Posttranslational modification of proteins, including phosphorylation, ubiquitylation, and acetylation, is an important means of signaling and functional specialization in the cell. The acetylation of lysine residues is best understood in histone proteins, in which histone acetyltransferases modify histone tail sequences to regulate chromatin structure. However, lysine acetylation has recently become more appreciated as a widely distributed phenomenon regulating diverse proteins and cellular systems (1). One such system is the microtubule (MT) cytoskeleton. MTs are highly dynamic tubular polymers assembled from protofilaments of α/β-tubulin dimers, and are essential for intracellular transport, architectural organization, and force production in eukaryotic cells (2). Acetylation of the conserved lysine 40 (K40) residue of α-tubulin, first reported more than 25 y ago (3), is one of several conserved posttranslational modifications found in the tubulin protein (2, 3). Although MTs generally function as highly dynamic polymers, α-tubulin K40 acetylation is associated with unusually stable MTs. These act as tracks for organelle and macromolecule transport in neurons, and form the structural scaffolds, called axonemes, at the core of beating cilia and flagella (4, 5). α-Tubulin K40 acetylation is catalyzed by a conserved α-tubulin acetyltransferase (α-TAT), but neither the detailed mechanism of tubulin acetylation nor its downstream functional consequences are well understood (6, 7). Now, in parallel studies presented in PNAS, Friedmann et al. (8) and Taschner et al. (9) describe the 3D structure of human α-TAT and the chemical basis for α-tubulin K40 acetylation. The two studies show how α-TAT selectively acetylates α-tubulin K40 and explore α-TAT’s catalytic mechanism through detailed enzymatic analysis (8, 9).
MTs are regulated by a host of posttranslational modifications such as polyglutamylation, polyglycylation, detyrosination, and tyrosination, most of which occur at the C termini of α- and β-tubulin exposed on the outer MT surface, and directly regulate MT association with motor proteins and so-called plus-end binding proteins (2). In contrast, α-tubulin K40 acetylation occurs on the inner or lumenal surface of the MT and probably affects MT structure and dynamics directly (Fig. 1) (2, 4). Structural modeling suggests that acetylation may stabilize lateral interactions between neighboring tubulin protofilaments, potentially explaining the increased stability of acetylated MTs observed in vivo (10). Acetylation-mediated modification of protofilament interactions may also explain why loss of α-TAT in Caenorhabditis elegans disrupts the tight regulation of MT protofilament number in touch receptor neurons, resulting in shorter MTs with a range of protofilament numbers (10, 11).
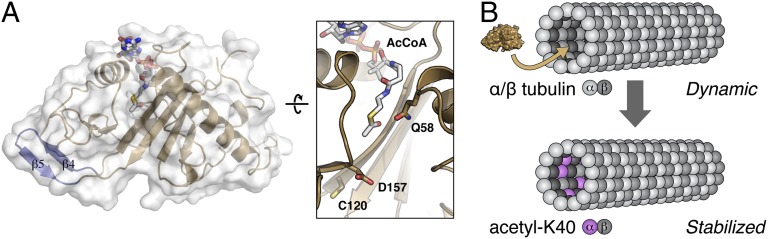
Fig. 1.
(A) Overview of the structure of human α-TAT (9). Right: Detailed view of the enzyme’s active site shows bound AcCoA cofactor and proposed general base catalytic residues Q58, C120, and D157. (B) α-TAT (brown) acetylates α-tubulin K40, located in the MT lumen, potentially altering its structure and dynamics.
α-TAT was identified as a component associated with intraflagellar Bardet–Biedl syndrome complex (i.e., BBsome), which is necessary for forming and stabilizing axonemal MTs (7, 12), and α-TAT orthologues are found in all organisms with MT axonemal structures such as cilia and flagella (7). Purified α-TAT is highly specific for α-tubulin K40; it possesses no detectable activity on substrates such as histones. α-TAT also shows a sixfold higher catalytic rate when acetylating α-tubulin K40 in preformed MTs vs. soluble α/β-tubulin dimers (7, 13). To better understand α-TAT mechanism and specificity, Taschner et al. (9) and Friedmann et al. (8) took a combined structural/biochemical approach. Both groups determined the 3D structure of the N-terminal two thirds of the protein (residues 1–195 of 333), which adopts a compact, roughly triangular shape and binds an acetyl CoA (AcCoA) cofactor in its active site (Fig. 1). The structures reveal that α-TAT is distantly related to the GCN5 family of histone acetyltransferases, with the closest structural similarities evident in the cores of the two proteins and the manner of their binding to AcCoA. Outside the conserved core, the most distinctive structural feature of α-TAT is a β-hairpin structure (β4–β5) that is unique to and strictly conserved within the α-TAT protein family (Fig. 1, blue).
Although the structures lack a direct visualization of α-tubulin binding, both studies (8, 9) converge on a plausible mechanism for α-TAT’s observed substrate specificity. α-TAT contains a conserved surface pocket close to the active site composed largely of hydrophobic and basic residues, which likely complement the acidic loop containing α-tubulin K40. Consistent with this idea, structure-based mutagenesis of residues in this pocket substantially reduces the enzyme’s catalytic activity (8, 9). Friedmann et al. (8) further identify two mutations in the conserved α-TAT–specific β4–β5 hairpin that actually enhance α-tubulin acetylation activity in vivo and in vitro, suggesting more extensive interactions that may further increase specificity for α-tubulin (Fig. 1).
The four known families of histone acetyltransferases—GCN5, Rtt109, CBP300, and MYST—share a common fold, yet they use distinct catalytic mechanisms (14). GCN5 is the most structurally similar to α-TAT and uses an acidic residue in the active site as a general base to deprotonate the incoming substrate lysine amine and catalyze transfer of the acetyl group (15). The other three families use two catalytic residues and catalyze acetylation through a “ping-pong” mechanism (16) or a “hit-and-run” mechanism (13, 17). In α-TAT, previous mutation of a highly conserved aspartic acid (D157), identified through sequence comparisons with GCN5, to asparagine resulted in an inactive enzyme (7, 13). Although this result suggested a close similarity to GCN5’s catalytic mechanism, the structures of α-TAT reveal a more complicated picture of catalysis. The protein’s active site contains several conserved residues that could potentially function as general bases in the reaction: glutamine 58 (Q58), cysteine (C120), and aspartic acid 157 (D157; Fig. 1). Based on their analysis of different active-site mutants, Friedmann et al. (8) suggest that C120 and D157 can both act as general bases. Consistent with this model, mutation of either C120 or D157 significantly compromises catalysis, but neither mutant is completely inactive (9). Taschner et al. (9), on the contrary, propose that Q58 may instead act as the general base in catalysis, based on its position close to the bound AcCoA and its coordination of a water molecule in the active site (8). Supporting this idea, enzymatic assays show that mutation of Q58 to alanine completely abolishes the enzyme’s activity in vitro (8). Although these results make a strong case for Q58’s importance for acetyl transfer, the final word on α-TAT’s catalytic mechanism must await more extensive biochemical analysis of active site mutants, as well as structural work to visualize α-TAT’s interaction with its substrate. Such studies
Friedmann et al. and Taschner et al. describe the 3D structure of human α-TAT and the chemical basis for α-tubulin K40 acetylation.
may also aid the design of α-TAT catalytic inhibitors, which could then be used to examine the in vivo functions of α-TAT in more detail.
The structural examination of α-TAT highlights outstanding questions and critical experiments regarding the roles of α-tubulin acetylation in specialized MT structures (8, 9). In particular, the effects of α-tubulin K40 acetylation on the structure and dynamics of MTs represent open questions that could be readily addressed in vitro with existing biochemical and biophysical tools. Additionally, it is not understood how α-TAT accesses the MT lumen to acetylate α-tubulin K40, and whether the enzyme functions alone or with one or more binding partners. Indeed, its original identification as part of the BBsome complex highlights the possibility of cooperation with other proteins in this complex. Finally, a remaining structural question is exactly how α-TAT recognizes its substrate. In this context, structural studies using X-ray crystallography or EM will be important to characterize α-TAT’s interactions with isolated tubulin and/or polymerized MTs. The answers to these and other questions will add critical detail to the emerging picture of MT regulation through diverse phenomena, including protein–protein interactions and posttranslational modifications, that together allow for the functional specialization of these important polymers in complex cellular systems.
Note added in proof:
In addition to the structures of human alpha-TAT, a structure of the enzyme from the zebrafish Danio rerio by Kormendi et al. has been published (J. Biol. Chem. published online, DOI: jbc.C112.421222), which largely agrees with the structural and functional conclusions described above.
Acknowledgments
This work was supported by National Institutes of Health Grant R00-GM8249 (to J.A.-B.) and the Ludwig Institute for Cancer Research (to K.D.C.).
Footnotes
References
- 1.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 2.Janke C, Bulinski JC. Post-translational regulation of the microtubule cytoskeleton: Mechanisms and functions. Nat Rev Mol Cell Biol. 2011;12(12):773–786. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- 3.L’Hernault SW, Rosenbaum JL. Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry. 1985;24(2):473–478. doi: 10.1021/bi00323a034. [DOI] [PubMed] [Google Scholar]
- 4.Li L, et al. MEC-17 deficiency leads to reduced α-tubulin acetylation and impaired migration of cortical neurons. J Neurosci. 2012;32(37):12673–12683. doi: 10.1523/JNEUROSCI.0016-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konno A, Setou M, Ikegami K. Ciliary and flagellar structure and function-their regulations by posttranslational modifications of axonemal tubulin. Int Rev Cell Mol Biol. 2012;294:133–170. doi: 10.1016/B978-0-12-394305-7.00003-3. [DOI] [PubMed] [Google Scholar]
- 6.Akella JS, et al. MEC-17 is an alpha-tubulin acetyltransferase. Nature. 2010;467(7312):218–222. doi: 10.1038/nature09324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc Natl Acad Sci USA. 2010;107(50):21517–21522. doi: 10.1073/pnas.1013728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedmann DR, Aguilar A, Fan J, Nachury MV, Marmorstein R. Structure of the α-tubulin acetyltransferase, αTAT1, and implications for tubulin-specific acetylation. Proc Natl Acad Sci USA. 2012;109:19655–19660. doi: 10.1073/pnas.1209357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taschner M, Vetter M, Lorentzen E. Atomic resolution structure of human α-tubulin acetyltransferase bound to acetyl-CoA. Proc Natl Acad Sci USA. 2012;109:19649–19654. doi: 10.1073/pnas.1209343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cueva JG, Hsin J, Huang KC, Goodman MB. Posttranslational acetylation of α-tubulin constrains protofilament number in native microtubules. Curr Biol. 2012;22(12):1066–1074. doi: 10.1016/j.cub.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Topalidou I, et al. Genetically separable functions of the MEC-17 tubulin acetyltransferase affect microtubule organization. Curr Biol. 2012;22(12):1057–1065. doi: 10.1016/j.cub.2012.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loktev AV, et al. A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev Cell. 2008;15(6):854–865. doi: 10.1016/j.devcel.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Jin H, et al. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141(7):1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Tang Y, Cole PA, Marmorstein R. Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: Implications for histone acetyltransferase evolution and function. Curr Opin Struct Biol. 2008;18(6):741–747. doi: 10.1016/j.sbi.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clements A, et al. Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Mol Cell. 2003;12(2):461–473. doi: 10.1016/s1097-2765(03)00288-0. [DOI] [PubMed] [Google Scholar]
- 16.Yan Y, Harper S, Speicher DW, Marmorstein R. The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat Struct Biol. 2002;9(11):862–869. doi: 10.1038/nsb849. [DOI] [PubMed] [Google Scholar]
- 17.Tang Y, et al. Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP. Nat Struct Mol Biol. 2008;15(9):998. doi: 10.1038/nsmb.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]