Abstract
Fully mature enamel is about 98% mineral by weight. While mineral crystals appear very early during its formative phase, the newly secreted enamel is a soft gel-like matrix containing several enamel matrix proteins of which the most abundant is amelogenin (Amelx). Histological analysis of mineralized dental enamel reveals markings called cross-striations associated with daily increments of enamel formation, as evidenced by injections of labeling dyes at known time intervals. The daily incremental growth of enamel has led to the hypothesis that the circadian clock might be involved in the regulation of enamel development. To identify daily rhythms of clock genes and Amelx, we subjected murine ameloblast cells to serum synchronization to analyze the expression of the circadian transcription factors Per2 and Bmal1 by real-time PCR. Results indicate that these key genetic regulators of the circadian clock are expressed in synchronized murine ameloblast cell cultures and that their expression profile follows a circadian pattern with acrophase and bathyphase for both gene transcripts in antiphase. Immunohistological analysis confirms the protein expression of Bmal and Cry in enamel cells. Amelx expression in 2-day postnatal mouse molars dissected every 4 hours for a duration of 48 hours oscillated with an approximately 24-hour period, with a significant approximately 2-fold decrease in expression during the dark period compared to the light period. The expression of genes involved in bicarbonate production (Car2) and transport (Slc4a4), as well as in enamel matrix endocytosis (Lamp1), was greater during the dark period, indicating that ameloblasts express these proteins when Amelx expression is at the nadir. The human and mouse Amelx genes each contain a single nonconserved E-box element within 10 kb upstream of their respective transcription start sites. We also found that within 2 kb of the transcription start site of the human NFYA gene, which encodes a positive regulator of amelogenin, there is an E-box element that is conserved in rodents and other mammals. Moreover, we found that Nfya expression in serum-synchronized murine ameloblasts oscillated with a strong 24-hour rhythm. Taken together, our data support the hypothesis that the circadian clock temporally regulates enamel development.
Keywords: circadian rhythms, enamel development, ameloblast cells, amelogenin
Enamel, the hardest and most mineralized tissue within the vertebrate body, is formed during development by ameloblast cells and is incapable of cell-mediated repair or remodeling. The temporal sequence for enamel formation comprises the secretory and maturation phases (Lacruz et al., 2012). In the secretory stage, newly secreted enamel is a soft gel-like matrix containing enamel-specific proteins (EMPs) and proteases that are essential for the proper growth of the thin enamel crystals (Paine and Snead, 1997; Paine et al., 2001). In the ensuing maturation stage, crystals expand in width and thickness as the extracellular matrix is removed (Smith, 1998). Fully matured enamel is about 98% mineral by weight.
Enamel crystallites are roughly organized into bundles, forming enamel prisms (also known as enamel rods), and under polarized light, the prisms show small transverse lines called cross-striations (Boyde, 1989, 1990; Lacruz and Bromage, 2006). These markings, which also show as varicosities and constrictions by scanning electron microscopy (SEM), have been associated with daily changes in the rate of production of enamel matrix (Boyde, 1964, 1979, 1989), suggesting a punctuated pattern of enamel fabrication. Vitally labeled enamel of various mammalian taxa shows that the number of cross-striations between fluorescent dyes correlates with the time interval between injections (Gysi, 1931; Mimura, 1939; Schour and Poncher, 1937; Dean, 1987; Bromage, 1991; Smith, 2006). Enamel formation is also dependent upon the concerted effort of several layers of epithelial cells that comprise the enamel organ (Butler, 1956). At the base of the forming tooth, cells of the inner enamel epithelium arise from the cervical loop, a reservoir of stem cells that develops into ameloblast precursors (Thesleff et al., 2007). Daily mitotic rhythms have been observed in epithelial cells at the apical loop of rat incisors (Kiely and Wilde, 1974; Gasser et al., 1972).
Because the circadian clock regulates a broad range of physiological processes (King and Takahashi, 2000; Reppert and Weaver, 2002; Schibler and Sassone-Corsi, 2002; DeBruyne et al., 2007), including that of other mineralized tissues influencing bone homeostasis and growth (Fu et al., 2005; Zvonic et al., 2007; Gafni et al., 2009), the periodicity observed in enamel suggests that the molecular underpinnings orchestrating enamel development may be influenced by the circadian clock (Boyde, 1989, 1990). It is known that ablation of the suprachiasmatic nucleus in rats results in a disrupted patterning of incremental lines in dentine (Andresen lines), supporting the involvement of the circadian clock in the formation of this tissue (Ohtsuka-Isoya et al., 2001). However, the association between the circadian clock and enamel development remains poorly defined. Immunohistochemical (IHC) studies have shown the expression of CLOCK, BMAL1, PER1, and PER2 proteins in the developing tooth organ (Zheng et al., 2011). Moreover, intravenous injections of 3H-methionine administered at different times of the day visualized daily variations of radiolabeled proteins secreted by ameloblasts (Smith and Nanci, 1996; Simmer et al., 2010). Xu et al. (2006), using in situ hybridization data and transcriptional analysis of the amelogenin promoter in mice, suggested a cyclic production pattern of amelogenin. Nevertheless, circadian rhythms of enamel-related gene expression have not yet been described.
The purpose of this study was to analyze the expression of circadian clock genes in ameloblast cells and to investigate whether enamel products oscillate in a circadian pattern. We focused our analysis on the expression of the circadian transcription factors Per2 and Bmal1 in serum-synchronized ameloblast cells, and we also investigated the expression of Amelx, a differentiation product of ameloblasts in 2- to 4-day postnatal mouse molars. Furthermore, we screened for the presence of conserved E-box sequence elements in the promoters of the rodent and human enamel-related genes including Nfya, a known positive regulator of amelogenin transcription (Xu et al., 2006). Taken together, our results strongly support the assertion that the circadian clock regulates enamel development.
MATERIALS AND METHODS
Cell Cultures
To characterize gene expression profiles in enamel-producing cells, the ameloblast-like cell line LS8 (Chen et al., 1992) was cultured in 10-mm cell culture plates grown in DMEM supplemented with 10% FBS, 5% CO2, at 37 °C. The circadian cycles of the cells were synchronized using a modified “serum shock” protocol based on that described by Balsalobre et al. (1998). Specifically, cells were treated for 2 hours in DMEM with a high serum concentration (50% FBS). This serum-rich medium was removed and replaced with serum-free medium. The synchronized cells were cultured for either 48 hours or 72 hours and harvested at regular intervals. Harvested cells were stored at −80 °C until the completion of the experiment and then processed for nucleic acid extraction.
Whole Mouse Molar Dissections
Timed-pregnant Swiss-Webster female mice were purchased from Taconic (Hudson, NY). Dams were housed in the University of Southern California vivarium in normal light-dark (LD) cycles with the light phase starting at 0600 h and the dark phase at 1800 h for 10 days prior to birth. Animals were collected starting at postnatal day 2 (2 DPN). Starting at 0800 h, a single pup was euthanized every 4 hours for 48 hours, and the first mandibular molars were immediately dissected free from the jaw. During the night phase, the pups were euthanized under dim red illumination within 2 minutes from entering the vivarium room, and the first mandibular molars were immediately dissected free from the jaw. Molars were placed into lysis buffer and stored at −80 °C until all samples had been collected and then processed. All animal manipulations complied with institutional and federal guidelines.
Real-Time PCR
Homogenized molars or synchronized cells were processed for RNA using a Qiagen RNeasy Mini kit (Hilden, Germany). Reverse-transcribed PCR was performed using an iScript cDNA Synthesis kit (Bio-Rad Laboratories, Hercules, CA). Real-time PCR reactions were performed using iQ SYBR Green Supermix (Bio-Rad Laboratories). Supplementary Table S1 provides all primer sequences. Triplicate samples for each time point and for each animal were processed. All values for the mRNA species were normalized to β-actin using methods described by Livak and Schmittgen (2001). One-way ANOVA was used to compare means of sampled genes.
Immunohistochemistry
C57BL/6J mice were sacrificed at 4 DPN, and the mandibles were dissected, fixed in 4% paraformaldehyde overnight, and embedded in paraffin to obtain sagittal sections at 5-μm thickness using standard techniques. Tissue sections were deparaffinized and rehydrated, and endogenous peroxidase was blocked with 0.3% H2O2 in deionized water. Sections were blocked with 1% bovine serum albumin (BSA) and incubated overnight with a primary polyclonal antibody against Bmal1 (AB4140, Millipore-Chemicon, Billerica, MA) using a dilution of 1:200 or a polyclonal antibody against Cry1 (sc-5953, Santa Cruz Biotechnology, Santa Cruz, CA) using a 1:500 dilution. After washing, sections were incubated with a biotinylated anti-rabbit IgG secondary antibody (Vector Laboratories, Burlingame, CA). Sections were then incubated with Histostain-Plus kit (AEC) (Zymed Laboratories, South San Francisco, CA) following manufacturer specifications. No counter-staining step was used. Negative control sections were prepared in which no primary antibody was applied, but all other steps were performed. Bmal1 was chosen as one protein target for this analysis because it forms a dimeric protein complex with Clock, constituting a positive regulatory transcription factor in the circadian oscillator (Reppert and Weaver, 2002). In contrast, Cryptochrome family members (Cry1-Cry2) were chosen because they are a critical component of the negative feedback leg of the circadian cycle and for their ability to form multimeric complexes with Period family proteins, thereby opposing the transcriptional activation conferred by the Clock/Bmal1 dimer (Schibler and Sassone-Corsi, 2002).
Analysis of E-Box Sequence Elements Upstream of Enamel-Related Genes
We screened for canonical E-box (5′-CACGTG-3′) and noncanonical E-box (5′-CACGTT-3′ or 5′-AACGTG-3′) (Yoo et al., 2005) sequence elements in the regions 10 kb upstream of the transcription start sites (TSS) of enamel-related genes in humans and rodents, based on sequences obtained from the UCSC Genome Browser (http://genome.ucsc.edu/) (Suppl. Table S2). TSS are based on Reference Sequence (RefSeq) records for Amelx (NM_001142 human, NM_009666.3 mouse, and NM_019154 rat), Ambn (NM_016519.5 human, NM_009664.1 mouse, and NM_019154.1 rat), Enam (NM_031889.2 human, NM_017468.3 mouse, and NM_001106001.1 rat), Mmp20 (NM_004771.3 human, NM_013903.2 mouse, and NM_001106800.1 rat), and Nfya (NM_002505.4 human, NM_001110832.1 mouse, and NM_012865.1 rat). As indicated in Supplementary Tables S1 and S2, human sequences were also mapped to additional mammalian genomes using the UCSC Genome Browser Blat tool (http://genome.ucsc.edu/cgi-bin/hgBlat). Sequences were based on chimpanzee genome build (CGSC 2.1.3, GCA_000001515.3), orangutan genome build (WUSTL version Pongo_ albelii-2.0.2), rhesus macaque genome build (v.1.0 Mmul_051212), marmoset genome build (WUGSC 3.2 GCA_000004665.1), horse genome build (UCSC version equCab2), and dog genome assembly v2.0. We obtained multiple alignments for these sequences using the ClustalW2 tool (http://www.ebi.ac.uk/Tools/msa/clustalw2/) with default parameters and settings (Larkin et al., 2007).
To evaluate the distribution of E-box sequence elements 10 kb upstream of TSS on a global level, we obtained all National Center for Biotechnology Information (NCBI) RefSeq records for human (GRCh37/hg19 assembly), mouse (NCBI37/mm9 assembly), and rat (Baylor 3.4/rn4 assembly) genomes using the UCSC Table Browser. We focused on RefSeq records with an unambiguous chromosome location and a unique NM designation indicating they correspond to an mRNA transcript. In cases where different RefSeq records have the same TSS in a given organism, we only considered the first RefSeq record based on alphabetical order. Using the UCSC Table Browser, we obtained sequences 10 kb upstream of the TSS of the filtered RefSeq records (21201 human, 21630 mouse, and 15852 rat records). We counted the number of canonical E-box (5′-CACGTG-3′) and noncanonical E-box (5′-CACGTT-3′ or 5′-AACGTG-3′) elements for each filtered RefSeq record using regular expression matching from the R project (http://www.r-project.org/). This information, along with the corresponding gene names and TSS, is available upon request.
RESULTS
Immunolocalization of Murine Bmal1 and Cry1
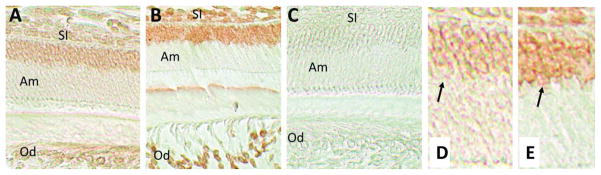
Bmal1 protein expression at 4 DPN was localized to the nuclei of ameloblasts and odontoblasts (Fig. 1A and 1E). Some staining was also evident in cells of the adjacent stratum intermedium and stellate reticulum, cells that form identifiable layers within the enamel organ, thus supporting the development of the ameloblast layer (Butler, 1956). This same cellular localization was observed for Cry1 protein immunolocalization, suggesting that both proteins are expressed in cells of the enamel organ (Fig. 1B and 1F). No staining was detected in the negative control (Fig. 1C).
Figure 1.
Immunohistochemical analysis of Bmal1 and Cry1 in 4 DPN mouse incisors. (A) Bmal1 is localized to the nuclei of ameloblasts (Am) and odontoblasts (Od). Cells of the stratum intermedium (SI) and stellate reticulum (SR) are also positively stained for Bmal1. (B) A similar immunohistochemical distribution among these cell populations was identified for Cry1. (C) Negative controls that omitted the primary antibody showed no immunostaining. (E, F) High magnification views of the nuclear staining observed in ameloblasts for Bmal1 (A) and Cry1 (B) antibodies, respectively.
Real-Time PCR
Cultured ameloblasts
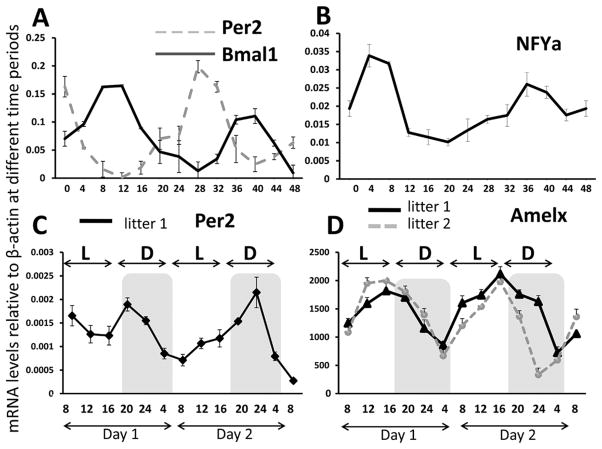
The profiles of Per2 and Bmal1 transcripts are shown in Figure 2A in which murine ameloblast cells were serum synchronized and RNA targets were quantitated every 4 hours for a duration of 48 hours. As expected, the abundance of mRNA for both circadian genes oscillated with a period of 24 hours over 48 hours of data acquisition. The circadian times of peak expression (acrophases) were 180° out of phase (i.e., bathyphase), a finding observed for other cell lines and tissues (Balsalobre et al., 2000; Zvonic et al., 2007; Gumz et al., 2009). One-way ANOVA was performed to compare Per2 mRNA and Bmal1 transcript abundance over the 48-hour period: abundance of both transcripts varied significantly over this period (p < 0.001), and post hoc analyses showed acrophase expression to be much larger than bathyphase expression for both transcripts (p < 0.001), demonstrating that each rhythm exhibited significant amplitude. The expression profile for Nfya transcripts (Fig. 2B) was obtained from these same samples and demonstrated a transcript abundance profile similar to that seen for Bmal1 (Fig. 2A). One-way ANOVA demonstrated Nfya transcript abundance varied significantly over 24 hours (p < 0.001), and post hoc analysis again showed acrophase expression to be significantly larger than bathyphase expression (p < 0.001). Nfya is a transcription factor that synergizes with the transcription proteins C/EBP-α and/or -δ family members to strongly activate amelogenin gene transcription (Xu et al., 2006, 2007). The circadian oscillations of transcription factors regulating Amelx transcription were confirmed in a second round of experiments with cells harvested every 3 hours for a 72-hour period following serum synchronization (Suppl. Fig. S1). The expression of Per2 and Cry1 oscillated in phase over 72 hours. The expression of Cry1 was significantly different between acrophase and bathyphase over the 72-hour period (one-way ANOVA, p < 0.05), and although the expression of Per2 continued to oscillate, acrophase and bathyphase were not significantly different after 48 hours (Suppl. Fig. S1).
Figure 2.
Real-time PCR analysis of circadian gene RNA transcript expression in serum-synchronized ameloblast-like LS8 cells (A, B) or whole molars (C, D). Per2 and Bmal1 mRNA expression (A) and Nfya mRNA expression (B) for synchronized LS8 cells were quantitated using reverse transcription followed by real-time PCR analysis. One-way ANOVA showed significant differences (p < 0.05) between the acrophase and bathyphase transcript abundance for Per2 and Bmal1. Real-time PCR of whole mouse molars collected every 4 hours for 48 hours from littermates revealed an approximately 24-hour oscillation in the Per2 (C) and Amelx (D) genes. The expression of Amelx decreases in the dark period. Values represent the mean of triplicate samples for the first lower molar from each animal at each collection time point. Standard deviations are shown for each sampled time point. For Amelx, one-way ANOVA showed a significant difference (p < 0.05) between the acrophase and bathyphase means for transcript values for each 24-hour period. L = light period; D = dark period. The y-axis of C and D indicates the time of the day at which tissue collection took place during the 2 days of harvesting molars.
Whole mouse molars
The expression levels of Per2 and Amelx transcripts recovered from whole postnatal molars every 4 hours over a 48-hour duration are shown in Figure 2C and 2D, respectively. Transcript abundance measurements showed that Per2 expression (Fig. 2C) is up-regulated during the dark period. In contrast, Amelx expression decreases in the dark period (Fig. 2D), exhibiting an approximately 2-fold decrease at the bathyphase (circadian time [CT] 22 for litter 1 and CT24 for litter 2) relative to acrophase expression. These differences were statistically significant (ANOVA, p < 0.05), with post hoc analysis showing acrophase expression to be significantly larger than bathyphase expression (p < 0.05).
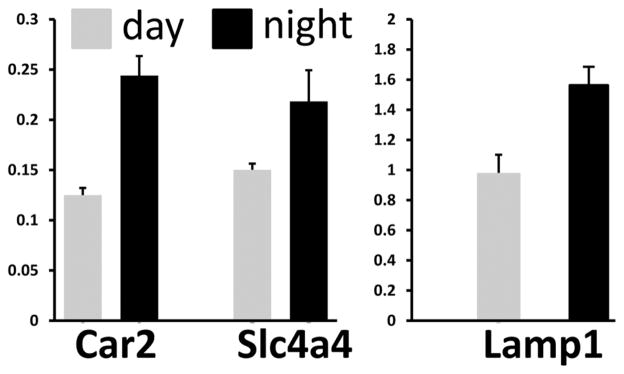
We further analyzed the expression of a number of genes involved in enamel development, including lysosomal associated membrane protein 1 (Lamp1) associated with endocytosis of enamel matrix proteins, carbonic anhydrase 2 (Car2), and sodium-bicarbonate cotransporter (Slc4a4). Car2 and Slca4 are associated with the production and transport of bicarbonate, respectively, and also participate in maintaining pH homeostasis during enamel biomineralization (Lacruz et al., 2010). The fold change in transcript abundance for Lamp1, Car2, and Slc4a4, comparing their expression levels at 1200 h and 2400 h, is shown in Figure 3 and indicates that their expression levels increase during the dark period relative to the light period in antiphase to Amelx expression levels.
Figure 3.
Real-time PCR of several genes involved with enamel development comparing day and night expression. Relative mRNA expression levels for carbonic anhydrase 2 (Car2), Slc4a4, and Lamp1 comparing RNA transcript levels at zeitgeber time (ZT) 2400 h relative to 1200 h. The expression of these 3 genes significantly increased during the night period relative to the light period (p < 0.05, one-way ANOVA).
In Silico Analysis of E-Boxes
To identify potential cis-acting elements responsible for the observed circadian rhythms of gene expression, we screened for canonical (5′-CACGTG-3′) and noncanonical (5′-CACGTT-3′ or 5′-AACGTG-3′) E-box elements within 10 kb upstream of the TSS of human, mouse, and rat enamel-specific genes. We focused on the classes of secreted enamel proteins that are most abundantly expressed during the secretory (matrix formative) stage of enamel formation, including amelogenin (Amelx), ameloblastin (Ambn), enamelin (Enam), and matrix metallopeptidase 20 (Mmp20) (Suppl. Table S2). The human and mouse Amelx sequences each contain one E-box element, although none were found in the rat. There are 3 E-box elements in the human and 2 in the rat Ambn upstream region; however, none were present in the mouse. Only the human Enam sequence had a single E-box element. All 3 species showed multiple E-box elements in the upstream regions of their Mmp20 and Nfya gene.
To evaluate if any of the upstream regions showed an unusual abundance of E-box elements, we determined the number of E-box elements (5′-CACGTG-3′ and 5′-CACGTT-3′) in the 10-kb region upstream of the TSS RefSeq records of 21201 for the human, 21630 for the mouse, and 15852 for the rat corresponding to protein-coding genes (see Materials and Methods). In our analysis of enamel-related genes (Suppl. Table S2), the 10-kb region upstream of the rat Nfya TSS had the highest number of E-box elements (n = 6), placing it within the top 92% of our filtered rat RefSeq records. However, 4.1% of other rat RefSeq records also showed 6 E-box elements in the 10-kb region upstream of their TSS. To place this in perspective, the maximum number of E-box elements in the 10-kb region upstream of the Period family of genes, critical components of the mammalian circadian clock in the suprachiasmatic nucleus of the hypothalamus and known to be expressed in a circadian pattern, never exceeded 8 in the species analyzed (Per1: 7 human, 7 mouse, 5 rat; Per2: 8 human, 3 mouse, 4 rat; Per3: 6 human, 1 mouse, 6 rat) and fluctuated greatly among orthologs. The data from the murine Per2 and Per3 genes indicate that a circadian pattern of expression is possible even if the 10-kb region upstream of the TSS contains a limited number of E-box elements.
To evaluate the conservation of the human E-box elements, we aligned the human sequences with orthologous genomic sequences from other primates (chimpanzees, orangutans, rhesus macaques, and marmosets). In only 2 cases (MMP20 positions −44 to −39 and NFYA positions −1839 to −1834) were the human E-box elements conserved in all primates surveyed (Suppl. Table S1). This MMP20 E-box element (−44 to −39) was conserved in horse but not in dog, mouse, or rat sequences. The human NFYA E-box element (−1839 to −1834) was conserved in all primates as well as other mammalian genomes surveyed (dog, horse, mouse, and rat). However, in all these species, the E-box element spans codons and the 5′-GT splice donor sequence of intron 1 of the UNC5CL (also known as ZUD) gene, which encodes an inhibitor of NF-κB activation, a possible alternative regulatory mechanism for NF-κB–mediated transcription (Zhang et al., 2004). This could provide an alternative explanation for the observed conservation of this E-box element that is independent of Nfya gene function.
DISCUSSION
We have investigated some aspects of the possible association between the circadian clock and enamel development. We have shown that the circadian clock genes Per2 and Bmal1 oscillate in cultured ameloblast LS8 cells in antiphase to one another over 48 hours. We have also confirmed the expression of Bmal1 and Cry1 proteins in ameloblasts of the developing enamel organ, with these 2 proteins showing a similar cellular distribution. Importantly, we detected daily oscillations in the expression of Amelx in the ameloblasts of whole developing mouse molars. Amelx is a differentiation-specific product of ameloblast cells. The expression of Amelx decreased markedly during the dark period relative to the light period. However, Amelx expression does not cease over the sampled periods but instead oscillates, as previously proposed (Boyde, 1990). These data confirm the histological analysis of mineralized enamel in which fast- and slow-growth periods were observed over 24 hours (Boyde, 1989) and also confirm previous in situ hybridization data showing changes in Amelx expression in whole mouse incisors (Xu et al., 2006). Our data also suggest that the expression of genes associated with other functions in the developing tooth such as matrix endocytosis (Lamp1), bicarbonate transport (Nbce1), and bicarbonate production (Car2) increases expression during the night, when Amelx is low. Bicarbonate is an essential component of mineralizing enamel (Smith, 1998; Lacruz et al., 2010). This suggests that ameloblasts segregate these functions according to the time of the day, with a possible critical dependency on Amelx production or reduction during cycles of fabrication. Given that the sampling of enamel organ cells shown here indicates that the mRNA of enamel- and none-namel-specific gene products changes between day and night, these data suggest that other enamel-specific genes (ameloblastin, enamelin) are also likely influenced by the circadian clock, but this remains to be tested.
The complex formed by CLOCK and BMAL1 proteins regulates gene expression by interacting with E-box elements present in gene regulatory regions (Gekakis et al., 1998; Ueda et al., 2005). Gene outputs can be either directly induced by the transcription factors of the circadian oscillator (CLOCK-BMAL1) or activated via specific effectors, which are responsive to the circadian transcription factors (Schibler et al., 2003). As shown in this study, the expression of Nfya, a recognized positive transcriptional activator of Amelx (Xu et al., 2006, 2007), oscillates in cultured ameloblasts with a daily rhythm (Fig. 2B). We identified 2 E-boxes proximal to the human NFYA TSS (Suppl. Table S2) and showed that one of these elements is conserved in the region upstream of the orthologous chimpanzee, orangutan, rhesus macaque, marmoset, dog, horse, rat, and mouse Nfya genes. However, we note that the conserved E-box element spans a splice junction of the proximal UNC5L gene, which could also partially or completely account for its cross-species conservation. Our analysis of E-box elements 10 kb upstream of the TSS of >15,000 human, mouse, and rat RefSeq records provides a context to evaluate the relationship between the number of E-box elements and the ability of the corresponding gene to show circadian expression. The murine Per2 and Per3 genes demonstrate that circadian expression is possible even if the 10-kb region upstream of the TSS contains few E-box elements. To more fully elucidate the relationship between various cis-acting elements and circadian expression, we believe that more in-depth genome-wide investigations of this nature are warranted.
We have previously shown that Amelx transcription is regulated by the CAAT enhancer binding protein (C/EBP) family, with C/EBP-α (Zhou and Snead, 2000) and -δ family members synergizing with NFY members to significantly up-regulate amelogenin promoter transcription several fold greater than with C/EBP alone (Xu et al., 2006, 2007). It is possible that circadian oscillations of NFY genes are more relevant than those of C/EBP family members. While there is an E-box in the C/EBP-α promoter (Kim et al., 2007), we have previously shown that the unpaired homeobox protein, Msx2, interacted with C/EBP-α via protein-to-protein interactions to suppress Amelx transcription rather than by binding to the amelogenin promoter (Zhou et al., 2000; Zhou and Snead, 2000). We suggest that by orchestrating the circadian abundance of Nfya (likely controlled synergistically by C/EBP-α) and Msx2, an oscillation of amelogenin expression can occur that is consistent with the histological observations of daily pulses of forming and matured enamel. Enamel is the only biomineralized tissue of ectodermal origin involved with feeding; thus, its correct development is tied to proper functioning of the organism’s metabolism. It has recently become apparent that there are strong links between metabolic functions and the circadian clock, with a relatively high percentage (20%) of the adipose transcriptome influenced by the circadian clock (Dallmann and Weaver, 2010; Johnston, 2012). Furthermore, the physiological functions of vital organs (e.g., lung, liver) are also dependent on or modulated by the circadian clock (Miller et al., 2007; Vollmers et al., 2009; Sukumaran et al., 2011). The extent of the involvement of the circadian clock in the development of healthy enamel is presently unclear. However, given the clinical relevance of Amelx in both normal enamel development and pathologies, which commonly result in a group of dental clinical disorders referred to as amelogenesis imperfecta (Wright, 2011), we suggest that the circadian clock is likely to impact the physiologically functional development of enamel. The analysis of the dentition of animal models defective in clock-related genes is therefore warranted.
CONCLUSION
This study has shown that the circadian clock genes Cry1, Per2, and Bmal1 are expressed in an ameloblast-like cell line and that their expressions oscillate in antiphase when their circadian rhythms are synchronized by serum. Amelx is the predominant enamel matrix protein secreted by ameloblast cells. We showed Amelx expression is down-regulated during the night period in developing mouse molars. The Nfya gene, which encodes a transcriptional activator of Amelx, contains a highly conserved E-box element near its TSS, suggesting that its transcription could be regulated by the circadian clock and thus providing a candidate mechanistic basis for the observed Amelx expression patterns. The numbers of E-boxes in necessary clock elements (e.g., Per, Bmal1), which are themselves expressed in circadian fashion, are comparable to the numbers of E-boxes in the enamel-related genes in this study. Thus, these data suggest that strong circadian regulation is possible even in the presence of limited numbers of E-boxes.
Supplementary Material
Acknowledgments
We thank Richard Pelikan (University of Southern California) for assistance in transcription factor binding site analysis. This research was funded by NIH/National Institute for Dental and Craniofacial Research grants to M.L.P. (DE 013404, DE 019629) and M.L.S. (DE 06988, DE 013045) and by NIH/NIGMS grants to J.G.H. (GM 072447). Research support provided to T.G.B. by the 2010 Max Planck Research Award, administered by the Max Planck Society and the Alexander von Humboldt Foundation with respect to the Hard Tissue Research Program in Human Paleobiomics. We are indebted to David Weaver for his support and for critically reviewing previous versions of this article. We also thank the anonymous reviewers whose comments helped improve the work presented here.
Footnotes
CONFLICT OF INTEREST STATEMENT
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
NOTE
Supplementary material is available on the journal’s website at http://jbr.sagepub.com/supplemental.
References
- Balsalobre A, Damiola F, Schibler U. A serum shock indices circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol. 2000;19:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- Boyde A. PhD Dissertation. London: University of London; 1964. The Structure and Development of Mammalian Enamel. [Google Scholar]
- Boyde A. Carbonate concentration, crystal centres, core dissolution, caries, cross striations, circadian rhythms and compositional contrast in SEM. J Dent Res. 1979;58:981–983. doi: 10.1177/00220345790580025101. [DOI] [PubMed] [Google Scholar]
- Boyde A. Enamel. In: Oksche A, Vollrath L, editors. Handbook of Microscopic Anatomy. Vol. 6. Berlin: Springer Verlag; 1989. pp. 309–473. [Google Scholar]
- Boyde A. Developmental interpretations of dental microstructure. In. In: de Rousseau J, editor. Primate Life History and Evolution. New York: Wiley-Liss; 1990. pp. 229–267. [Google Scholar]
- Bromage TG. Enamel incremental periodicity in the pig-tailed macaque: a polychrome fluorescent labelling study of dental hard tissues. Am J Phys Anthropol. 1991;86:205–214. [Google Scholar]
- Butler PM. The ontogeny of molar pattern. Biol Rev. 1956;31:3–70. [Google Scholar]
- Chen LS, Couwenhoven RI, Hsu D, Luo W, Snead ML. Maintenance of amelogenin gene expression by transformed epithelial cells of mouse enamel organ. Arch Oral Biol. 1992;37:771–778. doi: 10.1016/0003-9969(92)90110-t. [DOI] [PubMed] [Google Scholar]
- Dallmann R, Weaver DR. Altered body mass regulation in male mPeriod mutant mice on high-fat diet. Chronobiol Int. 2010;27:1317–1328. doi: 10.3109/07420528.2010.489166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean MC. Growth layers and incremental markings in hard tissues: a review of the literature and some preliminary observations about enamel structure of Paranthropus boisei. J Hum Evol. 1987;16:157–172. [Google Scholar]
- DeBruyne JP, Weaver DR, Reppert SM. Peripheral circadian oscillators require CLOCK. Curr Biol. 2007;17:538–539. doi: 10.1016/j.cub.2007.05.067. [DOI] [PubMed] [Google Scholar]
- Fu L, Patel MS, Bradley A, Wagner EF, Karsenty G. The molecular clock mediates leptin-regulated bone formation. Cell. 2005;122:803–815. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Gafni Y, Ptitsyn AA, Zilberman Y, Pelled G, Gimble JM, Gazit D. A circadian rhythm of osteoclacin in the maxillomandibular complex. J Dent Res. 2009;88:45–50. doi: 10.1177/0022034508328012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser RF, Scheving LE, Pauly J. Circadian rhythms in cell division rate of the inner enamel epithelium and in the uptake of 3H-Thymidine by the root tip of incisors. J Dent Res. 1972;51:740–746. doi: 10.1177/00220345720510030901. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Gumz ML, Stow LR, Lynch IJ, Greenlee MM, Rudin A, Cain BD, Weaver DR, Wingo CS. The circadian clock protein Period 1 regulates expression of the renal epithelial sodium channel in mice. J Clin Invest. 2009;119:2423–2434. doi: 10.1172/JCI36908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gysi A. Metabolism in adult enamel. Dent Dig. 1931;37:661–668. [Google Scholar]
- Johnston JD. Adipose circadian rhythms: translating cellular and animal studies to human physiology. Mol Cell Endocrinol. 2012;349:45–50. doi: 10.1016/j.mce.2011.05.008. [DOI] [PubMed] [Google Scholar]
- Kiely ML, Wilde JL. Circadian mitotic rhythms in the cervical tissues of rat maxillary incisor. J Dent Res. 1974;53:1432–1438. doi: 10.1177/00220345740530062401. [DOI] [PubMed] [Google Scholar]
- Kim JW, Monila H, Pandey A, Lane MD. Upstream stimulatory factors regulate the CEBPalpha gene during differentiation of 3T3-L1 adipocytes. Biochem Biophys Res Comm. 2007;354:517–521. doi: 10.1016/j.bbrc.2007.01.008. [DOI] [PubMed] [Google Scholar]
- King DP, Takahashi JS. Molecular genetics of circadian rhythms in mammals. Annu Rev Neurosci. 2000;23:713–742. doi: 10.1146/annurev.neuro.23.1.713. [DOI] [PubMed] [Google Scholar]
- Lacruz RS, Bromage TG. Appositional growth in molars of South African hominids. J Anat. 2006;209:13–20. doi: 10.1111/j.1469-7580.2006.00597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacruz RS, Nanci A, Kurtz I, Wright TJ, Paine ML. Regulation of pH during amelogenesis. Calcif Tissue Int. 2010;86:91–103. doi: 10.1007/s00223-009-9326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacruz RS, Smith CE, Hubbard MJ, Bringas P, Jr, Chen Y, Kurtz I, Snead ML, Paine ML. Identification of novel candidate genes involved in mineralization of dental enamel by genome-wide transcript profiling. J Cell Physiol. 2012;227:2264–2275. doi: 10.1002/jcp.22965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, Antoch MP, Walker JR, Esser KA, Hogenesch JB, Takahashi JS. Circadian and CLOCK-controlled regulation of the mouse transcrip-tome and cell proliferation. Proc Natl Acad Sci U S A. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura F. The periodicity of growth lines seen in enamel. Kobvo-shi. 1939;13:454–455. [Google Scholar]
- Ohtsuka-Isoya M, Hayashi H, Shinoda H. Effect of suprachiasmatic nucleus lesion on circadian dentin increments in rats. Am J Physiol. 2001;280:1364–1370. doi: 10.1152/ajpregu.2001.280.5.R1364. [DOI] [PubMed] [Google Scholar]
- Paine ML, Snead ML. Protein interactions during assembly of the enamel organic extracellular matrix. J Bone Miner Res. 1997;12:221–227. doi: 10.1359/jbmr.1997.12.2.221. [DOI] [PubMed] [Google Scholar]
- Paine ML, White SN, Luo W, Fong H, Sarikaya M, Snead ML. Regulated gene expression dictates enamel structure and tooth function. Matrix Biol. 2001;20:273–292. doi: 10.1016/s0945-053x(01)00153-6. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Schibler U, Ripperger J, Brown SA. Peripheral circadian oscillators in mammals: time and food. J Biol Rhythms. 2003;18:250–260. doi: 10.1177/0748730403018003007. [DOI] [PubMed] [Google Scholar]
- Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;11:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- Schour I, Poncher HG. Rate of apposition of enamel and dentine measured by the effect of acute fluorosis. Am J Dis Child. 1937;54:757–756. [Google Scholar]
- Simmer JP, Papagerakis P, Smith CE, Fisher DC, Rountrey AN, Zheng L, Hu JC. Regulation of dental enamel shape and hardness. J Dent Res. 2010;89:1024–1038. doi: 10.1177/0022034510375829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- Smith CE, Nanci A. Protein dynamics of amelogenesis. Anat Rec. 1996;245:186–207. doi: 10.1002/(SICI)1097-0185(199606)245:2<186::AID-AR7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Smith TM. Experimental determination of the periodicity of incremental features in enamel. J Anat. 2006;208:99–103. doi: 10.1111/j.1469-7580.2006.00499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran S, Jusko WJ, Dubois DC, Almon RR. Light-dark oscillations in the lung transcriptome: implications for lung homeostasis, repair, metabolism, disease and drug action. J Appl Physiol. 2011;110:1732–1747. doi: 10.1152/japplphysiol.00079.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thesleff I, Wang XP, Suomalainen M. Regulation of epithelial stem cells in tooth regeneration. C R Biol. 2007;330:561–564. doi: 10.1016/j.crvi.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clock. Nat Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright TJ. Amelogenesis imperfecta. Eur J Oral Sci. 2011;119:338–341. doi: 10.1111/j.1600-0722.2011.00933.x. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhou Y, Gonzalez FJ, Snead ML. C/EBPdelta maintains amelogenin expression in the absence of C/EBPalpha, in vivo. J Biol Chem. 2007;282:29882–29889. doi: 10.1074/jbc.M702097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhou YL, Luo W, Zhu QS, Levy D, MacDougald OA, Snead ML. NF-Y and CCAAT/enhancer-binding protein alpha synergistically activate the mouse amelogenin gene. J Biol Chem. 2006;281:16090–16098. doi: 10.1074/jbc.M510514200. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Ko CH, Lowrey PL, Buhr ED, Song EJ, Chang S, Yoo OJ, Yamazaki S, Lee C, Takahashi JS. A non canonical E-box enhance drives mouse Period2 circadian oscillations in vivo. Proc Natl Acad Sci U S A. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xu LG, Han KJ, Shu HB. Identification of a ZU5 and death domain-containing inhibitor of NF-kappaB. J Biol Chem. 2004;279:17819–17825. doi: 10.1074/jbc.M310737200. [DOI] [PubMed] [Google Scholar]
- Zheng L, Papagerakis S, Schnell SD, Hoogerwerf WA, Papagerakis P. Expression of clock proteins in developing tooth. Gene Expr Patterns. 2011;11:202–206. doi: 10.1016/j.gep.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YL, Lei Y, Snead ML. Functional antagonism between Msx2 and CCAAT/enhancer-binding protein alpha in regulating the mouse amelogenin gene expression is mediated by protein-protein interaction. J Biol Chem. 2000;275:29066–29075. doi: 10.1074/jbc.M002031200. [DOI] [PubMed] [Google Scholar]
- Zhou YL, Snead ML. Identification of CCAAT/enhancer-binding protein alpha as a transactivator of the mouse amelogenin gene. J Biol Chem. 2000;275:12273–12280. doi: 10.1074/jbc.275.16.12273. [DOI] [PubMed] [Google Scholar]
- Zvonic S, Ptitsyn AA, Kilroy G, Wu X, Conrad SA, Scott LK, Guilak F, Pelled G, Gazit D, Gimble JM. Circadian oscillation of gene expression in murine cal-varial bone. J Bone Miner Res. 2007;22:357–365. doi: 10.1359/jbmr.061114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.