts (4s, 8s) per 100 epithelial cells in foxO+ (A4 line) and control polyps (nepithelial cells = ∼1,000; n = 3 replicates). Asterisks indicate significant changes in cell numbers (t test); P values: 1s + 2s = 0.0016, 4s = 0.0078, 8s = 0.0474. (H) Expression levels of stem cell genes in ectodermal tissue layer of foxO+ polyps (A4 and C2 line) analyzed by qRT-PCR (n = 3 replicates). Asterisks indicate significant changes in expression levels (t test); P values: cnvas1A4 line = 0.0238, cnvas1C2 line = 0.0031. (I–P) Douple in situ hybridization of cniwi and mc17 in (I–L) control and (M–P) foxO+ (A4 line) polyps. (I and J) Mc17-positive nematoblasts and cniwi-positive interstitial stem cells. (K) Mc17-positive nematoblasts. (L) Cniwi-negative nematocyte. (M–O) Coexpression of mc17 and cniwi in nematoblasts. (P) Cniwi-positive nematocyte. (Scale bars: B–D, K, and O—10 µm; J and N—20 µm; L and P—5 µm.) cyt, cytoplasm; ic, interstitial stem cells; nb, nematoblasts; nc, nematocyst.
As shown previously (19–22), interstitial stem cells express cniwi and cnvas1 whereas nematoblast precursors specifically express the minicollagen mc17. We therefore asked next if foxO overexpression in interstitial cells could affect the expression of stem cell genes. qRT-PCR results show that the level cnvas1 and cniwi expression is substantially increased in polyps of both transgenic lines overexpressing foxO in their interstitial stem cells (Fig. 3H). As assessed by double in situ hybridization in polyps transfected with the control construct, nematocyte precursors express minicollagen mc17 whereas interstitial stem cells express cniwi (Fig. 3 I–K). Mature nematocytes never express cniwi (Fig. 3L). In transgenic polyps overexpressing foxO in the interstitial cell lineage, we observe not only a higher density of nematoblast progenitor cells (Fig. 3M), but also a large number of nematoblast precursors expressing both cniwi and mc17 simultaneously (Fig. 3 M–O). Most strikingly, cniwi transcripts can even be found in terminally differentiated nematocytes such as stenoteles or isorhizas in both transgenic lines (Figs. 3P and Fig. S3). Thus, overexpression of foxO appears to induce expression of stem cell genes in terminally differentiated somatic cells. This finding resembles earlier observations in C. elegans that ectopic expression of DAF-16/foxO in somatic tissue also induces the ectopic expression of germ-line genes (23) and may indicate that foxO overexpression transfers stem cell character to terminally differentiated cells in the interstitial cell lineage.
FoxO Silencing in Epithelial Cells Results in Enhanced Terminal Differentiation and a Slow Growth Phenotype.
To further investigate functions of FoxO in Hydra stem cells, we knocked down its expression in ectodermal and endodermal epithelial cells using a stable hairpin transcription approach (Fig. 4A). We have analyzed six independent transgenic lines (three endodermal, two ectodermal, and one ecto- and endodermal). Levels of foxO transcript are significantly and specifically reduced in all lines as assessed by qRT-PCR (Fig. 4B and Fig. S4). foxO silencing is associated with a severe enlargement of the foot and stalk region, as assessed by in situ staining with the stalk-specific Pedibin gene (24) (Fig. 4 C–F and Fig. S5A). By examining the expression patterns of endodermal transcription factors known to be important for foot formation, we observed that both nk2 homeobox (nk2)2 and erythroblast transformation-specific 2 (ets2) are strongly expressed in the enlarged stalk structures (Fig. S5B). Measuring of the expression field of pedibin revealed (Fig. 4G) that all six transgenic lines have an increased number of pedibin-expressing cells, suggesting that silencing FoxO function results in an increase of terminally differentiated foot cells. The finding that mc17-expressing progenitor cells, which normally are never present in foot tissue, are absent from the enlarged stalks of transgenic polyps (Fig. S5B) supports this view. foxO silencing and the associated increase in terminally differentiated foot cells are accompanied by defects in growth rate. Quantification of growth rate under standard culture conditions shows that polyps with foxO-deficient endoderm have a reduced population growth rate, with a doubling time of 6 d compared with 4 d for the control transgenic line (Fig. 4H) due to a 2-d prolonged bud attachment to the mother polyp (Fig. 4I).
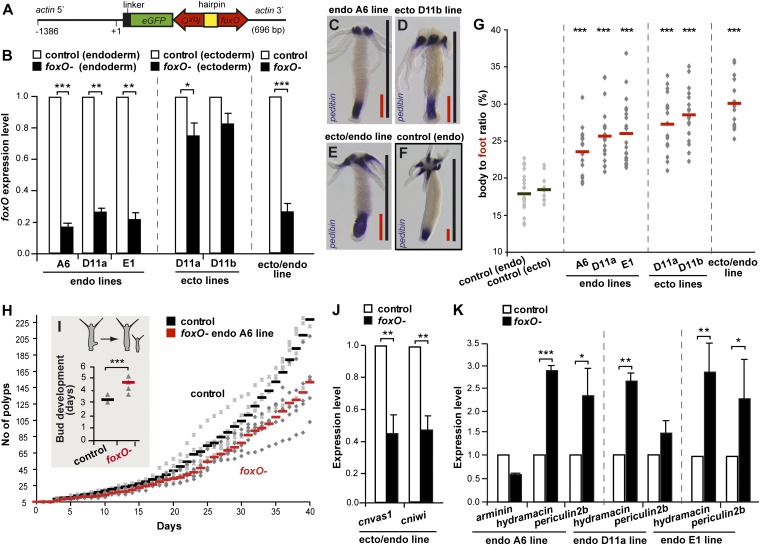
Fig. 4.
foxO down-regulation. (A) Construct used for foxO down-regulation. (B) foxO expression levels in endodermal tissue layer of foxO− endo polyps, ectodermal tissue layer of foxO− ecto polyps, and total tissue of foxO− ecto/endo polyps, analyzed by qRT-PCR (n = 3 replicates). Asterisks indicate significant changes in expression levels (t test); P values: endo A6 line = <0.0001, endo D11a line = 0.0087, endo E1 line = 0.0032, ecto D11a line = 0.0123, and ecto/endo line = 0.0008. (C–F) In situ hybridization of pedibin in FoxO− endo, foxO− ecto, foxO− ecto/endo and control polyps. (G) Foot length compared with body length in foxO− and control polyps, as determined by pedibin labeling (ncontrol endo = 21, ncontrol ecto = 12, nendo A6 line = 17, nendo D11a line + nendo E1 line = 22, necto D11a line + necto D11b line = 18, and necto/endo line = 15). Asterisks indicate significant differences in foot length compared with control (t test); P values: all <0.0001. (H) Growth curves (n = 10 replicates à five polyps each) and (I) time of bud development (n = 10 replicates) of foxO− endo (A6 line) and control polyps. Asterisks indicate significant differences in growth (t test); P value = <0.0001. (J) Expression levels of stem cell genes in foxO− ecto/endo polyps analyzed by qRT-PCR (n = 3 replicates). Asterisks indicate significant changes in expression levels (t test); P values: cnvas1 = 0.0021, cniwi = 0.0054. (K) Expression levels of AMPs in foxO− endo polyps analyzed by qRT-PCR (n = 3 replicates). Asterisks indicate significant changes in expression levels (t test); P values: hydramacinA6 line = 0.0001, periculin2bA6 line = 0.0316, hydramacinD11a line = 0.0031, hydramacinE1 line = 0.0011, and periculin2bE1 line = 0.0136.
As indicated above (Fig. 2B), genes strongly expressed in all three stem cell lineages include cniwi and cnvas1. We therefore wondered if FoxO is required for the expression of these genes in epithelial tissue. As shown in Fig. 4J, in polyps with foxO-silenced ecto- and endoderm, both cnvas1 and cniwi are significantly down-regulated. These findings indicate once more that cnvas1 and cniwi are FoxO target genes and resemble earlier observations in C. elegans (23). Taken together, these observations raise the possibility that foxO-deficient epithelial cells are driven into terminal differentiation, indicating that FoxO function is necessary for stem cell self-renewal and continuous growth.
Silencing FoxO Activity in Epithelial Cells Causes Severe Changes in the Functionality of Hydra’s Innate Immune System.
Our findings are intriguing in light of prior data showing that FoxO plays a key role in controlling life span and stress resistance in both C. elegans and flies (10, 11, 13). Because previous studies reported drastic changes of the immune system with age (“immuno senescence”; for review see ref. 25), we tested whether silencing of foxO might also cause changes in the immune status of polyps by comparing the expression patterns of genes known to play important roles in Hydra’s immune system, which is predominantly located in the endoderm (2, 26–30). foxO down-regulation in endodermal epithelial cells of three independent lines reduces the expression of the AMP gene arminin and causes severe overexpression of the AMPs hydramacin and periculin2b (Fig. 4K). Analysis of the hydramacin, periculin2b, and arminin promoters reveals the presence of numerous FoxO-binding sites (Fig. S6), indicating that FoxO can directly bind to these regulatory regions, acting either as repressor or as transcriptional activator (31). Because AMPs are key players in Hydra’s innate immunity and not only important defense molecules but also regulators of the host–microbe interaction (28, 32), our observations indicate that down-regulation of foxO results in significant changes in the functionality of Hydra’s innate immune system. This is consistent with prior results from Drosophila (33) that showed that AMP genes are activated in response to nuclear dFOXO activity. Thus, in addition to its importance in stem cell homeostasis, FoxO appears to have an unexpected role in controlling innate immune genes in Hydra.
Discussion
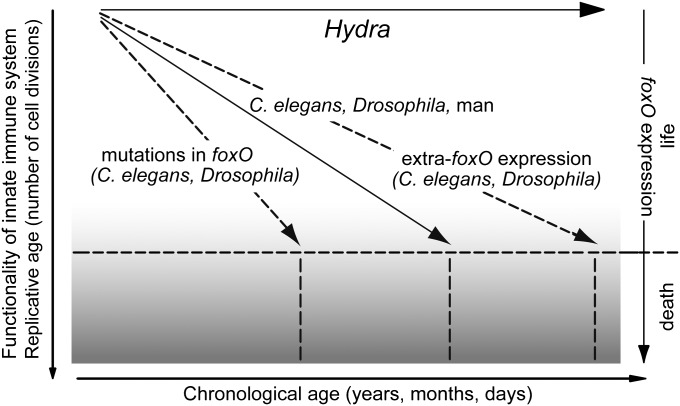
By the following three lines of evidence we have established that FoxO controls stem cell behavior in Hydra (summarized in Fig. S7A and schematized in Fig. S7B): (i) foxO is strongly expressed in all three stem cell lineages; (ii) overexpression of foxO in interstitial stem cells stimulates stem cell and progenitor cell proliferation and confers stemness by activation of stem cell genes to terminally differentiated cells such as nematocytes; and (iii) silencing of foxO in epithelial cells increases the number of terminally differentiated foot cells at the cost of growth rate and causes down-regulation of stem cell genes as well as changes in expression of genes controlling the functionality of the innate immune system. Based on the findings in Hydra we propose a general model in which FoxO plays a key role in controlling longevity (Fig. 5). According to this model, in immortal Hydra the high expression of foxO in all three stem cell lineages is crucial for the continuous self-renewal capacity and unlimited life span as well as the continuous maintenance of the functionality of the innate immune system. In contrast, the aging process of most organisms is caused by a reduction of foxO activity, which decreases functions of the immune system and stem cells. Aging in stem cell systems from flies to humans is associated with progressive loss of stem cell number and activity (23). Experimental evidence in flies, worms, and mice indicates that changes in life span indeed can occur through changes in foxO expression. Mutations in daf16/foxO in C. elegans prevent the longevity effect whereas overexpression increases it (23). Moreover, specific mutations in the sequence of foxO3a significantly increase human life span (9–13, 34).
Fig. 5.
Model of the role of FoxO in controlling longevity. Decline of foxO expression results in aging and death. Mutations in DAF-16 and dfoxO reduce life span in both C. elegans and flies. Increase of foxO expression delays aging by maintaining stem cell self-renewal and functionality of the immune system. Universally expressed foxO in Hydra results in a continuous self-renewal capacity of stem cells and immortality.
Our results also delineate a role for FoxO in innate immunity. We show that silencing FoxO activity results in significant changes in the expression of AMPs. AMPs in both plants and animals are activated through pattern recognition receptors in response to microbe-associated molecular patterns. Although our observations do not elucidate the precise genetic network responsible for FoxO-mediated AMP activation, they are consistent with previous observations in flies (33) and make a significant prediction: control of stem cell self-renewal may be tightly coupled to innate immune pathways, and aging may be the consequence of both reduced stem cell function and altered innate immune defense. This may be a common feature in plants and animals because stem cells in the shoot apical meristem of plants express high levels of a peptide controlling immune signaling and microbe interaction (35).
Taken together, studies of FoxO in Hydra have several important implications. They not only reveal FoxO as a molecular factor that has contributed to the early evolution of stem cells, but also highlight intriguing similarities between Hydra and other multicellular organisms including humans, in the mechanisms that maintain stemness (14–17) and control life span (9, 12, 13, 23, 36, 37). Thus, the work furthers our understanding of stem cell self-renewal at the beginning of animal evolution and also has implications for regenerative medicine and cellular aging.
Materials and Methods
Animals and Culture Conditions.
Experiments were carried out using H. vulgaris strain AEP Animals were cultured according to standard procedures (38).
Generation of Transgenic H. vulgaris Strain AEP.
foxO overexpression was achieved by generating transgenic polyps expressing foxO-eGFP transgene under the control of actin promoter. The stable knockdown of foxO was achieved by generating transgenic polyps expressing a foxO hairpin construct under control of actin promoter. As control, transgenic lines expressing eGFP in the endodermal and ectodermal epithelial cell and interstitial cell lineage, respectively, were used (3, 4). SI Materials and Methods include more information.
BrdU Labeling and Detection.
For analysis of cell proliferation, animals were exposed for 6 h to BrdU (39), macerated (40), and subjected to BrdU detection (39) as described previously.
In Situ Hybridization.
Gene expression analysis in foxO gain-of-function and loss-of-function strains was done by in situ hybridization and double in situ hybridization as described previously (3, 41). GenBank accession numbers of the analyzed genes can be found in SI Materials and Methods.
qRT-PCR.
Gene expression analysis in wild-type and foxO gain-of-function and loss-of-function strains was analyzed by qRT-PCR using the QuantiTect Probe RT-PCR Kit (QIAGEN) and the 7300 real-time PCR system (ABI) according to the manufacturer’s protocols (for primers see Table S1). GenBank accession numbers of the analyzed genes can be found in SI Materials and Methods.
Phylogenetic Analysis.
Phylogenetic analyses were based on 31 amino acid sequences of the forkhead domain, aligned using ClustalW (42). The most parsimonious tree was found using MEGA 4 (43). Maximum-parsimony and Maximum-likelihood bootstrap values were calculated based on 1,000 replicates. Bayesian posterior probabilities were calculated using MrBayes version 3.1.2 (44, 45) with two independent runs of 3,500,000 generations each, sampled every 1,000 generations with four chains. SI Materials and Methods includes more information.
Supplementary Material
Acknowledgments
This work was supported by Grant Bo 848/15-1 from the Deutsche Forschungsgemeinschaft (DFG) and by several grants from the DFG Excellence Initiative (to T.C.G.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JX118843–JX118848).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209714109/-/DCSupplemental.
References
- 1.Martínez DE. Mortality patterns suggest lack of senescence in hydra. Exp Gerontol. 1998;33(3):217–225. doi: 10.1016/s0531-5565(97)00113-7. [DOI] [PubMed] [Google Scholar]
- 2.Bosch TC. Hydra and the evolution of stem cells. Bioessays. 2009;31(4):478–486. doi: 10.1002/bies.200800183. [DOI] [PubMed] [Google Scholar]
- 3.Khalturin K, et al. Transgenic stem cells in Hydra reveal an early evolutionary origin for key elements controlling self-renewal and differentiation. Dev Biol. 2007;309(1):32–44. doi: 10.1016/j.ydbio.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TC. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc Natl Acad Sci USA. 2006;103(16):6208–6211. doi: 10.1073/pnas.0510163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch TC, Anton-Erxleben F, Hemmrich G, Khalturin K. The Hydra polyp: Nothing but an active stem cell community. Dev Growth Differ. 2010;52(1):15–25. doi: 10.1111/j.1440-169X.2009.01143.x. [DOI] [PubMed] [Google Scholar]
- 6.Fujisawa T, Sugiyama T. Genetic analysis of developmental mechanisms in Hydra. IV. Characterization of a nematocyst-deficient strain. J Cell Sci. 1978;30:175–185. doi: 10.1242/jcs.30.1.175. [DOI] [PubMed] [Google Scholar]
- 7.Hemmrich G, Bosch TC. Compagen, a comparative genomics platform for early branching metazoan animals, reveals early origins of genes regulating stem-cell differentiation. Bioessays. 2008;30(10):1010–1018. doi: 10.1002/bies.20813. [DOI] [PubMed] [Google Scholar]
- 8.Hemmrich G, et al. Molecular signatures of the three stem cell lineages in hydra and the emergence of stem cell function at the base of multicellularity. Mol Biol Evol. 2012;29(11):3267–3280. doi: 10.1093/molbev/mss134. [DOI] [PubMed] [Google Scholar]
- 9.Flachsbart F, et al. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci USA. 2009;106(8):2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jünger MA, et al. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol. 2003;2(3):20. doi: 10.1186/1475-4924-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenyon C. A pathway that links reproductive status to lifespan in Caenorhabditis elegans. Ann N Y Acad Sci. 2010;1204:156–162. doi: 10.1111/j.1749-6632.2010.05640.x. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, et al. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009;18(24):4897–4904. doi: 10.1093/hmg/ddp459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol. 2008;20(2):126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyamoto K, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1(1):101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Tothova Z, Gilliland DG. FoxO transcription factors and stem cell homeostasis: Insights from the hematopoietic system. Cell Stem Cell. 2007;1(2):140–152. doi: 10.1016/j.stem.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 16.Renault VM, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5(5):527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paik JH, et al. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell. 2009;5(5):540–553. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, et al. FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nat Cell Biol. 2011;13(9):1092–1099. doi: 10.1038/ncb2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seipel K, Yanze N, Schmid V. The germ line and somatic stem cell gene Cniwi in the jellyfish Podocoryne carnea. Int J Dev Biol. 2004;48(1):1–7. doi: 10.1387/ijdb.15005568. [DOI] [PubMed] [Google Scholar]
- 20.Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T. Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Dev Genes Evol. 2001;211(6):299–308. doi: 10.1007/s004270100156. [DOI] [PubMed] [Google Scholar]
- 21.Adamczyk P, et al. Minicollagen-15, a novel minicollagen isolated from Hydra, forms tubule structures in nematocysts. J Mol Biol. 2008;376(4):1008–1020. doi: 10.1016/j.jmb.2007.10.090. [DOI] [PubMed] [Google Scholar]
- 22.David CN, et al. Evolution of complex structures: Minicollagens shape the cnidarian nematocyst. Trends Genet. 2008;24(9):431–438. doi: 10.1016/j.tig.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Curran SP, Wu X, Riedel CG, Ruvkun G. A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature. 2009;459(7250):1079–1084. doi: 10.1038/nature08106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grens A, Shimizu H, Hoffmeister SA, Bode HR, Fujisawa T. The novel signal peptides, pedibin and Hym-346, lower positional value thereby enhancing foot formation in hydra. Development. 1999;126(3):517–524. doi: 10.1242/dev.126.3.517. [DOI] [PubMed] [Google Scholar]
- 25.Pawelec G. Immunosenescence comes of age. Symposium on Aging Research in Immunology: The Impact of Genomics. EMBO Rep. 2007;8(3):220–223. doi: 10.1038/sj.embor.7400922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosch TC, et al. Uncovering the evolutionary history of innate immunity: The simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol. 2009;33(4):559–569. doi: 10.1016/j.dci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Augustin R, et al. Activity of the novel peptide arminin against multiresistant human pathogens shows the considerable potential of phylogenetically ancient organisms as drug sources. Antimicrob Agents Chemother. 2009;53(12):5245–5250. doi: 10.1128/AAC.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Augustin R, Fraune S, Bosch TC. How Hydra senses and destroys microbes. Semin Immunol. 2010;22(1):54–58. doi: 10.1016/j.smim.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Augustin R, Siebert S, Bosch TC. Identification of a kazal-type serine protease inhibitor with potent anti-staphylococcal activity as part of Hydra’s innate immune system. Dev Comp Immunol. 2009;33(7):830–837. doi: 10.1016/j.dci.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Jung S, et al. Hydramacin-1, structure and antibacterial activity of a protein from the basal metazoan Hydra. J Biol Chem. 2009;284(3):1896–1905. doi: 10.1074/jbc.M804713200. [DOI] [PubMed] [Google Scholar]
- 31.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24(50):7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 32.Augustin R, Fraune S, Franzenburg S, Bosch TC. Where simplicity meets complexity: Hydra, a model for host-microbe interactions. Adv Exp Med Biol. 2012;710:71–81. doi: 10.1007/978-1-4419-5638-5_8. [DOI] [PubMed] [Google Scholar]
- 33.Becker T, et al. FOXO-dependent regulation of innate immune homeostasis. Nature. 2010;463(7279):369–373. doi: 10.1038/nature08698. [DOI] [PubMed] [Google Scholar]
- 34.Zeng Y, et al. Effects of FOXO genotypes on longevity: A biodemographic analysis. J Gerontol A Biol Sci Med Sci. 2010;65(12):1285–1299. doi: 10.1093/gerona/glq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee H, Chah OK, Sheen J. Stem-cell-triggered immunity through CLV3p-FLS2 signalling. Nature. 2011;473(7347):376–379. doi: 10.1038/nature09958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greer EL, Brunet A. FOXO transcription factors in ageing and cancer. Acta Physiol (Oxf) 2008;192(1):19–28. doi: 10.1111/j.1748-1716.2007.01780.x. [DOI] [PubMed] [Google Scholar]
- 37.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366(6454):461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 38.Lenhoff HM, Brown RD. Mass culture of hydra: An improved method and its application to other aquatic invertebrates. Lab Anim. 1970;4(1):139–154. doi: 10.1258/002367770781036463. [DOI] [PubMed] [Google Scholar]
- 39.Holstein TW, Hobmayer E, David CN. Pattern of epithelial cell cycling in hydra. Dev Biol. 1991;148(2):602–611. doi: 10.1016/0012-1606(91)90277-a. [DOI] [PubMed] [Google Scholar]
- 40.David CN. A quantitative method for maceration of Hydra tissue. Wilhelm Roux’ Archiv. 1973;171:259–268. doi: 10.1007/BF00577724. [DOI] [PubMed] [Google Scholar]
- 41.Hansen GN, Williamson M, Grimmelikhuijzen CJ. Two-color double-labeling in situ hybridization of whole-mount Hydra using RNA probes for five different Hydra neuropeptide preprohormones: Evidence for colocalization. Cell Tissue Res. 2000;301(2):245–253. doi: 10.1007/s004410000240. [DOI] [PubMed] [Google Scholar]
- 42.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 44.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 45.Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294(5550):2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.