Abstract
Mutations affecting ciliary components cause a series of related genetic disorders in humans, including nephronophthisis (NPHP), Joubert syndrome (JBTS), Meckel-Gruber syndrome (MKS), and Bardet-Biedl syndrome (BBS), which are collectively termed “ciliopathies.” Recent protein–protein interaction studies combined with genetic analyses revealed that ciliopathy-related proteins form several functional networks/modules that build and maintain the primary cilium. However, the precise function of many ciliopathy-related proteins and the mechanisms by which these proteins are targeted to primary cilia are still not well understood. Here, we describe a protein–protein interaction network of inositol polyphosphate-5-phosphatase E (INPP5E), a prenylated protein associated with JBTS, and its ciliary targeting mechanisms. INPP5E is targeted to the primary cilium through a motif near the C terminus and prenyl-binding protein phosphodiesterase 6D (PDE6D)-dependent mechanisms. Ciliary targeting of INPP5E is facilitated by another JBTS protein, ADP-ribosylation factor-like 13B (ARL13B), but not by ARL2 or ARL3. ARL13B missense mutations that cause JBTS in humans disrupt the ARL13B–INPP5E interaction. We further demonstrate interactions of INPP5E with several ciliary and centrosomal proteins, including a recently identified ciliopathy protein centrosomal protein 164 (CEP164). These findings indicate that ARL13B, INPP5E, PDE6D, and CEP164 form a distinct functional network that is involved in JBTS and NPHP but independent of the ones previously defined by NPHP and MKS proteins.
Keywords: photoreceptor degeneration, retinitis pigmentosa, leber congenital amaurosis, polydactyly, cystic kidney
Primary cilia are microtubule-based cell surface projections that emanate from the centrosome. This subcellular organelle functions as an antenna, sensing and transducing extracellular signals into the cell, and plays an essential role in regulating multiple cellular processes including the cell cycle, embryonic development, and tissue homeostasis (1–3). Mutations affecting ciliary and centrosomal components underlie a group of related human disorders such as Joubert syndrome (JBTS), Meckel-Gruber syndrome (MKS), nephronophthisis (NPHP), and Bardet-Biedl syndrome (BBS), collectively termed ciliopathies (1–3). Recent protein–protein interaction studies have identified several functional modules or networks involved in these ciliopathies (4). For example, BBS proteins and intraflagellar transport (IFT) proteins form multiprotein complexes, the BBSome and the IFT complexes, respectively, and these complexes are involved in transporting ciliary proteins. Likewise, NPHP and MKS proteins form a distinct modular complex at the transition zone of primary cilia and regulate ciliary membrane compositions (5–9). However, there are many ciliary and centrosomal proteins [e.g., inositol polyphosphate-5-phosphatase E (INPP5E) and ADP-ribosylation factor-like 13B (ARL13B)] that have not been linked to any of the known functional networks and their precise functions remain to be elucidated.
INPP5E encodes an enzyme that hydrolyzes the 5-phosphate of PtdIns(3,4,5)P3 and PtdIns(4,5)P2 and localizes to primary cilia. Mutations in this gene cause JBTS in humans (10, 11). In mice, loss of Inpp5e activity results in cystic kidney, bilateral anophthalmia, polydactyly, skeletal defects, cleft palate, and cerebral developmental defects (11). Inactivation of Inpp5e in adult mice results in obesity and photoreceptor degeneration. Interestingly, many proteins that localize to cilia, including INPP5E, RPGR, PDE6 α and β subunits, GRK1 (Rhodopsin kinase), and GNGT1 (Transducin γ chain), are prenylated (either farnesylated or geranylgeranylated), and mutations in these genes or genes involved in their prenylation (e.g., AIPL1 and RCE1) lead to photoreceptor degeneration in vertebrates and humans (12–18). Understanding the molecular mechanisms by which prenylated proteins are transported to primary cilia would have significant ramifications in developing therapeutic strategies to treat blindness and other ciliopathy phenotypes associated with these genes.
In this study, we sought to determine the functional network of INPP5E and the mechanisms by which INPP5E is targeted to primary cilia. We determined the ciliary targeting sequence (CTS) of INPP5E, the small GTPase responsible for its ciliary targeting, and an interaction network that connects INPP5E to other known ciliopathy genes and new candidates.
Results
CTS of INPP5E.
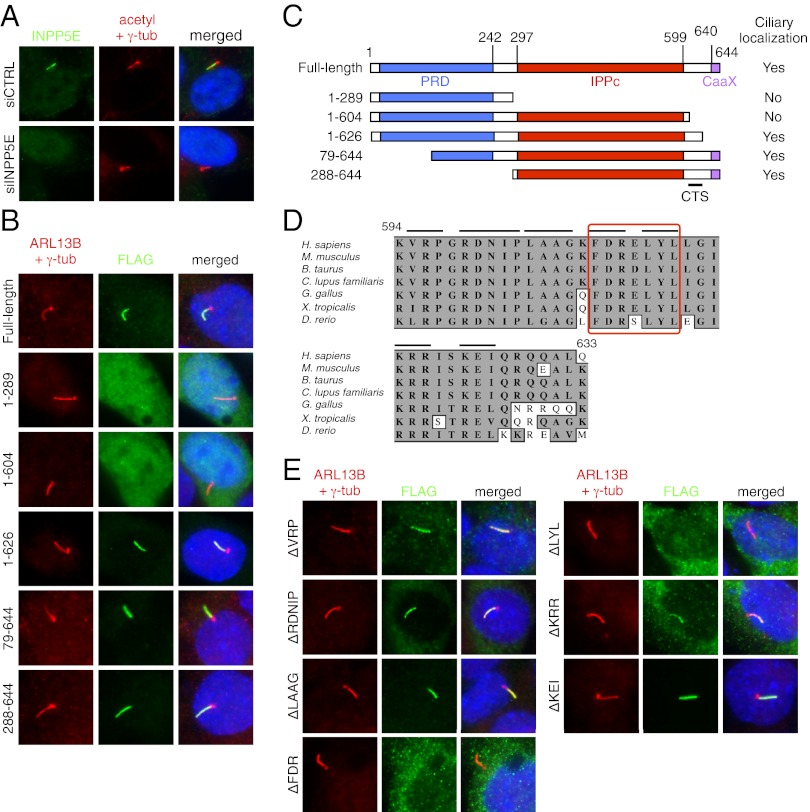
Previously, INPP5E was shown to localize to primary cilia (10, 11). We confirmed its ciliary localization in hTERT–RPE1 and IMCD3 cells by detecting endogenous and transfected FLAG-tagged INPP5E (Fig. 1 A and B). Ablation of INPP5E expression by siRNA-mediated gene knockdown verified the specificity of the antibody staining. To map the CTS of INPP5E, we generated a series of deletion mutants with N-terminal FLAG tags and evaluated their localization in IMCD3 cells (Fig. 1 B and C). The constructs with N-terminal 289 or 604 residues showed diffused localization throughout the cell including the nucleus. In contrast, constructs with the C-terminal portion of INPP5E (amino acids 79–644 and 288–644) showed specific ciliary localization. Consistent with the previous observation (11), the N-terminal 626 residues of INPP5E were sufficient to localize to cilia, suggesting that the CTS of INPP5E is between the phosphatase catalytic domain and the CaaX motif, which is a prenylation signal, and that prenylation itself is not necessary for INPP5E ciliary targeting.
Fig. 1.
Ciliary targeting sequence of INPP5E. (A) INPP5E localizes to the primary cilium in hTERT–RPE1 cells. Antibodies against acetylated tubulin and γ-tubulin (red) were used to mark cilia and centrosomes, respectively. Endogenous INPP5E (green) was detected within the primary cilium. siRNA against INPP5E was transfected to verify the specificity of the staining. Merged images are shown (Right) with DAPI staining for the nucleus (blue). (B) Localization of INPP5E deletion mutants. FLAG-tagged INPP5E deletion mutants were transfected into IMCD3 cells and their localization was probed with anti-FLAG antibody (green). Cilia and centrosomes (red) were labeled with ARL13B and γ-tubulin antibodies, respectively. Numbers indicate the amino acid residues transfected. (C) Summary of INPP5E immunolocalization study. Proline-rich domain (PRD), inositol polyphosphate phosphatase catalytic (IPPc) domain, and CaaX motif were marked in blue, red, and purple boxes, respectively. The location of ciliary targeting sequence (CTS) is highlighted (Bottom). (D) Conservation of amino acid sequences around the CTS in vertebrates. The numbers above the sequence denote the amino acid position in human INPP5E. Gray box marks conserved (identical or similar) amino acids. Black bars represent the mutated residues in each construct. Red box highlights the CTS determined in E. (E) Fine mapping of the CTS in INPP5E. Mutated amino acids are shown (Left).
The amino acid sequence in this region (amino acids 605–626) is highly conserved in vertebrates (Fig. 1D). For fine mapping of the CTS in this region, we generated small deletion mutants (three to five residue deletions from the full-length construct) spanning this region and evaluated their localization (Fig. 1 D and E). Immunofluorescence microscopy results indicate that a motif consisting of seven residues (FDRELYL; amino acids 609–615) is critical for INPP5E ciliary targeting. Mutations in VRP residues (amino acids 595–597), which resemble the ciliary targeting VxPx motif found in rhodopsin, polycystin-1, and polycystin-2 (19–21), did not affect INPP5E ciliary localization. Mutations in KRR residues (amino acids 609–611) resulted in a partial disruption of ciliary targeting: weak ciliary localization with a significant accumulation within the cytoplasm. Substitution of FDRELYL residues with alanines (FDR to AAA and LYL to AAA) also caused INPP5E mislocalization (Fig. S1A). Finally, we tested whether this motif is sufficient for ciliary targeting by fusing the C-terminal portion of INPP5E to GFP. Fusion of 39 C-terminal residues (amino acids 606–644) or 66 residues (amino acids 579–644) of INPP5E, including the CaaX motif, to the C terminus of GFP was not sufficient to target GFP to primary cilia (Fig. S1B). This insufficiency may be due to the inability of the tested peptides to form native structures on their own. Alternatively, additional motif(s) within the phosphatase catalytic domain may be necessary for INPP5E ciliary targeting in addition to this motif.
ARL13B Mediates Ciliary Trafficking of INPP5E.
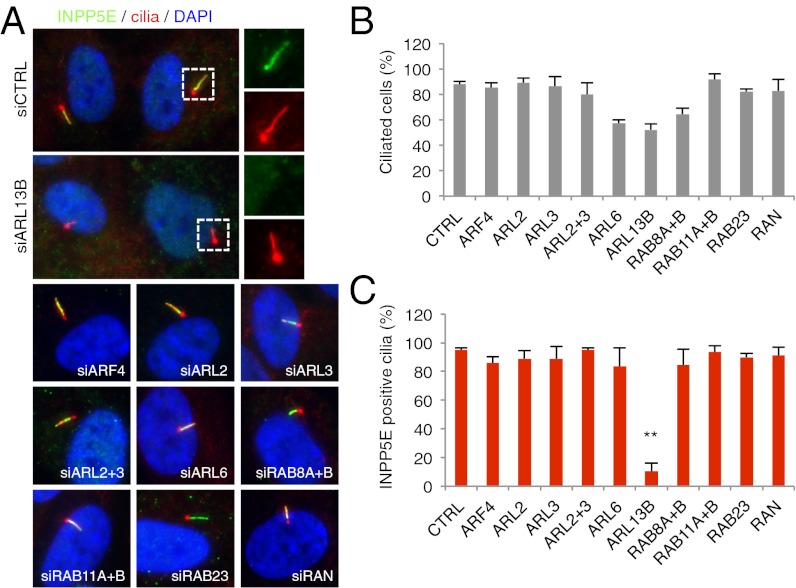
Protein trafficking to primary cilia is mediated by various small GTPases. We sought to determine the GTPase that targets INPP5E to cilia. INPP5E has a CaaX motif at its C terminus, and phosphodiesterase 6D (PDE6D) is known to bind to prenyl groups (22, 23). We confirmed that PDE6D associates with INPP5E (see below). Additionally, ARL2 and ARL3 are known to bind to PDE6D (22, 24) and regulate PDE6D–prenyl group interactions (23). ARL3 was further shown to facilitate ciliary targeting of NPHP3 (25). Therefore, we initially tested the requirement of ARL2 and ARL3 for INPP5E ciliary targeting. We blocked expression of ARL2 and ARL3 in hTERT–RPE1 cells by transfecting siRNAs against these genes and evaluated localization of endogenous INPP5E. Efficient knockdown was confirmed by immunoblotting and quantitative real-time PCR (Fig. S2). To our surprise, depletion of ARL2 and ARL3, either individually or combined, had no impact on INPP5E ciliary targeting (Fig. 2), suggesting that ciliary targeting of INPP5E is mediated through a different mechanism. Therefore, we expanded our search to include all small GTPases that are known to be involved in ciliary trafficking (ARF4, ARL2, ARL3, ARL6, ARL13B, RAB8A, RAB8B, RAB11A, RAB11B, RAB23, and RAN). In this experiment, we found that ARL13B, a protein associated with JBTS (26–30), is specifically required for ciliary targeting of INPP5E (Fig. 2). In contrast, knockdown of other small GTPases did not abolish INPP5E ciliary targeting, although some of them are required for efficient ciliogenesis (e.g., ARL6, ARL13B, RAB8A, and RAB8B) (Fig. 2B).
Fig. 2.
ARL13B mediates ciliary targeting of INPP5E. (A) ARL13B is required for ciliary localization of INPP5E. Expression of indicated genes was blocked by siRNA transfection and ciliary localization of INPP5E (green) was examined in hTERT–RPE1 cells. RAB8A and RAB8B were depleted simultaneously, as were RAB11A and RAB11B. Cilia and centrosomes were marked by acetylated tubulin and γ-tubulin antibodies (red). Enlarged images of boxed area are shown (Right). DAPI (blue) was used to stain the nuclei. (B) Percentage of ciliation in small GTPase-depleted cells. Graphs represent average of at least two independent experiments and bars, SEs. Depleted small GTPases are shown in the abscissa. (C) Percentage of INPP5E-positive cilia in small GTPase-depleted cells. One-way ANOVA analysis followed by Tukey test indicates that INPP5E-positive cilia are significantly reduced in ARL13B-depleted cells compared with control (P < 0.01).
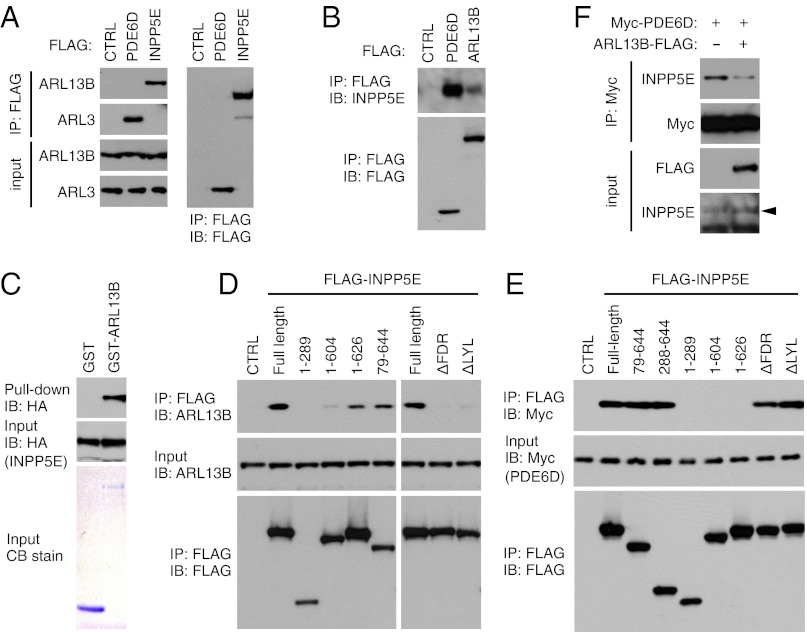
We next examined physical interactions between INPP5E, ARL13B, and PDE6D. When immunoprecipitated with anti-FLAG antibodies, endogenous ARL13B was efficiently coprecipitated with FLAG–INPP5E (Fig. 3A). Contrary to ARL13B (but consistent with the localization study), ARL3 did not show any physical interaction with INPP5E. We also tested whether ARL13B interacts with PDE6D. Whereas we could readily detect the ARL3–PDE6D interaction, we were not able to detect physical interactions between ARL13B and PDE6D, even when both proteins were overexpressed (Fig. 3A and Fig. S3). In reciprocal immunoprecipitation, both ARL13B and PDE6D coprecipitated with endogenous INPP5E (Fig. 3B). GST pull-down assay using purified GST–ARL13B and in vitro translated INPP5E indicated that ARL13B directly interacts with INPP5E (Fig. 3C). In interaction domain mapping experiments, INPP5E mutants that possess the FDRELYL motif and localize to cilia (i.e., 1–626 and 79–644) maintained their ability to interact with ARL13B (Fig. 3D). Mutant variants that failed to localize to cilia were not able to interact with ARL13B. On the other hand, PDE6D–INPP5E interaction was dependent on the presence of the C-terminal CaaX motif but independent of the FDRELYL motif (Fig. 3E). The facts that INPP5E interacts with both ARL13B and PDE6D but an ARL13B–PDE6D interaction is not detectable and that ARL13B and PDE6D bind to close parts of INPP5E prompted us to test whether ARL13B can regulate the INPP5E–PDE6D interaction. Indeed, overexpression of ARL13B decreased the INPP5E–PDE6D interaction (Fig. 3F). Taken together, our data suggest that ARL13B facilitates ciliary targeting of INPP5E by directly interacting with INPP5E and promoting its release from PDE6D, rather than by interacting with PDE6D as proposed for the ARL3–PDE6D interaction (23).
Fig. 3.
Physical interactions between ARL13B, INPP5E, and PDE6D. (A and B) INPP5E interacts with ARL13B and PDE6D but not with ARL3. HEK293T cells were transfected with FLAG-tagged PDE6D, INPP5E, or ARL13B and cell lysates were subjected to immunoprecipitation (IP) using anti-FLAG antibody. Coprecipitation of endogenous ARL13B, ARL3, and INPP5E was examined. Untransfected cells were used as a negative control (CTRL). IB, immunoblotting. (C) ARL13B directly interacts with INPP5E. GST pull-down assay was conducted using in vitro translated HA-INPP5E and purified GST and GST–ARL13B proteins. GST and GST–ARL13B proteins were detected by Coomassie Blue (CB) staining and HA-INPP5E, by immunoblotting. (D and E) ARL13B- and PDE6D-interaction domain mapping in INPP5E. Various deletion mutants of FLAG-INPP5E were transfected into HEK293T cells and their interactions with endogenous ARL13B (D) and Myc-PDE6D (E) were examined. (F) Overexpression of ARL13B decreases INPP5E–PDE6D interaction. HEK293T cells were transfected as indicated and lysates were subjected to IP using anti-Myc antibodies. Arrowhead marks INPP5E.
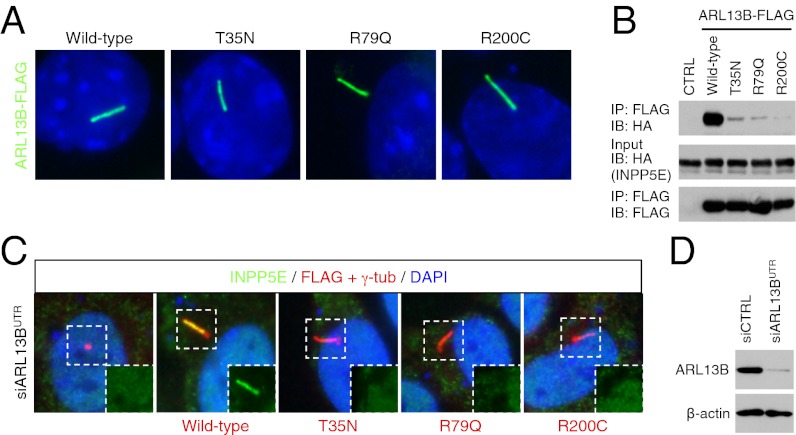
We then examined the impact of mutations found in human JBTS patients. Because all missense mutations found in INPP5E are within the catalytic domain and none of them disrupt INPP5E ciliary targeting (10), we focused our efforts on ARL13B. To date, two missense mutations in ARL13B, R79Q and R200C, have been found in JBTS patients (26). Whereas the precise molecular consequence of R200C mutation is unknown, the R79Q mutation is known to interfere with the GTP-binding activity of ARL13B. Neither of these mutations disrupted ciliary localization of ARL13B (Fig. 4A). We also generated an ARL13B variant with T35N mutation, which locks ARL13B in the GDP-bound form. Consistent with previous observations (28, 31), T35N mutation did not affect ARL13B ciliary localization. However, all of these mutations disrupted ARL13B–INPP5E interaction (Fig. 4B). To test whether these ARL13B mutants can mediate INPP5E ciliary trafficking, we blocked endogenous ARL13B expression using siRNA targeting its 3′ untranslated region (UTR) and then transfected FLAG-tagged ARL13B variants. As shown in Fig. 4C, wild-type ARL13B was able to restore ciliary localization of INPP5E, whereas mutant variants with T35N, R79Q, and R200C mutations failed to rescue INPP5E ciliary localization. These results indicate that although GDP-bound ARL13B localizes to cilia, only the GTP-bound form can interact with INPP5E and mediate its ciliary targeting. These findings also suggest that a trafficking defect of INPP5E is at least part of the molecular etiology of JBTS associated with ARL13B mutations.
Fig. 4.
ARL13B mutations that cause JBTS are not able to mediate INPP5E ciliary targeting. (A) Localization of ARL13B mutant variants. FLAG-tagged ARL13B variants (green) were transfected into IMCD3 cells and their localization was probed with anti-FLAG antibody. (B) GDP-locked ARL13B and JBTS-associated missense mutants show greatly reduced ability to bind to INPP5E. (C) ARL13B mutants fail to mediate INPP5E (green) ciliary targeting. Expression of endogenous ARL13B was ablated by siRNA targeting ARL13B 3′ UTR and indicated ARL13B variants with FLAG tags (red) were transfected. Insets show the green channel images of the boxed area. (D) Suppression of endogenous ARL13B expression by siRNA targeting ARL13B 3′ UTR used in C. β-actin was used as a loading control.
Identification of INPP5E Interacting Proteins.
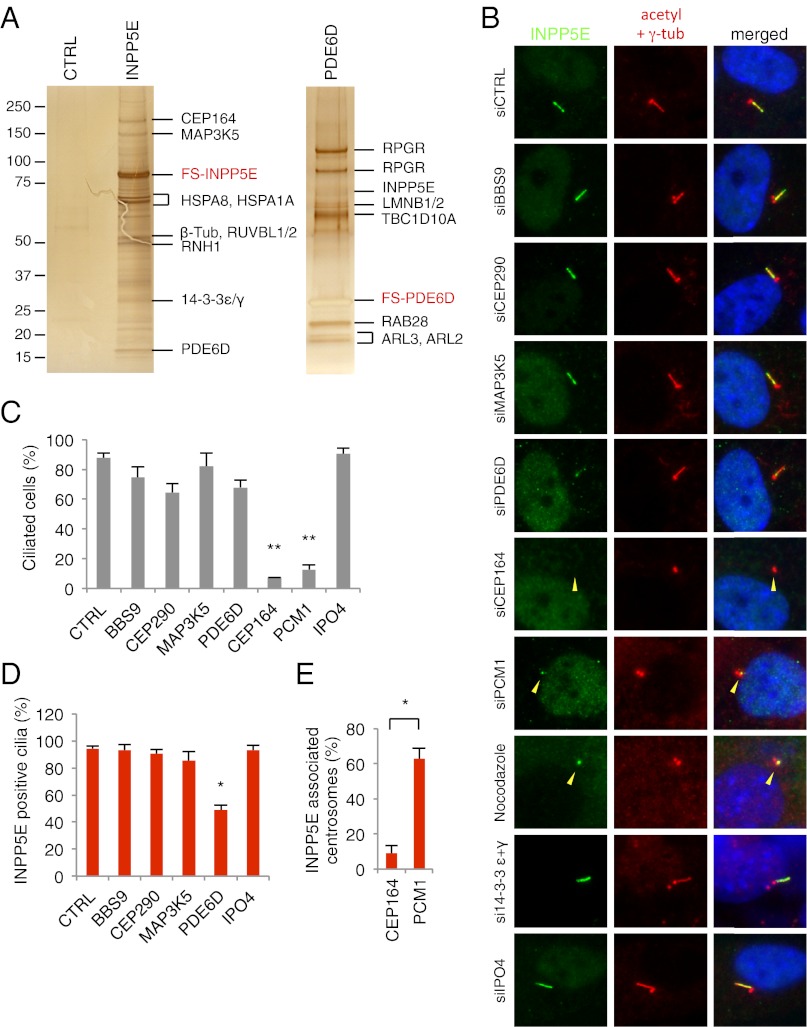
To better understand the protein interaction network around INPP5E and to identify other proteins involved in the same biological pathway, we performed tandem affinity purification (TAP) using INPP5E as bait. We generated a stable cell line expressing FLAG- and S-tagged INPP5E (FS-INPP5E) in HEK293T cells and purified INPP5E interacting proteins (Fig. 5A). Mass spectrometry analysis identified several proteins that are already known to localize to cilia and/or centrosomes (Dataset S1). For example, we found that centrosomal protein 164 (CEP164), RUVBL1, RUVBL2, and 14-3-3ε/γ proteins interact with INPP5E. CEP164 is a centriole distal appendage protein required for primary cilia formation (32, 33). CEP164 also localizes to the nucleus where it is involved in DNA damage response (34, 35). More recently, CEP164 was identified as one of the NPHP-related ciliopathy genes (36). RUVBL1 and RUVBL2 have been found in proteomic studies for mammalian cilia but their roles in centrosomes and/or cilia are unknown (37, 38). The 14-3-3 proteins were previously shown to interact with INPP5E and localize to cilia (11, 39), but their roles in cilia are also unknown. Additionally, we found that MAP3K5 and PDE6D were copurified with INPP5E. In a reciprocal TAP using PDE6D as bait, several prenylated proteins including RPGR (two isoforms), INPP5E, LMNB1, LMNB2, and RAB28 were copurified (Fig. 5A and Dataset S1). Membrane association and subcellular localization of these proteins are likely to be regulated by PDE6D. In addition to these prenylated proteins, small GTPases, ARL2 and ARL3, which are known to bind and regulate PDE6D (22, 24), were copurified. Finally, TBC1D10A, a GTPase-activating protein for RAB27A and RAB35 (40, 41), was copurified with PDE6D.
Fig. 5.
Identification of INPP5E interacting proteins and their roles in INPP5E ciliary targeting. (A) Tandem affinity purification (TAP) of INPP5E and PDE6D. Lysates from HEK293T cells stably expressing FLAG- and S-tagged INPP5E (FS-INPP5E) and PDE6D (FS-PDE6D) were subjected to TAP. Parental cells were used as a negative control. Purified proteins were visualized by silver staining. (B) Requirement of CEP164 and PDE6D for INPP5E ciliary targeting. RPE1 cells were transfected with siRNAs as indicated and localization of endogenous INPP5E (green) was evaluated. Yellow arrowheads indicate the location of centrosomes. (C) Quantitation of ciliated cells in B. Results are averages of at least two independent experiments (mean ± SEM). Double asterisks indicate statistically significant decreases compared with control (P < 0.01, one-way ANOVA). (D) Quantitation of INPP5E-positive cilia in B. Asterisk indicates a statistically significant decrease compared with control (P < 0.05, one-way ANOVA). (E) Comparison of INPP5E-associated centrosomes in CEP164 and PCM1 depleted, unciliated cells. Asterisk indicates statistical significance (P < 0.05, Student t test).
Roles of INPP5E Interacting Proteins in Ciliogenesis and INPP5E Ciliary Targeting.
Among the identified INPP5E interacting partners, we chose five proteins (CEP164, MAP3K5, PDE6D, IPO4, and 14-3-3ε) for further analyses. CEP164, 14-3-3ε, and PDE6D were chosen based on their known localization to cilia and/or centrosomes and interaction with INPP5E (32, 33, 39, and this study). IPO4, which is an importin β family member, was chosen for testing because recent data suggest that some of the importin β family proteins are involved in ciliary trafficking (42, 43). Recent studies also found that several kinases (e.g., LKB1, MEK1/2, ERK1/2, and AKT) localize to cilia and/or centrosomes and regulate various signaling events (44–46). Thus, we included MAP3K5 in our analyses to test whether it affects INPP5E ciliary localization.
First, we verified their interactions with INPP5E using coimmunoprecipitation (Fig. S4). Next, we examined the subcellular localization patterns of the INPP5E interacting partners (Fig. S5). Consistent with previous reports (32, 33), both endogenous and FLAG-tagged CEP164 specifically localized to the distal appendage of the mother centriole and often appeared as a doughnut-shaped structure, slightly larger than the centriole (Fig. S5 A and B). MAP3K5 and PDE6D were detected throughout the cell but specific enrichment was found around the centrosome (Fig. S5 C and D). Contrary to UNC119B, which is structurally similar to PDE6D and binds to the myristoyl group of NPHP3 to mediate its ciliary trafficking (25), we were not able to detect ciliary localization of PDE6D in hTERT–RPE1 cells. Consistent with the previous observation (39), we found a subset of 14-3-3 proteins localize to the primary cilium (Fig. S5E). IPO4 was found throughout the cytoplasm and we were not able to detect ciliary or centrosomal pools of IPO4 (Fig. S5F).
Next, we examined the requirement of INPP5E interacting partners for ciliogenesis and INPP5E ciliary targeting. Efficient suppression of gene expression by siRNA transfection was verified at the protein or mRNA level (Fig. S2). Depletion of MAP3K5, 14-3-3ε/γ, and IPO4 in hTERT–RPE1 cells did not cause noticeable changes in cilia formation or INPP5E localization (Fig. 5 B–D). However, depletion of PDE6D significantly decreased ciliary localization of INPP5E. Within the remaining INPP5E-positive cilia, the intensity of INPP5E staining was noticeably lower than in control siRNA transfected cells. Ciliogenesis itself was not significantly affected by PDE6D depletion. The requirement of PDE6D for INPP5E ciliary targeting was interesting because the N-terminal 626 residues of INPP5E, which do not contain the CaaX motif and do not interact with PDE6D, were sufficient to target INPP5E to cilia (Fig. 1). In addition, INPP5E amino acids 1–626 mutant localized to cilia in a PDE6D-independent manner (Fig. S1C), suggesting that PDE6D is required for ciliary targeting of full-length, prenylated INPP5E but not for solubilized INPP5E.
Contrary to PDE6D, ablation of CEP164 almost completely blocked cilia formation (Fig. 5 B and C). This complete loss of cilia made it difficult to evaluate the direct requirement of CEP164 for INPP5E ciliary targeting. Therefore, we compared INPP5E localization in CEP164-depleted cells with two other conditions where cilia formation is suppressed. Previously, it was shown that depletion of PCM1, which is essential for centriolar satellite formation (47, 48), blocks ciliogenesis in RPE1 cells (49, 50). Also, 2-h nocodazole treatment (20 μM) destabilizes cilia and causes cilia resorption in RPE1 cells (50). INPP5E was still found associated with one centriole of the centrosome in most PCM1-depleted, unciliated cells (∼63%) and essentially every cell in nocodazole-treated, unciliated cells (∼96%) (Fig. 5 B and E). In contrast, INPP5E was not associated with the centrosome in CEP164-depleted, unciliated cells (∼9%). Therefore, CEP164 appears to be required for ciliary targeting of INPP5E (potentially for INPP5E docking to the transition fiber) in addition to ciliogenesis. Consistent with the ARL6 (also known as BBS3) results (Fig. 2), knockdown of BBS9 did not affect INPP5E ciliary localization, suggesting that INPP5E ciliary targeting is not mediated by BBS proteins. Likewise, ablation of CEP290 did not affect INPP5E ciliary targeting.
Discussion
Recent studies have shown that many proteins involved in NPHP, MKS, and JBTS localize to the ciliary transition zone and function as modular components of a ciliary gate, regulating protein entry and exit to cilia (5–9). However, JBTS proteins INPP5E and ARL13B have not been linked to any of the known ciliary transition zone complexes, and their relationships with other ciliopathy proteins have been elusive. In this study, we demonstrate that INPP5E, ARL13B, PDE6D, and CEP164 form a functional interaction network that converges on INPP5E ciliary targeting. This interaction network is distinct from previously defined protein networks composed of transition zone-localizing ciliopathy proteins that likely form a supramolecular complex. Indeed, INPP5E and ARL13B localize to the primary cilium, CEP164 to the distal appendage, and PDE6D to the cytoplasm and near the centrosome. This indicates that ARL13B, CEP164, and PDE6D are involved in different steps of INPP5E ciliary targeting and that mutations in these genes cause JBTS and NPHP through different mechanisms than ciliary gating defects. Because loss of ARL13B, PDE6D, and CEP164 commonly causes a loss of INPP5E in cilia, reduction of INPP5E activity within cilia is likely to be a part of a common molecular etiology underlying the ciliopathies associated with these genes.
Based on our findings, we propose that INPP5E is targeted to the primary cilium by sequential interactions with PDE6D, CEP164, and ARL13B (Fig. S6). In this model, farnesylated INPP5E is initially confined to the donor membrane (e.g., endoplasmic reticulum, Golgi, or transport vesicles). PDE6D is required to extract INPP5E from the donor membrane and the PDE6D–INPP5E complex is transported to the basal body by a currently unknown mechanism. CEP164 may facilitate the docking of the INPP5E–PDE6D complex to the centrosomal distal appendage/transition fiber. ARL13B, including the GDP-bound form, predominantly localizes to cilia. Therefore, ARL13B may be activated by a currently unknown GEF within cilia. Activated ARL13B would bind to INPP5E and promote its release from PDE6D and subsequent ciliary entry.
This model of INPP5E ciliary targeting is in general similar to that of NPHP3, a myristoylated protein targeted to cilia by UNC119B-, ARL3-, and RP2-dependent mechanisms (25), but also shows several differences. First, whereas ARL3 regulates ciliary targeting of NPHP3 by interaction with UNC119B, ARL13B directly binds to the cargo INPP5E rather than the prenyl binding protein PDE6D. Interestingly, although ARL2 and ARL3 interact with PDE6D, ablation of their expression has no impact on INPP5E ciliary localization. Second, whereas N-terminal myristoylation of NPHP3 appears to be essential for its ciliary targeting, prenylation is dispensable for INPP5E ciliary targeting. For example, whereas C-terminally tagged NPHP3 can localize properly to cilia, N-terminally tagged NPHP3, myristoylation of which would be blocked by the tag, failed to localize to cilia (25). In contrast, removal of the CaaX motif from INPP5E does not prevent its ciliary localization, indicating that prenylation is not necessary for ciliary targeting. This also suggests that ciliary targeting of INPP5E is mainly mediated by its interactions with ARL13B and that PDE6D is required to extract farnesylated INPP5E from the donor membranes but not for ciliary targeting itself.
Although this model is the most likely, alternative models are conceivable. For example, although we do not observe obvious ARL13B accumulation in any potential donor membranes, we cannot rule out the possibility that a small subset of ARL13B localizes to the donor membrane, sorts INPP5E, and mediates INPP5E trafficking from the donor membrane to the ciliary base. Another interesting possibility is related to INPP5E interacting proteins involved in the DNA damage response (DDR) pathway (CEP164, DDB1, IPO4, RUVBL1, and RUVBL2) (36, 51). Mutations in CEP164 cause defects in the DDR pathway in addition to ciliogenesis defects. Interestingly, INPP5E phosphorylation is regulated by DNA damage (52) and some of the deletion mutants of INPP5E show nuclear localization. Although it is currently unknown whether a subset of endogenous INPP5E translocates into the nucleus in response to DNA damage and is involved in the DDR pathway, interactions of INPP5E with CEP164 and other DDR-related proteins may have relevance to this pathway.
Several other prenylated proteins localize to the primary cilia and the photoreceptor outer segment (e.g., RPGR, PDE6A, PDE6B, and GNGT1), and loss of these proteins or perturbation of their ciliary trafficking results in photoreceptor degeneration (12–18). Mechanisms that target these proteins to cilia and to the photoreceptor outer segment remain to be determined. Our study provides a model for ciliary targeting of prenylated proteins. Our study also presents a protein–protein interaction network of INPP5E that is linked to JBTS and NPHP and identifies previously unknown ciliopathy candidate genes.
Materials and Methods
Protein–protein interaction studies were performed using HEK293T cells and immunolocalization studies were conducted in hTERT–RPE1 and IMCD3 cells. Tandem affinity purification, coimmunoprecipitation, and immunofluorescence microscopy were conducted as previously described with minor modifications (49, 53). Antibodies were purchased from the following sources: Mouse monoclonal antibodies against acetylated tubulin (6-11B-1), γ-tubulin (GTU-88), FLAG (M2), and β-actin (AC-15) and rabbit polyclonal antibodies for BBS9 (HPA021289), CEP164 (HPA037605), γ-tubulin (T3559), and PCM1 (HPA023374) are from Sigma. Rabbit polyclonal antibodies against ARL3 (10961), ARL6 (12676), ARL13B (17711), INPP5E (17797), IPO4 (11679), RAB8A (55296), and RAB11A (20229) are from Proteintech Group. Other antibodies used are for MAP3K5 (Abgent; AJ1472b), 14-3-3 ε (Cell Signaling Technology; 9635), and CEP290 (Bethyl Laboratories; IHC-00365). Quantitative PCR primer sequences are shown in Table S1. For more details, please see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. G. Pazour and K. Kontani for providing RAB8A and ARL13B expression plasmids. This work was supported by the Pediatric Ophthalmology Career-Starter Research grant from the Knights Templar Eye Foundation (to S.S.) and National Institutes of Health Grants R01EY022616 (to S.S.) and R01EY110298 and R01EY017168 (to V.C.S.). V.C.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210916109/-/DCSupplemental.
References
- 1.Goetz SC, Anderson KV. The primary cilium: A signalling centre during vertebrate development. Nat Rev Genet. 2010;11(5):331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singla V, Reiter JF. The primary cilium as the cell’s antenna: Signaling at a sensory organelle. Science. 2006;313(5787):629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 3.Mockel A, et al. Retinal dystrophy in Bardet-Biedl syndrome and related syndromic ciliopathies. Prog Retin Eye Res. 2011;30(4):258–274. doi: 10.1016/j.preteyeres.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 4.van Reeuwijk J, Arts HH, Roepman R. Scrutinizing ciliopathies by unraveling ciliary interaction networks. Hum Mol Genet. 2011;20(R2):R149–R157. doi: 10.1093/hmg/ddr354. [DOI] [PubMed] [Google Scholar]
- 5.Williams CL, et al. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol. 2011;192(6):1023–1041. doi: 10.1083/jcb.201012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craige B, et al. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J Cell Biol. 2010;190(5):927–940. doi: 10.1083/jcb.201006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Gonzalo FR, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43(8):776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sang L, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145(4):513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chih B, et al. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol. 2012;14(1):61–72. doi: 10.1038/ncb2410. [DOI] [PubMed] [Google Scholar]
- 10.Bielas SL, et al. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat Genet. 2009;41(9):1032–1036. doi: 10.1038/ng.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacoby M, et al. INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nat Genet. 2009;41(9):1027–1031. doi: 10.1038/ng.427. [DOI] [PubMed] [Google Scholar]
- 12.Meindl A, et al. A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3) Nat Genet. 1996;13(1):35–42. doi: 10.1038/ng0596-35. [DOI] [PubMed] [Google Scholar]
- 13.Roepman R, et al. Positional cloning of the gene for X-linked retinitis pigmentosa 3: Homology with the guanine-nucleotide-exchange factor RCC1. Hum Mol Genet. 1996;5(7):1035–1041. doi: 10.1093/hmg/5.7.1035. [DOI] [PubMed] [Google Scholar]
- 14.Huang SH, et al. Autosomal recessive retinitis pigmentosa caused by mutations in the alpha subunit of rod cGMP phosphodiesterase. Nat Genet. 1995;11(4):468–471. doi: 10.1038/ng1295-468. [DOI] [PubMed] [Google Scholar]
- 15.McLaughlin ME, Sandberg MA, Berson EL, Dryja TP. Recessive mutations in the gene encoding the beta-subunit of rod phosphodiesterase in patients with retinitis pigmentosa. Nat Genet. 1993;4(2):130–134. doi: 10.1038/ng0693-130. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto S, Sippel KC, Berson EL, Dryja TP. Defects in the rhodopsin kinase gene in the Oguchi form of stationary night blindness. Nat Genet. 1997;15(2):175–178. doi: 10.1038/ng0297-175. [DOI] [PubMed] [Google Scholar]
- 17.Sohocki MM, et al. Mutations in a new photoreceptor-pineal gene on 17p cause Leber congenital amaurosis. Nat Genet. 2000;24(1):79–83. doi: 10.1038/71732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christiansen JR, Kolandaivelu S, Bergo MO, Ramamurthy V. RAS-converting enzyme 1-mediated endoproteolysis is required for trafficking of rod phosphodiesterase 6 to photoreceptor outer segments. Proc Natl Acad Sci USA. 2011;108(21):8862–8866. doi: 10.1073/pnas.1103627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazelova J, et al. Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. EMBO J. 2009;28(3):183–192. doi: 10.1038/emboj.2008.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward HH, et al. A conserved signal and GTPase complex are required for the ciliary transport of polycystin-1. Mol Biol Cell. 2011;22(18):3289–3305. doi: 10.1091/mbc.E11-01-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geng L, et al. Polycystin-2 traffics to cilia independently of polycystin-1 by using an N-terminal RVxP motif. J Cell Sci. 2006;119(Pt 7):1383–1395. doi: 10.1242/jcs.02818. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, et al. Photoreceptor cGMP phosphodiesterase delta subunit (PDEdelta) functions as a prenyl-binding protein. J Biol Chem. 2004;279(1):407–413. doi: 10.1074/jbc.M306559200. [DOI] [PubMed] [Google Scholar]
- 23.Ismail SA, et al. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat Chem Biol. 2011;7(12):942–949. doi: 10.1038/nchembio.686. [DOI] [PubMed] [Google Scholar]
- 24.Linari M, Hanzal-Bayer M, Becker J. The delta subunit of rod specific cyclic GMP phosphodiesterase, PDE delta, interacts with the Arf-like protein Arl3 in a GTP specific manner. FEBS Lett. 1999;458(1):55–59. doi: 10.1016/s0014-5793(99)01117-5. [DOI] [PubMed] [Google Scholar]
- 25.Wright KJ, et al. An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev. 2011;25(22):2347–2360. doi: 10.1101/gad.173443.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantagrel V, et al. International Joubert Syndrome Related Disorders Study Group Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am J Hum Genet. 2008;83(2):170–179. doi: 10.1016/j.ajhg.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12(5):767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Duldulao NA, Lee S, Sun Z. Cilia localization is essential for in vivo functions of the Joubert syndrome protein Arl13b/Scorpion. Development. 2009;136(23):4033–4042. doi: 10.1242/dev.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cevik S, et al. Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J Cell Biol. 2010;188(6):953–969. doi: 10.1083/jcb.200908133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Wei Q, Zhang Y, Ling K, Hu J. The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J Cell Biol. 2010;189(6):1039–1051. doi: 10.1083/jcb.200912001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hori Y, Kobayashi T, Kikko Y, Kontani K, Katada T. Domain architecture of the atypical Arf-family GTPase Arl13b involved in cilia formation. Biochem Biophys Res Commun. 2008;373(1):119–124. doi: 10.1016/j.bbrc.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Graser S, et al. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179(2):321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sillibourne JE, et al. Assessing the localization of centrosomal proteins by PALM/STORM nanoscopy. Cytoskeleton (Hoboken) 2011;68(11):619–627. doi: 10.1002/cm.20536. [DOI] [PubMed] [Google Scholar]
- 34.Sivasubramaniam S, Sun X, Pan YR, Wang S, Lee EY. Cep164 is a mediator protein required for the maintenance of genomic stability through modulation of MDC1, RPA, and CHK1. Genes Dev. 2008;22(5):587–600. doi: 10.1101/gad.1627708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan YR, Lee EY. UV-dependent interaction between Cep164 and XPA mediates localization of Cep164 at sites of DNA damage and UV sensitivity. Cell Cycle. 2009;8(4):655–664. doi: 10.4161/cc.8.4.7844. [DOI] [PubMed] [Google Scholar]
- 36.Chaki M, et al. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell. 2012;150(3):533–548. doi: 10.1016/j.cell.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Q, et al. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6(8):1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishikawa H, Thompson J, Yates JR, 3rd, Marshall WF. Proteomic analysis of mammalian primary cilia. Curr Biol. 2012;22:414–419. doi: 10.1016/j.cub.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan S, et al. Polarity proteins control ciliogenesis via kinesin motor interactions. Curr Biol. 2004;14:1451–1461. doi: 10.1016/j.cub.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 40.Itoh T, Fukuda M. Identification of EPI64 as a GTPase-activating protein specific for Rab27A. J Biol Chem. 2006;281(42):31823–31831. doi: 10.1074/jbc.M603808200. [DOI] [PubMed] [Google Scholar]
- 41.Hsu C, et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 2010;189(2):223–232. doi: 10.1083/jcb.200911018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dishinger JF, et al. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol. 2010;12(7):703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hurd TW, Fan S, Margolis BL. Localization of retinitis pigmentosa 2 to cilia is regulated by Importin beta2. J Cell Sci. 2011;124(Pt 5):718–726. doi: 10.1242/jcs.070839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boehlke C, et al. Primary cilia regulate mTORC1 activity and cell size through Lkb1. Nat Cell Biol. 2010;12(11):1115–1122. doi: 10.1038/ncb2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider L, et al. PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Curr Biol. 2005;15:1861–1866. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 46.Christensen ST, Clement CA, Satir P, Pedersen LB. Primary cilia and coordination of receptor tyrosine kinase (RTK) signalling. J Pathol. 2012;226(2):172–184. doi: 10.1002/path.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159(2):255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopes CA, et al. Centriolar satellites are assembly points for proteins implicated in human ciliopathies, including oral-facial-digital syndrome 1. J Cell Sci. 2011;124(Pt 4):600–612. doi: 10.1242/jcs.077156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seo S, et al. A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and Smoothened. PLoS Genet. 2011;7(11):e1002358. doi: 10.1371/journal.pgen.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J, Krishnaswami SR, Gleeson JG. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum Mol Genet. 2008;17(23):3796–3805. doi: 10.1093/hmg/ddn277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, et al. CCAAT/enhancer binding protein delta (C/EBPdelta, CEBPD)-mediated nuclear import of FANCD2 by IPO4 augments cellular response to DNA damage. Proc Natl Acad Sci USA. 2010;107(37):16131–16136. doi: 10.1073/pnas.1002603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 53.Seo S, et al. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc Natl Acad Sci USA. 2010;107(4):1488–1493. doi: 10.1073/pnas.0910268107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.