Abstract
Glycoprotein E (gE) of HSV plays a key role in cell-to-cell spread and virus-induced cell fusion. Here, we report that this function of gE requires the cooperation of tegument proteins UL11, UL16, and UL21. We found that the four proteins come together with very high efficiency to form a complex in transfected cells and in a manner that is regulated and coordinated. In particular, the inefficient interaction of UL16 with each membrane protein (UL11 and gE) observed in pairwise transfections became efficient when other binding partners were present. The significance of these interactions was revealed in studies of viral mutants, which showed that each of these tegument proteins is critical for processing, transport, and biological activity of gE. These findings provide insights into the mechanisms of how gE executes its function and also have implications in understanding HSV assembly and budding.
HSV uses direct cell-to-cell transmission to spread laterally across cell junctions, and this is critical for efficient establishment of latent infections in neuronal cells (reviewed in refs. 1, 2). The molecular mechanism of this mode of transmission is poorly understood, but it is known to share some mechanistic details with the entry of extracellular virions (1), namely the use of the essential core fusion machinery, consisting of glycoproteins B, D, H, and L (gB, gD, gH, and gL, respectively). Moreover, it requires glycoproteins E and I (gE and gI, respectively) (1), which are not necessary for “cell-free” transmission (3). Consequently, HSV mutants lacking gE/gI are severely restricted for spread to neuronal cells (3–11). Remarkably, along with the core entry proteins, the gE/gI heterodimer is also required for HSV-induced cell fusion to produce syncytia (3, 12, 13). In cell cultures, WT HSV-1 rarely induces syncytia; however, mutations in genes encoding gB, gK, UL20, or UL24 can give rise to this property (14). In addition to a single syn mutation, syncytia formation requires the core fusion machinery, gE/gI, and membrane proteins gM and UL45 (13, 15). As a key mediator in cell-to-cell spread and cell fusion, the mechanism of how gE executes its function remains elusive. Here we show that tegument proteins UL11, UL16, and UL21 are also required.
Tegument proteins are those viral proteins that are located between the capsid and the viral envelope, and many are known to interact with the cytoplasmic tails of envelope glycoproteins (2, 16). There are three tegument proteins known to bind to gE: UL11, UL16, and VP22 (17–20). UL11 and UL16 were investigated here because they bind each other as well.
UL11 is a small (96 aa), peripherally bound membrane protein (21–23) (Fig. 1A) that is a critical player in the cytoplasmic envelopment of capsids (24). Its interaction with the gE tail is direct and efficient in vitro and in vivo. Moreover, these proteins depend on each other for their incorporation into virion particles (19).
Fig. 1.
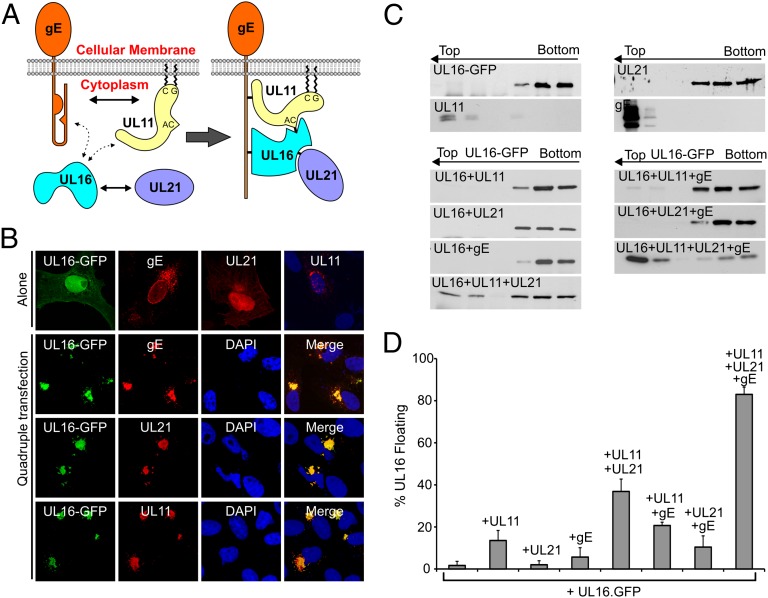
UL11, UL16, UL21 and gE form a complex in transfected cells. (A) Summary of the known interactions among UL11, UL16, UL21, and gE that have been identified by in vitro assays (Left). The dotted lines represent weak or inefficient interactions whereas the solid lines represent efficient interactions in pairwise transfection assays. The four proteins are proposed to assemble into a complex in mammalian cells (Right). (B) Top row: subcellular distribution of UL16-GFP, gE, UL21, or UL11 when each is expressed alone. The lower three rows show the locations of proteins in quadruple cotransfections. (C) Membrane flotation analysis of UL16. Vero cells expressing UL16-GFP alone or in different combinations with UL11, UL21, and gE were osmotically disrupted, and the ability of the proteins to float to the upper fractions of sucrose step gradients during centrifugation was examined. Representative immunoblots are shown. The tops and bottoms of the gradients are indicated, along with the direction of flotation (arrows). (D) Densitometry was used to quantitate immunoblots from three different experiments, and the results are shown as the percentage of floating proteins (top three fractions) relative to the total proteins (all fractions).
UL16 is found in the cytoplasm and nucleus of infected cells, as well as on the cytoplasmic capsids (25). This protein was the first to be identified as a binding partner of UL11 (26) (Fig. 1A). Although the interaction is robust in vitro (e.g., in pull-down assays with GST-UL11), UL16 poorly recognizes UL11 in cotransfected cells (26). Specifically, most of the UL16 molecules continue to be distributed throughout the cytoplasm and nucleus with only a small population relocalized to the site of UL11 accumulation. Like UL11, UL16 also interacts with the gE tail but at a different binding site (18). The UL16–gE interaction is also highly inefficient in vivo, and there is negligible colocalization when these two proteins are coexpressed, although interactions can be detected by in vitro binding assays (18).
We hypothesize that the disruption of cells triggers UL16 to undergo conformational changes that enable binding to UL11 or gE in vitro, but these interactions are normally subject to regulation in vivo. Indeed, the interactions of UL16 with gE and UL11 are dramatically enhanced upon removal of a large C-terminal segment of UL16 that appears to negatively regulate binding (18, 27). Thus, it seemed likely that other viral proteins might serve as the normal activator to enable these interactions. One such candidate is tegument protein UL21 (Fig. 1A), another binding partner of UL16 that is also distributed throughout the infected cells and found to be associated with capsids (28, 29). The experiments described here show that the assembly of UL11, UL16, and UL21 onto the tail of gE is an orderly process that is essential for syncytia formation.
Results
UL11, UL16, UL21 and gE Form a Quadruple Complex in Transfected Vero Cells.
To test whether UL11, UL16, UL21, and gE form a complex in vivo, plasmids encoding these proteins were transfected into Vero cells individually or all together (Fig. 1B). Individually, UL16-GFP and UL21 were distributed throughout the cell but mostly in the nucleus. In contrast, gE was associated with the nuclear membrane and speckles in the cytoplasm that resemble the Golgi and ER compartments, whereas UL11 accumulated at the TGN, as expected. In contrast, when all four were coexpressed, they colocalized. UL16 and UL21 underwent massive redistribution. They were completely absent from the nucleus and were relocalized to a juxtanuclear position. Dramatic redistribution was also observed for gE, which moved away from the nuclear membrane and accumulated at the same juxtanuclear location as the other three proteins. Quadruple staining of individual cells was not possible because of the lack of suitable reagents. The affinity of UL11, UL16, UL21, and gE for one another was also emphasized by the high efficiency with which UL16 was associated with membranes as measured by membrane flotation experiments (Fig. 1 C and D). These data suggest that the four proteins have a remarkable capacity to assemble in vivo.
UL21 Activates UL11–UL16 Interaction.
In stark contrast to the quadruple transfection, UL11 only partially colocalized with UL16-GFP in doubly transfected Vero cells (26) (Fig. 2 A and B), suggesting that UL21, gE, or both is necessary for activation of this interaction. UL21 seemed to be a good candidate because it has been shown to interact with UL16 but not UL11 in immunoprecipitation and GST pull-down assays (29). Indeed, in the triple transfection, UL16 and UL21 were efficiently relocalized to the cytoplasmic location of UL11 (Fig. 2C). Neither protein was found in the nucleus, and membrane-associated UL16 was greatly enhanced (Fig. 1 C and D).
Fig. 2.
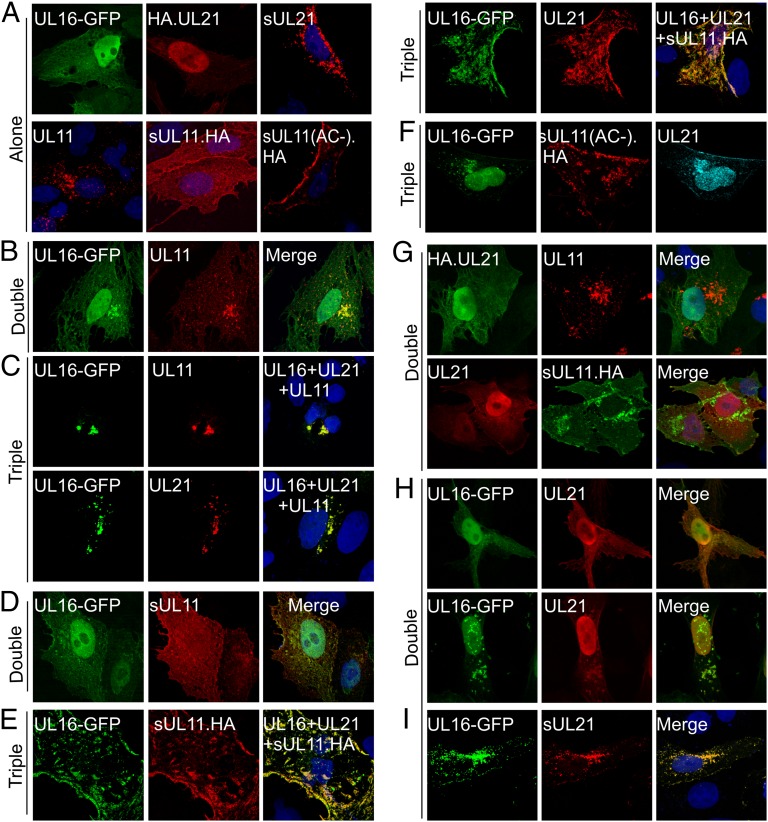
UL21 activates the UL11–UL16 interaction. Vero cells were transfected to express the indicated proteins, either alone (A), pairwise (B, D, G, H, and I), or in combinations of three (C, E, and F). At 16 to 18 h after transfection, the cells were fixed, stained with the appropriate antibodies, and examined by confocal microscopy. For double transfections, the merged images are shown in the right panels. For triple transfections, the right panels indicate which proteins were coexpressed and show the images for the two being analyzed.
The recruitment of UL16 to membranes was further measured by a relocalization assay with sUL11.HA, a chimera of UL11 that has the 12-aa membrane anchor from the Src oncoprotein at its N terminus. This peptide targets UL11 to the plasma membrane (22) (Fig. 2A). Accumulation is more evident when the endocytosis signal of UL11, namely the acidic cluster (AC), which also is the site for UL16 binding (26), is removed [sUL11(AC−).HA; Fig. 2A]. As expected, sUL11.HA did not relocalize UL16-GFP to the plasma membrane (Fig. 2D); however, in the presence of UL21, UL16 was completely colocalized with sUL11.HA (Fig. 2E). Moreover, UL16 and UL21 were no longer found in the nucleus. In contrast, UL16-GFP failed to respond to sUL11(AC-).HA (Fig. 2F), demonstrating that relocalization is dependent on its specific recognition of UL11 and not the Src peptide.
UL21 Activates UL11–UL16 Interaction Through Binding to UL16.
We presumed that activation of UL16 requires binding to UL21 rather than UL11 based on the results from in vitro assays (29), but these interactions have never been tested in vivo. Cotransfections confirmed that neither UL11 nor sUL11.HA could relocalize UL21 (Fig. 2G). Unfortunately, coexpression of UL16 and UL21 gave little insight into how well they find each other because the two proteins are normally localized to the same compartments, but in a small portion (10–20%) of cells, UL16-GFP and UL21 were found to aggregate in the nucleus and cytoplasm to form puncta containing both proteins (Fig. 2H). To better visualize the interaction, UL21 was tagged at its N terminus with the Src membrane anchor. This prevented UL21 from accumulating in the nucleus, and it was instead found in the cytoplasm (Fig. 2A). When UL16-GFP was coexpressed, it was completely relocalized to the sites where sUL21 accumulated (Fig. 2I), indicating a very efficient interaction between the two proteins. Thus, it appears that binding to UL21 induces the change in UL16 that reveals its UL11-binding site.
UL11, but Not UL21, Is Required to Activate gE-UL16 Interaction.
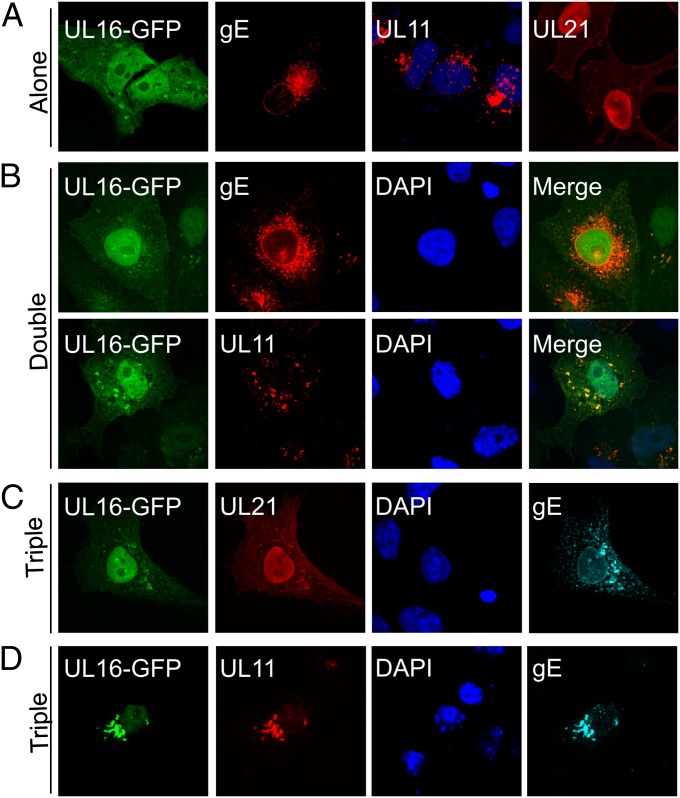
We hypothesized that the structural change induced by UL21 would also enable the interaction of UL16 with gE, which is otherwise highly inefficient in vivo (Fig. 3B). Surprisingly, this was not the case. In the triple transfection, only a very small fraction of UL16-GFP was relocalized to gE, with the majority remaining in the nucleus (Fig. 3C). Having previously shown that UL11 interacts efficiently with gE (19), we tested the hypothesis that this would enable UL16 to be recruited. Indeed, UL16-GFP was relocalized to a juxtanuclear location (Fig. 3D), even though it poorly interacts with UL11 or gE in pairwise transfections (Fig. 3B). Colocalization was not as robust as that seen for the induction of UL11-binding by UL21, as some UL16 was still visible in the nucleus. The membrane-association efficiency of UL16 was only a slightly higher than that in UL11–UL16 double transfections (Fig. 1 C and D), suggesting that the complex may not be very stable during centrifugation. In any case, it is clear that UL11 is needed to activate the UL16-gE interaction.
Fig. 3.
UL11 is required to activate the gE–UL16 interaction. Vero cells were transfected to express the indicated proteins alone (A), pairwise (B), or in combinations of three (C and D). At 16 to 18 h after transfection, the cells were fixed, stained with the appropriate antibodies, and examined by confocal microscopy. For triple transfections, the panels show the images of individual proteins being analyzed in the same cell.
Tegument Protein Mutants Fail to Induce Syncytia in Vero Cells.
Because gE is required for virus-induced cell fusion (3, 12, 13), we hypothesized that this function would be inhibited in the absence of UL11, UL16, or UL21. To test this, we first converted the HSV KOS strain into a syncytial mutant by introducing a substitution (A855V) in the cytoplasmic tail of gB to generate virus gBsyn (13, 30) and then constructed gE-null and gE tail-deletion mutants (ΔgE/gBsyn and gEΔCT/gBsyn) on this genetic background. Consistent with previous reports (13), virus gBsyn induced extensive cell fusion in Vero cells, and this phenotype was abolished by deletion of gE (ΔgE/gBsyn; Fig. 4A). Furthermore, the cytoplasmic tail of gE was also essential as its removal resulted in a block to syncytia formation. When the tail was restored, the syncytia phenotype was rescued.
Fig. 4.
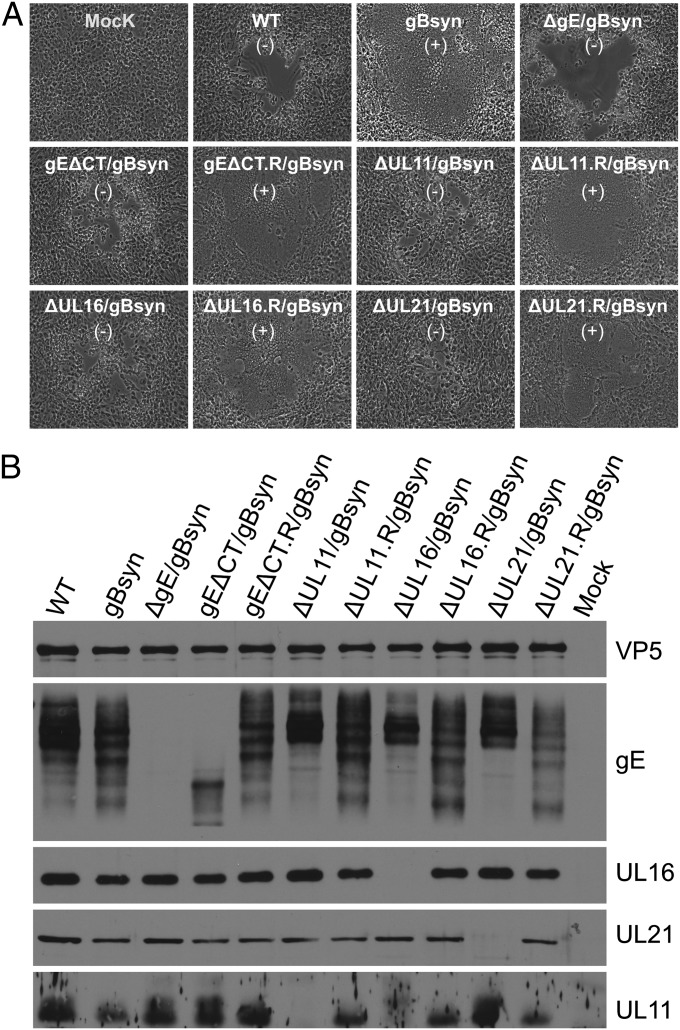
UL11, UL16, and UL21 are needed for fusion of Vero cells by a gBsyn mutant. (A) Vero cells were infected at a multiplicity of infection (MOI) of 0.01 with WT HSV or Syn mutants. Virus-induced cytopathic effect was monitored daily and recorded by an inverted microscope. (B) Vero cells were infected with WT HSV or mutants at an MOI of 5. At 18 to 24 h after infection, the cells were harvested, lysed in sample buffer, subjected to electrophoresis in denaturing gels, and immunoblotted with indicated antibodies. Capsid protein VP5 served as a loading control. The presence (+) and absence (−) of syncytia is indicated.
To test the importance of the three tegument proteins in cell fusion, we individually removed each coding sequence from the gBsyn virus to create mutants ΔUL11/gBsyn, ΔUL16/gBsyn, and ΔUL21/gBsyn. On Vero cells, these mutants behaved like the gE tail mutant (gEΔCT/gBsyn), inducing lytic infections rather than syncytia (Fig. 4A). Based on the analysis of more than 200 sites of infection for each mutant, we found that all were essentially unable to make syncytia (reduced 99.9% for ΔUL16/gBsyn, 99.7% for ΔUL11/gBsyn, and 99.0% for ΔUL21/gBsyn). When the missing coding sequences were inserted back into these mutants (ΔUL11.R/gBsyn, ΔUL16.R/gBsyn, and ΔUL21.R/gBsyn), the Syn phenotype was fully restored (Fig. 4A). Interestingly, the phenotypes exhibited by these mutant viruses were also reflected in an altered processing pattern of gE in infected Vero cells (Fig. 4B). The UL11, UL16, and UL21 deletion mutants, all of which produced lytic infections, behaved like WT HSV in exhibiting an abundance of slower migrating, mature forms of gE, whereas the revertants behaved like the gBsyn parent (Fig. 4B).
UL11–UL16–UL21 Complex Is Critical for Accumulation of gE on Plasma Membrane of Infected Vero Cells.
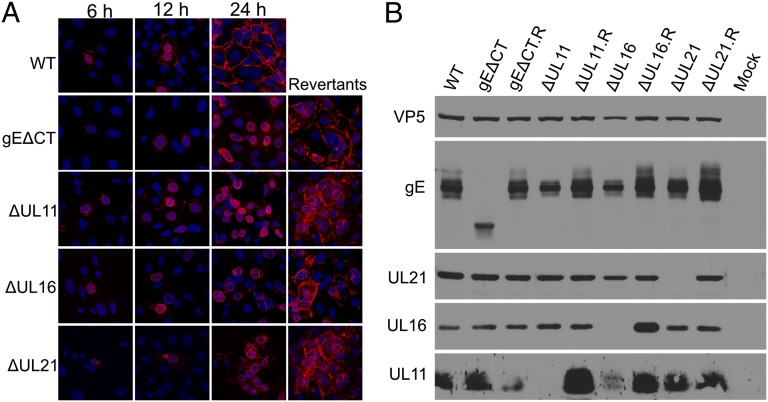
Syncytia formation and cell-to-cell spread have been suggested to require expression of gE at cell junctions, which occurs at the later stages of HSV infection (31) (Fig. 5A). Moreover, this occurs even when virion production is blocked (31) (Fig. S1). In Vero cells, surface accumulation was found to require the cytoplasmic tail, and, in its absence (i.e., mutant gEΔCT), gE was instead found on the nuclear membrane (Fig. 5A). Replacement of the tail (i.e., gEΔCT.R) confirmed that a mutation elsewhere in the genome was not responsible.
Fig. 5.
gE fails to accumulate on the surface of infected Vero cells in the absence of its binding partners. (A) Vero cells were infected at an MOI of 0.01 with WT or the indicated mutant and revertant viruses. At 18 to 24 h after infection, the cells were fixed and reacted with a mouse monoclonal antibody (clone 3114) to gE before microscopy. (B) Vero cells were infected with WT or mutants at an MOI of 5. At 18 to 24 h after infection, the cells were harvested, lysed in sample buffer, subjected to electrophoresis in denaturing gels, and immunoblotted with indicated antibodies. Capsid protein VP5 was used as the loading control.
To test whether the triplex of binding partners is required for gE accumulation on the cell surface, individual null mutants (ΔUL11, ΔUL21, ΔUL16), and the corresponding revertant viruses (ΔUL11.R, ΔUL21.R, ΔUL16.R) were constructed. These mutants all express WT gB. In contrast to the dramatic differences in gE processing observed in the context of the gBsyn background (Fig. 4B), those observed in the context with WT gB were much less obvious in Vero cells infected by mutants lacking UL11, UL16, or UL21 (Fig. 5B). However, the mutants exhibited a dramatic failure to accumulate gE on the cell surface as determined by immunofluorescence analysis (Fig. 5A) and flow cytometry analyses (Fig. S2). This was most striking for ΔUL11 and ΔUL16, but the ΔUL21 mutant also exhibited greatly reduced cell surface expression of gE. Cell surface accumulation of gE was also examined for the above mutants in the context of the gBsyn background, and the results were the same (Fig. S3A).
We also examined the subcellular localization of each of the three tegument proteins. In strong support for their active role in cell fusion, UL11, UL16, and UL21 (like gE) were all found on the WT HSV-infected Vero cell surface or at cell junctions (Fig. S4A). Moreover, these tegument proteins were responsive to the absence of the gE tail and to each other in a manner leading to dramatically reduced cell surface expression of UL11 and increased nuclear retention of UL16 and UL21. We conclude that the four proteins work together, and the reduced accumulation of gE on the surface of Vero cells might explain the absence of syncytia.
Tegument Binding Partners Are Needed for Cell Fusion Even When gE Is Surface-Expressed.
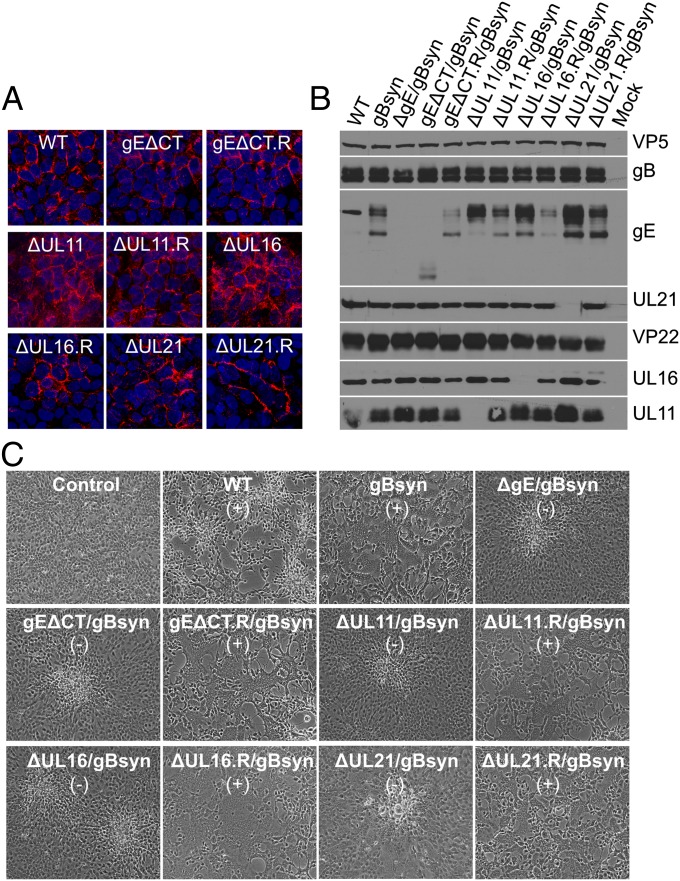
In HaCaT cells, gE accumulates at cell junctions even when its cytoplasmic tail is absent (32). This was also found to be true for viruses gEΔCT (Fig. 6A) and gEΔCT/gBsyn (Fig. S3B), as described here. However, gEΔCT/gBsyn still failed to induce cell–cell fusion of HaCaT cells (Fig. 6C), suggesting that the tail is required for gE function and not merely for trafficking. Therefore, we predicted that the triplex of tegument proteins would also be needed.
Fig. 6.
The Syn phenotype is lost in HaCaT cells even though gE accumulates on the cell surface when its binding partners are absent. (A) HaCaT cells grown on coverslips were infected with WT or the indicated mutant viruses at an MOI of 0.01. At 24 h after infection, the cells were fixed and reacted with a mouse monoclonal antibody (clone 3114) to gE before microscopy. (B) HaCaT cells were infected with WT or mutants viruses at an MOI of 5. At 18 to 24 h after infection, the cells were harvested, lysed in sample buffer, subjected to electrophoresis in denaturing gels, and reacted with the indicated antibodies. VP5 was used as a loading control. (C) HaCaT cells grown in six-well plates were infected with WT or Syn mutant viruses at low MOI. Virus-induced cytopathic effect was monitored daily and recorded by an inverted microscope. The presence (+) and absence (−) of syncytia is indicated.
UL11, UL16, and UL21 were not required for accumulation of gE on the surface of HaCaT cells (Fig. 6A). The substitution in gB to create gBsyn had no effect either (Fig. S3B). Similar to what was observed in Vero cells, these tegument proteins required each other and gE for correct and efficient localization (Fig. S4B). However, despite having gE at cell junctions (Fig. 6A), all the mutants still failed to induce syncytia but instead produced a small plaque phenotype of cell rounding and clumping (Fig. 6C) at >99% of the sites of infection. Concurrently, the gE processing pattern was also affected in the absence of each tegument protein (Fig. 6B). These results suggest that the interactions with the tegument protein partners are necessary for the function of gE.
Discussion
Because UL11 and UL16 (and perhaps UL21) have homologues in all the herpesviruses (33), studies of this interaction network are highly relevant to our understanding of these important pathogens. The experiments described here have revealed two salient findings: (i) these three tegument proteins and gE come together to form a highly efficient complex in vivo through coordinated interactions and (ii) the function of gE requires assembly of UL11, UL16, and UL21 on its cytoplasmic tail. The relevant implications are discussed in the subsequent sections.
Insights into the Regulation of Cell Fusion and Cell-to-Cell Spread.
HSV-induced cell fusion is a complex process. It also is highly related to cell-to-cell spread in that both require strikingly similar viral components and occur at cell interfaces. The mechanisms of these two processes are undoubtedly far more complicated than currently appreciated. In particular, the core fusion machinery provided by gB, gD, and gH/gL is fully capable of fusing cells to produce syncytia when expressed alone via transfection (34), but not within the context of virus infection, suggesting the existence of a regulatory network. All previously identified viral proteins required for the Syn phenotype are membrane proteins (13, 14). This report shows that non–membrane-bound tegument proteins (e.g., UL16 and UL21) are also required. Furthermore, these tegument proteins appear to modulate the function of gE.
The regulation of gE was seen at multiple levels. Beginning with synthesis, we observed that the oligosaccharide processing of gE was affected by disruption of the UL11–UL16–UL21 triplex. This was particularly evident in the context of the Syn background. The mechanism and significance of this are unclear, but the effects on glycosylation suggest that the four proteins travel together as a complex along the secretory pathway. The tegument proteins were also found to be needed for the accumulation of gE on the plasma membrane, although in a cell type-dependent manner. It is not clear how these binding partners promote surface expression in Vero cells because gE was found only on internal membranes in the quadruple transfections. Whether the triplex plays an active role in transporting gE to the cell surface or in blocking endocytosis, it is clear that yet other viral proteins must also be involved. In any case, UL11, UL16, and UL21 seem to play an active role in the cell fusion mechanism. When any of these was absent, the production of syncytia was blocked in HaCaT cells, where gE still accumulated at cell junctions. However, we cannot rule out the possibility that the triplex of tegument proteins works independently of gE by forming complexes on the tails of other viral membrane proteins. For example, there is an unconfirmed report that UL11 binds to the tail of gD (17). If so, these three tegument proteins might directly regulate the core fusion machinery.
Whether the triplex of tegument proteins triggers a change in the external domain of gE or provides an additional function in the cytoplasm is unclear. Previous studies have shown that antibodies against gE can block cell fusion induced by syncytial strains (12). Also, changes in the external domain of gE have been found that block cell-to-cell spread (8). On the contrary, in the prevailing model for HSV cell-to-cell spread (1, 2), vesicles carrying mature virions are specifically transported to lateral cell junctions. The limiting membranes of these vesicles are thought to contain gE, with its tail extended into the cytoplasm. Alterations in the tail of gE reduce the numbers of virions at cell junctions, and they instead accumulate on apical surfaces (2, 6, 17, 35). This misrouting results in smaller plaques and reduces epithelial cell-to-cell spread in vivo and in vitro. UL11, UL16, and UL21 may play a role in trafficking of vesicles by binding the gE tail. Consistent with this idea, mutants lacking UL11, UL16, or UL21 form small plaques similar to gEΔCT in Vero and HaCaT Cells. Moreover, Rich1, a host protein that has been proposed to be part of a sorting mechanism that enables transport of vesicles from recycling endosomes to tight junctions (36), has been found to be associated with native UL16 complexes (18).
Implications for HSV Assembly and Budding.
This report reveals a striking example of the highly efficient and coordinated assembly of tegument proteins (UL11, UL16, and UL21) on the tail of a single glycoprotein (gE). Moreover, previous studies have extended this interaction network to the capsid (25, 28). Notably, all four proteins have been shown to be involved in secondary envelopment (2, 16) and are interdependent for their incorporation into virion particles (19, 28). Our results suggest a model for how these proteins might contribute to virus assembly and egress. UL16 and UL21 have been found to be associated with cytoplasmic capsids (25, 28), and we propose that their interaction induces a conformational change of UL16 to expose the UL11-binding site located within its N-terminal sequence. Consistent with this, deletion of C-terminal residues 156 to 373 enables the remaining N-terminal fragment of UL16 to bind to UL11 in the absence of UL21 (27). An interaction of UL21 with microtubules has been proposed to aid transport of capsids to cytoplasmic membranes for budding (37). As capsids approach the membrane, the activated UL16 molecules are ready to bind to UL11. In this model, UL11 binds to the tail of gE to expose its distinct UL16-binding site. Thus, capsid-bound UL16 becomes linked to two different membrane proteins. Alternatively, the four proteins may assemble as a complex on membranes before capsids arrive. In either case, the data suggest an orchestrated mechanism of assembly that warrants vigorous investigation.
Materials and Methods
Recombinant HSV mutants were generated by using a bacterial artificial chromosome (BAC) containing the HSV-1 KOS strain genome. The detailed procedures have been reported previously (19). Details on the construction of specific viruses and plasmids, transfections, antibodies, FACS analysis, immunofluorescence, and membrane flotation assay are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. David C. Johnson, Harvey M. Friedman, Richard Courtney, and David A. Leib for providing reagents; the Penn State Hershey College of Medicine core facilities for flow cytometry, confocal microscopy, and oligo synthesis; and Carol Wilson for excellent technical assistance. This study was supported by National Institutes of Health (NIH) Grant AI071286 (to J.W.W.) and NIH Training Grant T32 CA60395 (to J.L.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.W.E. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212900109/-/DCSupplemental.
References
- 1.Johnson DC, Huber MT. Directed egress of animal viruses promotes cell-to-cell spread. J Virol. 2002;76(1):1–8. doi: 10.1128/JVI.76.1.1-8.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson DC, Baines JD. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol. 2011;9(5):382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 3.Balan P, et al. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J Gen Virol. 1994;75(Pt 6):1245–1258. doi: 10.1099/0022-1317-75-6-1245. [DOI] [PubMed] [Google Scholar]
- 4.Dingwell KS, et al. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol. 1994;68(2):834–845. doi: 10.1128/jvi.68.2.834-845.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dingwell KS, Doering LC, Johnson DC. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J Virol. 1995;69(11):7087–7098. doi: 10.1128/jvi.69.11.7087-7098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson DC, Webb M, Wisner TW, Brunetti C. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J Virol. 2001;75(2):821–833. doi: 10.1128/JVI.75.2.821-833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGraw HM, Friedman HM. Herpes simplex virus type 1 glycoprotein E mediates retrograde spread from epithelial cells to neurites. J Virol. 2009;83(10):4791–4799. doi: 10.1128/JVI.02341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polcicova K, Goldsmith K, Rainish BL, Wisner TW, Johnson DC. The extracellular domain of herpes simplex virus gE is indispensable for efficient cell-to-cell spread: Evidence for gE/gI receptors. J Virol. 2005;79(18):11990–12001. doi: 10.1128/JVI.79.18.11990-12001.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saldanha CE, et al. Herpes simplex virus type 1 glycoprotein E domains involved in virus spread and disease. J Virol. 2000;74(15):6712–6719. doi: 10.1128/jvi.74.15.6712-6719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F, et al. Herpes simplex virus type 1 glycoprotein E is required for axonal localization of capsid, tegument, and membrane glycoproteins. J Virol. 2005;79(21):13362–13372. doi: 10.1128/JVI.79.21.13362-13372.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, et al. Herpes simplex virus type 2 glycoprotein E is required for efficient virus spread from epithelial cells to neurons and for targeting viral proteins from the neuron cell body into axons. Virology. 2010;405(2):269–279. doi: 10.1016/j.virol.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatterjee S, Koga J, Whitley RJ. A role for herpes simplex virus type 1 glycoprotein E in induction of cell fusion. J Gen Virol. 1989;70(Pt 8):2157–2162. doi: 10.1099/0022-1317-70-8-2157. [DOI] [PubMed] [Google Scholar]
- 13.Davis-Poynter N, Bell S, Minson T, Browne H. Analysis of the contributions of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J Virol. 1994;68(11):7586–7590. doi: 10.1128/jvi.68.11.7586-7590.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spear PG. Membrane fusion induced by herpes simplex virus. In: Bentz J, editor. Viral Fusion Mechanisms. Boca Raton, FL: CRC; 1993. pp. 201–232. [Google Scholar]
- 15.Haanes EJ, Nelson CM, Soule CL, Goodman JL. The UL45 gene product is required for herpes simplex virus type 1 glycoprotein B-induced fusion. J Virol. 1994;68(9):5825–5834. doi: 10.1128/jvi.68.9.5825-5834.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mettenleiter TC, Klupp BG, Granzow H. Herpesvirus assembly: An update. Virus Res. 2009;143(2):222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 17.Farnsworth A, Wisner TW, Johnson DC. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J Virol. 2007;81(1):319–331. doi: 10.1128/JVI.01842-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeh PC, et al. Direct and specific binding of the UL16 tegument protein of herpes simplex virus to the cytoplasmic tail of glycoprotein E. J Virol. 2011;85(18):9425–9436. doi: 10.1128/JVI.05178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Chadha P, Meckes DG, Jr, Baird NL, Wills JW. Interaction and interdependent packaging of tegument protein UL11 and glycoprotein E of herpes simplex virus. J Virol. 2011;85(18):9437–9446. doi: 10.1128/JVI.05207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stylianou J, Maringer K, Cook R, Bernard E, Elliott G. Virion incorporation of the herpes simplex virus type 1 tegument protein VP22 occurs via glycoprotein E-specific recruitment to the late secretory pathway. J Virol. 2009;83(10):5204–5218. doi: 10.1128/JVI.00069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baird NL, Yeh PC, Courtney RJ, Wills JW. Sequences in the UL11 tegument protein of herpes simplex virus that control association with detergent-resistant membranes. Virology. 2008;374(2):315–321. doi: 10.1016/j.virol.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loomis JS, Bowzard JB, Courtney RJ, Wills JW. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J Virol. 2001;75(24):12209–12219. doi: 10.1128/JVI.75.24.12209-12219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacLean CA, Clark B, McGeoch DJ. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J Gen Virol. 1989;70(Pt 12):3147–3157. doi: 10.1099/0022-1317-70-12-3147. [DOI] [PubMed] [Google Scholar]
- 24.Baines JD, Roizman B. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J Virol. 1992;66(8):5168–5174. doi: 10.1128/jvi.66.8.5168-5174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meckes DG, Jr, Wills JW. Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J Virol. 2007;81(23):13028–13036. doi: 10.1128/JVI.01306-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loomis JS, Courtney RJ, Wills JW. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J Virol. 2003;77(21):11417–11424. doi: 10.1128/JVI.77.21.11417-11424.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chadha P, Han J, Starkey JL, Wills JW. Regulated interaction of tegument proteins UL16 and UL11 from herpes simplex virus. J Virol. 2012;86(21):11886–11898. doi: 10.1128/JVI.01879-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meckes DG, Jr, Marsh JA, Wills JW. Complex mechanisms for the packaging of the UL16 tegument protein into herpes simplex virus. Virology. 2010;398(2):208–213. doi: 10.1016/j.virol.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harper AL, et al. Interaction domains of the UL16 and UL21 tegument proteins of herpes simplex virus. J Virol. 2010;84(6):2963–2971. doi: 10.1128/JVI.02015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gage PJ, Levine M, Glorioso JC. Syncytium-inducing mutations localize to two discrete regions within the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B. J Virol. 1993;67(4):2191–2201. doi: 10.1128/jvi.67.4.2191-2201.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wisner TW, Johnson DC. Redistribution of cellular and herpes simplex virus proteins from the trans-Golgi network to cell junctions without enveloped capsids. J Virol. 2004;78(21):11519–11535. doi: 10.1128/JVI.78.21.11519-11535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wisner T, Brunetti C, Dingwell K, Johnson DC. The extracellular domain of herpes simplex virus gE is sufficient for accumulation at cell junctions but not for cell-to-cell spread. J Virol. 2000;74(5):2278–2287. doi: 10.1128/jvi.74.5.2278-2287.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mettenleiter TC. Budding events in herpesvirus morphogenesis. Virus Res. 2004;106(2):167–180. doi: 10.1016/j.virusres.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Turner A, Bruun B, Minson T, Browne H. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J Virol. 1998;72(1):873–875. doi: 10.1128/jvi.72.1.873-875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farnsworth A, Johnson DC. Herpes simplex virus gE/gI must accumulate in the trans-Golgi network at early times and then redistribute to cell junctions to promote cell-cell spread. J Virol. 2006;80(7):3167–3179. doi: 10.1128/JVI.80.7.3167-3179.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wells CD, et al. A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell. 2006;125(3):535–548. doi: 10.1016/j.cell.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 37.Takakuwa H, et al. Herpes simplex virus encodes a virion-associated protein which promotes long cellular processes in over-expressing cells. Genes Cells. 2001;6(11):955–966. doi: 10.1046/j.1365-2443.2001.00475.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.