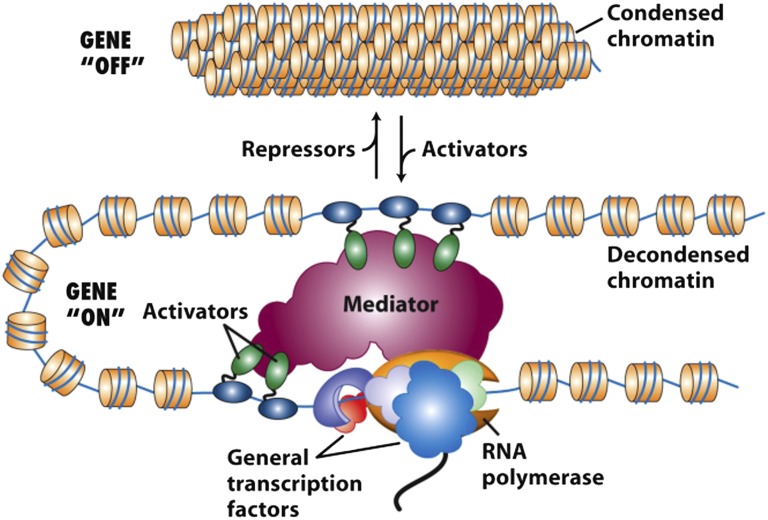
Mediator is an ∼30-subunit multiprotein complex approximately twice the size of the 12-subunit RNA polymerase (i.e., RNA polymerase II [Pol II]) that transcribes most human genes. Mediator functions as a molecular bridge between Pol II bound at transcription start sites (TSSs) and activation and repression domains of regulatory sequence-specific DNA binding transcription factors (TFs) bound to enhancers and promoter proximal transcription control sequences (1–5) (Fig. 1). In PNAS, Zhou et al. (6) report a significant advance in understanding how mutations in one Mediator subunit, MED12, result in two types of X-linked intellectual disability known as FG and Lujan syndromes. Studies of the mutant Mediators reveal how MED12 interactions with different components of the transcriptional machine produce opposing activating and inhibitory influences necessary for the proper level of transcription required for normal development.
Fig. 1.
Diagram representing simultaneous binding of several regulatory TF activation domains to different surfaces of the Mediator. dsDNA is represented by a thin blue line and histone octamers by orange discs. Regulatory TFs bind to specific DNA sequences at promoter proximal regions (bottom left of Mediator) and distal enhancers (top of Mediator) via their DNA-binding domains (blue), which are linked through flexible regions of polypeptide to activation domains (green). In response to interactions with activation domains, Mediator binds to RNA Pol II and promotes binding of the polymerase and its general TFs to the TSS. [Reprinted with permission from W. H. Freeman and Company (21).]
Interactions between the Mediator complex and regulatory TFs control several processes that regulate gene transcription, including chromatin modification in the region of the promoter, which influences binding of TFs to DNA (7–9) (interconversion between condensed and decondensed chromatin, Fig. 1), the assembly at the TSS of a preinitiation complex composed of Pol II and its five initiation factors (called general TFs) (10–12), the frequency of reinitiation by Pol II and its general TFs (1–5), and, at a large fraction of promoters in multicellular animals, release from a pause in transcription by elongating Pol II in the promoter proximal region ∼50 bp from the TSS (13). In current models, the Mediator complex integrates positive and negative signals of varying strength from multiple regulatory TFs bound to specific sites in enhancer and promoter-proximal DNA control elements to determine the rate of initiation by RNA Pol II, and hence the amount of mRNA expressed and translated into protein. In the simplest model, regulatory domains of TFs bind simultaneously to various surfaces of the large mediator complex to influence the rate of transcription (Fig. 1). However, it is equally possible that signals from sequential interactions between Mediator and activators and/or repressors are integrated to control transcription initiation and elongation.
The importance of Mediator function for transcription control raises the expectation that mutations in genes encoding human Mediator subunits might have profound effects on embryonic development. Accordingly, complete KO of mouse Mediator subunits results in embryonic lethality. In the earliest study in mammals, KO of the mouse Mediator subunit Med22, which is highly conserved in Mediator complexes from all eukaryotes (14), and essential for viability of yeast (15), caused embryonic lethality at the early blastocyst stage, consistent with a profound defect in transcription (16). In contrast, KO of subunits such as Med1 (17) and Med23 (18), which have diverged considerably in sequence during the evolution of multicellular organisms (14), were not incompatible with cell viability because KO embryos grew to tens of thousands of cells. However, the mutant embryos die at about embryonic day 9.5, when they become large enough to require a functioning circulatory system to provide oxygen and nutrients throughout the embryo (17, 18). These Mediator subunits are not required for expression of genes in common with the unicellular ancestor of all eukaryotic cells that are needed to sustain cellular life. Rather, they are required for the fine control of gene expression needed to execute the genetic program that underlies embryological development. KO of Med1 leads to profound defects in embryonic development, including an abnormally functioning heart, resulting in embryonic death (17). Med23 KO embryos have less profound developmental defects, but defects in remodeling of the vasculature prevent the normal flow of red blood cells, probably accounting for death of the embryo (18).
Although these extreme mutations in Mediator subunits result in embryonic lethality, more subtle mutations in humans have been discovered because they allow development of viable fetuses to term, but present clinically after birth as a result of developmental abnormalities (19) (Table 1). Zhou et al. (6) report on experiments with cells cultured from patients with FG syndrome or Lujan syndrome that begin to explain the transcriptional abnormalities in these patients and also reveal how an interplay between positive and negative inputs to the Mediator result in the appropriate level of transcription required for normal development.
Table 1.
Human Mediator subunit mutations linked to developmental disorders
| Disorder | Associated mutation |
| X-linked mental retardation syndromes | |
| FG syndrome | MED12 R961W |
| Lujan syndrome | MED12 N1007S |
| Infantile cerebral and cerebellar atrophy | MED17 L371P |
| Autosomal recessive axonal Charcot–Marie–Tooth disease | MED25 A335V |
| Congenital retinal folds, microcephaly, and mental retardation | CDK19 haploinsufficiency |
| Transposition of the great arteries | MED13L haploinsufficiency; MED13L (E251G; R1872H; S2023G) |
References and in-depth discussion are provided by Spaeth et al. (19).
FG and Lujan syndromes are rare syndromes characterized by mental retardation, dysgenesis of the corpus callosum, deafness, seizures, and behavioral disturbances, and are often associated with anorectal and urogenital malformations and congenital heart defects in FG syndrome. Genetic analysis of the recessive mutations revealed that FG and Lujan syndromes are associated with amino acid substitutions R961W and N1007S, respectively, in Mediator subunit MED12. The MED12 subunit is required for normal gene control in response to signaling from neighboring cells by Notch, Wnt, and Sonic hedgehog (SHH) protein ligands. These ligands bind to regulatory receptors that span the cell membrane and elicit intracellular responses to the extracellular signals. Notch, Wnt, and SHH have critical functions in central nervous system development, from controlling the development of brain morphology and neuronal cell differentiation to regulation of the plasticity of synaptic connections between differentiated neurons. Earlier research by Ding et al (8) had shown that the FG and Lujan MED12 mutations disrupt repression of genes by the TF REST through dimethylation, followed by trimethylation of histone H3 lysine 9 (H3K9me3). H3K9me3 is a binding site for heterochromatin proteins (e.g., HP1s), and generates a chromatin structure that blocks access of TFs to their binding sites in DNA (Fig. 1, “Condensed chromatin”). As a consequence, REST is a master regulator of neuronal cell differentiation by inhibiting expression of genes that induce neuronal differentiation in proliferating neural progenitor cells and in differentiated nonneuronal cells.
Zhou et al (6) report that the MED12 amino acid substitutions associated with FG and Lujan syndromes also lead to misregulation by the GLI3 TF in response to SHH signaling. MED12 is a component of a four-subunit Mediator “kinase module” including MED12, MED13, CDK8, and cyclin C. CDK8, the protein kinase subunit of the module, interacts with MED12. GLI3, a TF whose activity is regulated by SHH signaling, was shown earlier by the same group to bind directly to MED12 (20). Zhou et al. (6) realized that several of the phenotypes of the FG and Lujan syndromes, such as corpus callosum defects, are also observed in patients with mutations in GLI3. Consequently, they analyzed the impact of the FG and Lujan MED12 mutations on transcription of genes activated by SHH signaling and on the association of CDK8 with several promoters through ChIP assays. They find that GLI3 target genes, but not genes regulated by several other signal transduction pathways, are greatly overexpressed in response to SHH signaling in cells from patients with FG and Lujan syndrome (6). ChIP assays showed that CDK8 was depleted at GLI3-regulated promoters, but not at other promoters. Moreover, in experiments with cells in which the endogenous CDK8 or MED12 were depleted by siRNAs, CDK8 enzymatic kinase activity and the WT MED12 sequence were required to constrain transcriptional activation by GLI3 in response to SHH signaling. These observations lead to a model in which the proper level of transcriptional activation by SHH signaling results from a balance of activating influences from the binding of the GLI3 activation domain to MED12 and inhibition by CDK8 phosphorylation of some component of the multiprotein transcription machine. The MED12 mutations in FG and Lujan syndromes greatly diminish the inhibitory influence of CDK8, resulting in abnormally high transcriptional activation of genes targeted by the SHH signal transduction pathway. This overactivation of SHH signaling likely contributes to the developmental abnormalities observed in these syndromes.
We can hope that further mapping of human mutations resulting in developmental abnormalities will uncover additional mutations in Mediator subunits, and that careful analysis of the mutant Mediators will be as revealing about unexpected mechanisms of Mediator function as the FG and Lujan syndrome MED12 mutants have been (6, 8).
Footnotes
The author declares no conflict of interest.
See companion article on page 19763.
References
- 1.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30(5):235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Conaway RC, Conaway JW. Function and regulation of the Mediator complex. Curr Opin Genet Dev. 2011;21(2):225–230. doi: 10.1016/j.gde.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11(11):761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larivière L, Seizl M, Cramer P. A structural perspective on Mediator function. Curr Opin Cell Biol. 2012;24(3):305–313. doi: 10.1016/j.ceb.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 6.Zhou H, et al. MED12 mutations link intellectual disability syndromes with dysregulated GLI3-dependent Sonic Hedgehog signaling. Proc Natl Acad Sci USA. 2012;109:19763–19768. doi: 10.1073/pnas.1121120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malik S, Roeder RG. Epigenetics? Mediator does that too! Mol Cell. 2008;31(3):305–306. doi: 10.1016/j.molcel.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 8.Ding N, et al. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol Cell. 2008;31(3):347–359. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding N, et al. MED19 and MED26 are synergistic functional targets of the RE1 silencing transcription factor in epigenetic silencing of neuronal gene expression. J Biol Chem. 2009;284(5):2648–2656. doi: 10.1074/jbc.M806514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantin GT, Stevens JL, Berk AJ. Activation domain-mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proc Natl Acad Sci USA. 2003;100(21):12003–12008. doi: 10.1073/pnas.2035253100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399(6736):609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 12.Li XY, Virbasius A, Zhu X, Green MR. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;399(6736):605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 13.Conaway RC, Conaway JW. The Mediator complex and transcription elongation. Biochim Biophys Acta. 2012;S1874-9399(12):00161–00167. doi: 10.1016/j.bbagrm.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36(12):3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hengartner CJ, et al. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 1995;9(8):897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 16.Tudor M, Murray PJ, Onufryk C, Jaenisch R, Young RA. Ubiquitous expression and embryonic requirement for RNA polymerase II coactivator subunit Srb7 in mice. Genes Dev. 1999;13(18):2365–2368. doi: 10.1101/gad.13.18.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol Cell. 2000;5(4):683–693. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- 18.Balamotis MA, et al. Complexity in transcription control at the activation domain-mediator interface. Sci Signal. 2009;2(69):ra20. doi: 10.1126/scisignal.1164302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spaeth JM, Kim NH, Boyer TG. Mediator and human disease. Semin Cell Dev Biol. 2011;22(7):776–787. doi: 10.1016/j.semcdb.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou H, Kim S, Ishii S, Boyer TG. Mediator modulates Gli3-dependent Sonic hedgehog signaling. Mol Cell Biol. 2006;26(23):8667–8682. doi: 10.1128/MCB.00443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lodish H, et al. Molecular Cell Biology. 6th Ed. New York: Freeman; 2008. [Google Scholar]