Summary
Background
Among the Aristotelian senses, the subcellular and molecular mechanisms involved in the sense of touch are the most poorly understood.
Results
We demonstrate that specialized sensory neurons, the class II and class III multidendritic (md) neurons, are gentle touch sensors of Drosophila larvae. Genetic silencing of these cells significantly impairs gentle touch responses, optogenetic activation of these cells triggers behavioral touch-like responses, and optical recordings from these neurons show that they respond to force. The class III neurons possess highly dynamic dendritic protrusions rich in F-actin. Genetic manipulations that alter actin dynamics indicate that the actin-rich protrusions (termed sensory filopodia) on the class III neurons are required for behavioral sensitivity to gentle touch. Through a genome-wide RNAi screen of ion channels, we identified Ripped Pocket (rpk), No Mechanoreceptor Potential C (nompC), and NMDA Receptors 1 and 2 (Nmdars) as playing critical roles in both behavioral responses to touch and in the formation of the actin-rich sensory filopodia. Consistent with this requirement, reporters for rpk and nompC show expression in the class III neurons. A genetic null allele of rpk confirms its critical role in touch responses.
Conclusions
Output from class II and III md neurons of the Drosophila larvae is necessary and sufficient for eliciting behavioral touch responses. These cells show physiological responses to force. Actin-rich organelles in the Class III neurons are required for gentle touch detection. Ion channels in several force-sensing gene families are required in these cells, both for behavioral sensitivity to touch, and for the formation of the actin-rich sensory filopodia.
Introduction
The sense of touch is critical to the existence of life. Indeed, virtually all animals use mechanosensory input to explore the textures of the world. Our mechanical senses allow us to detect forces that range from the tiny pressures of the mosquito that lands upon our skin, to the painful sensations experienced by the boxer in the ring. The features that endow mechanosensory neurons with the ability to detect such a wide range of force are essentially unknown.
Mechanosensory neurons show extremely rapid ionic influx in response to force which suggests that these neurons detect force through ion channels that directly sense the force in the absence of any upstream signaling or second messengers [1, 2]. The putative metazoan mechanotransduction channels that have been identified to date fall into several distinct gene families: the Degenerin Epithelial Sodium Channel (DEG/ENaC) family [3–9], the Transient Receptor Potential (TRP) family [10–18], the Transmembrane Channel Like (TMC) family [19], the TREK channel family [20], and most recently, the Piezo family [21–23].
Despite the many channels that have been implicated in force sensation important mechanistic questions remain unsolved. For instance, how does the mechanosensory threshold of a particular type of neuron relate to the intrinsic properties of mechanosensory channels that detect forces relevant to the cell [3]? Do highly sensitive touch neurons express a set of exquisitely sensitive mechanotransduction channels in comparison to neurons that are tuned to detect stronger forces? Or alternatively, do other features of the neurons (such as morphological specialization) play an important role?
We are attempting to answer these questions through the investigation of force sensing mechanisms in Drosophila larvae. The functions of most neuron types found in the Drosophila larval body wall remain unknown. Neuronal silencing, optogenetic activation, and thermogenetic activation experiments all indicate that the class IV multidendritic (md) neurons function as nociceptive neurons, detecting noxious heat and noxious mechanical stimuli [6, 9, 18]. Other evidence suggests that the class I and the bipolar md neurons function as proprioceptors required for coordinated larval locomotion [24]. The chordotonal neurons also function as propriocieptors and are additionally thought to play a role in responses to gentle touch [25].
Here, we identify the first known function for the class II and class III md neurons.
Results
Class II and III md neurons contribute to larval response to gentle touch
Although previous evidence suggests the gentle touch responses are in part mediated by internal stretch-receptive chordotonal neurons [25], the complete repertoire of sensory neurons that contribute to the larval gentle touch response is as yet unknown. Thus, to investigate the relative contributions of various sensory neuron classes in gentle touch responses, we used specific GAL4 driver lines to drive expression of the UAS-Tetanus toxin light chain (UAS-TNT-E) to silence neuronal activity [26] in distinct subclasses of larval sensory neurons. We then investigated behavioral responses to gentle touch in the transgenic animals using previously described gentle touch assays [6, 9, 12, 17, 25]. In this assay, larvae are gently stroked with the tip of an eyelash along the thoracic segments (Figure 1A) and the larval behavioral response is given a score of 1 to 4. Each larva is stimulated four times giving a total score that ranges from 0 to 16.
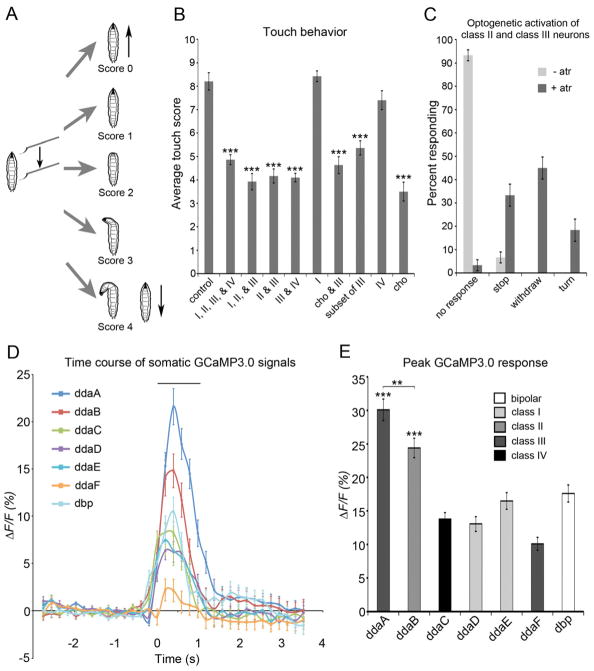
Figure 1. Class II and III md neurons are necessary and sufficient for the larval gentle touch response and they are activated by a force stimulus.
(A) Scheme of larval gentle touch assay. Scores were based on 5 categories of behavioral response. 0 = no response, 1 = pause of locomotion, 2 = withdrawal of head, 3 = turn of < 90 degrees, 4 = turn of > 90 degrees or reverse locomotion. (B) Expression of tetanus toxin in class II or class III neurons significantly reduced the larval response to gentle touch. Genotypes were w; UAS-TNT-E/+ (control), w; md-GAL4 tsh-GAL80/UAS-TNT-E (I, II, III, & IV), w; UAS-TNT-E/+; c161-GAL4/+ (I, II, & III)), w; UAS-TNT-E/+; 1003.3-GAL4/+ (II & III), w; UAS-TNT-E/tsh-GAL80; c755-GAL4/+ (III & IV), w; UAS-TNT-E/+; 2-21-GAL4/+ (I), w; nompC-GAL4/UAS-TNT-E (cho & III), w; tutl-GAL4/UAS-TNT-E (Class III (but not ddaF)), w; ppk-GAL4/UAS-TNT-E (IV), and w; 117y-GAL4/UAS-TNT-E (cho) (N = 60 larvae of each genotype). See also Figure S1. (C) Optogenetic activation (w;UAS-ChR2-YFP/1003.3-GAL4) of class II and III neurons was sufficient to elicit the gentle touch response (N=60 larvae of each genotype). See also Movie S1. (D and E) Optical recordings showed ddaB class II and ddaA class III neurons responded to force. (D) Traces showing average (+/− s.e.m) ΔF/F0 % of md neurons expressing GCaMP3.0 before, during, and after mechanical stimulation. The bar above the trace shows the duration of the average mechanical stimulus (1.02 ± 0.03 sec). (N = 66 force stimuli.) (E) Average peak ΔF/F0 % of each class of md neurons expressing GCaMP3.0. ***P < 0.001 when ddaC, ddaD, ddaE, ddaF, or dbp were compared to ddaA or ddaB. (N = 66 force stimuli.) Genotype was md-GAL4/UAS-GCaMP3.0. The differences of color of bars indicate types of class neurons. Statistical analyses were performed using a one-way analysis of variance (ANOVA) with Tukey’s HSD post-hoc for pair-wise comparisons. Unless otherwise indicated, all data are means ± s.e.m. ***P < 0.001, **P < 0.01 compared with control.
Confirming earlier findings, when UAS-TNT-E was expressed in chordotonal neurons, larvae were indeed strongly insensitive to gentle touch (cho in Figure 1B); However, in these animals residual touch responses remained suggesting the existence of other sensors.
Since non-ciliated md neurons highly innervate the larval epidermis [27–29], we hypothesized that these neurons were likely candidates to play a role in this process. To test this, we used the GAL4 109(2)80 driver (md-GAL4) which is expressed in all four classes of md neurons to drive UAS-TNT-E (Supplementary Figure 1A). To reduce the expression of TNT-E in the abdominal and thoracic ganglion, the strain utilized teashirt-GAL80 (tsh-GAL80) in combination with the md-GAL4 driver. Indeed, compared to controls, larvae expressing UAS-TNT-E under the control of md-GAL4 tsh-GAL80 were less sensitive to gentle touch (I, II, III, & IV in Figure 1B). This finding indicated that, like chordotonal neurons, non-ciliated md neurons act as gentle touch detecting mechanosensory neurons.
To determine the precise class of md neuron that was required for the gentle touch response, we used a set of GAL4 drivers that were expressed in specific subsets of md neurons (Supplementary Figure 1). As shown in Figure 1B, drivers targeting class III neurons all resulted in significant touch insensitivity when driving UAS-TNT-E. Touch insensitivity was seen with silencing of class I, II, and III neurons (C161-GAL4), class II and class III neurons (1003.3-GAL4), class III and IV (c755-GAL4 with tsh-GAL80), class III and chordotonal neurons (nompC-GAL4), or class III neurons alone (turtle-GAL4). In contrast, when we specifically inactivated class I neurons using the 2-21-GAL4 driver (Supplementary Figure 1E) or class IV neurons using the ppk-GAL4 driver (Supplementary Figure 1H), larvae showed a normal sensitivity to gentle touch compared to controls (Figure 1B). These data combined, indicated that the output of the class III neurons is required for normal behavioral responses to gentle touch. These data are also consistent with a potential role for class II neurons in gentle touch responses as all of the drivers targeting class II neurons also showed an impaired touch response. For example, C161-GAL4 is strongly expressed in Class I and II neurons but only weakly expressed in a stochastic population of Class III neurons.
Optogenetic activation of class II and III neurons elicits a gentle touch response
Given the suggested role for class II and class III neurons in gentle touch responses, we next assessed whether activation of the class II and class III neurons, or the class III neurons alone, would be sufficient to elicit a gentle touch response. Efficient optogenetic activation of larval neurons can be achieved through expression of Channelrhodopsin-2::YFP (Chr2::YFP), a light-activated cation channel [6, 9, 30, 31]. Expression of Chr2::YFP in the class II and class III neurons (using 1003.3-GAL4 driver) indeed resulted in gentle touch like responses upon blue light illumination. Quickly after illumination with blue light, larvae withdrew their anterior region or turned their head (Figure 1C, Movie S1). This response depended on the feeding of all trans retinal (atr) as most of the ChR2-expressing larvae that were fed yeast paste free of atr did not show gentle touch-like responses upon exposure to blue light (Figure 1C and Movie S1). These data demonstrate that activation of class II neurons combined with activation of class III neurons produced gentle touch-like responses. Driving Chr2::YFP specifically in subset of class III neurons (ddaA, ldaB, v’pda, and vdaD), under control of tutl-GAL4 did not produce touch like behaviors in response to blue light (data not shown). This result is consistent with a likely combinatorial role of class II and class III neurons in eliciting gentle touch responses. Alternatively, the output of the ddaF neuron (which is not targeted by tutl-GAL4) is absolutely required for the generation of optogenetically triggered touch responses.
Optical recordings reveal that class II and class III neurons respond to force
To directly test which of the md neurons showed physiological responses to force, we developed a preparation for imaging Ca2+ responses in these cells. In this preparation, the md neurons expressing GCaMP3.0 [32] were imaged through the transparent cuticle using high-speed time-lapse confocal microscopy while being exposed to 1mN force stimuli (see Experimental Procedures). Multidendritic neurons imaged in this preparation showed rapidly increasing GCaMP3.0 signals during the initial application of force and this signal rapidly declined (Figure 1D). The greatest GCaMP3.0 increases were indeed observed in the ddaB class II and the ddaA class III neurons (Figure 1D, E). Interestingly, the Class III ddaF neuron showed relatively smaller changes. The stronger response of the ddaA neurons relative to ddaF was likely in part due to the fact that the mechanical stimulus was applied to the dendritic field of ddaA. Consistent with this, when the dendritic field of ddaF class III neurons was stimulated, the GCaMP3.0 fluorescence in these cells was also seen to more strongly increase (data not shown).
Dendritic sensory filopodia on class III neurons are actin-rich structures that are dynamic in early third instar
The class III neurons are characterized by protrusions from their primary dendritic branches [27, 33]. These protrusions are rich in F-actin as they are strongly labeled by the F-actin binding protein, moesin::GFP [27, 33, 34] (Figure 2A-2D). These structures have been previously referred to either as dendritic filopodia [33], or as dendritic “spikes [27]”. Here, we refer to these actin-rich protrusions as “sensory filopodia” in order to distinguish them from the immature post-synaptic dendritic filopodia that have been described on vertebrate neurons.
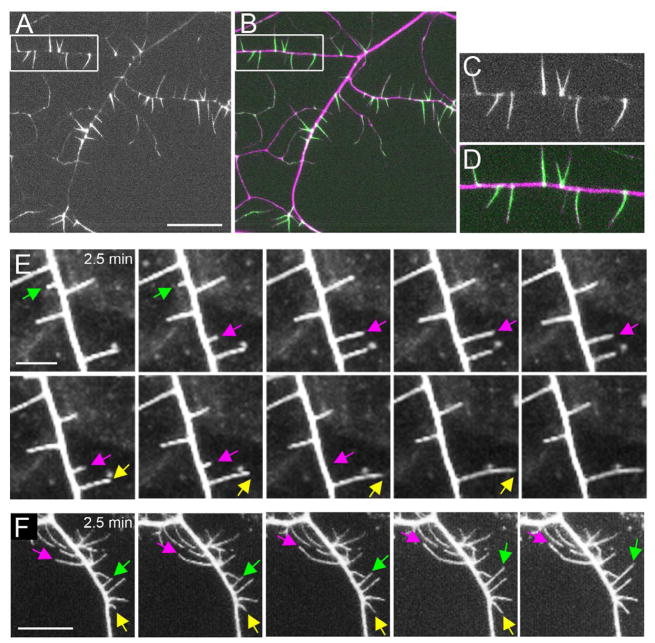
Figure 2. Sensory filopodia of class III neurons are dynamic structures rich in F-actin.
(A–D) Subcellular localization of moesin::GFP in class III neurons. F-actin-binding GFP moesin::GFP localized to the dendritic sensory filopodia of class III neurons (A and C, green in B and D). Neuronal plasma membrane was labeled with mCD8::DsRed (magenta in B and D). C and D are high magnification images of structures highlighted by the white boxes in A and B, respectively. (E) Time-lapse imaging of wild-type class III neurons shows the dynamic nature of dendritic sensory filopodia in early third instar. (F) Time-lapse imaging of neurons expressing Rac1 in class III neurons shows that sensory filopodia remain dynamic with this manipulation. Images shown were taken at 2.5-minute intervals. Each arrow highlights dynamic sensory filopodia. Scale bar in A = 20 μm, E = 5μm, and F = 10 μm. In this and all subsequent figures, anterior is to the left and dorsal is at the top. Genotypes were md-GAL4 UAS-mCD8::DsRed/UAS-moesin::GFP (A–D), 1003.3-GAL4 UAS-mCD8::GFP/+ (E), and w; UAS-Rac1 (WT)/+; 1003.3-GAL4 UAS-mCD8::GFP/+ (F). See also Movie S2.
It has been reported that the sensory filopodia are morphologically dynamic [33]. To better characterize these dynamics, we developed a time-lapse imaging preparation that allowed for stable imaging of the class III neurons for 30 minutes periods in anesthetized whole-mount larvae. Consistent with the prior report, we found that while some filopodia were stable over the time period, others were dynamic – newly forming or completely disappearing over the course of 30 minutes (Figure 2E and Supplementary Movie S2).
The number of sensory filopodia correlates with the strength of touch responses
We next investigated whether the actin cytoskeleton in the filopodia played an important role in mechanosensory responses. To achieve this, we manipulated actin through the expression of Rho family GTPases (which are well known regulators of actin) in the class III cells [35].
First, we investigated the effects of overexpressing Rac1 which activates the Arp2/3 complex and nucleates branched growth from existing actin filaments [35]. This manipulation has been reported to cause increased branching in class I [36], class III neurons [33], and class IV neurons [37, 38]. Similarly, we observed dramatic increases in the number of dendritic sensory filopodia in class III ddaA neurons overexpressing Rac1 relative to control neurons (Figure 3A, 3E and 3F). These filopodia were dynamic, and their stability was not changed compared to controls (Figure 2F and Supplementary Movie S2). Interestingly, behavioral tests performed on these larvae showed that they also became hypersensitive to gentle touch (Figure 3B).
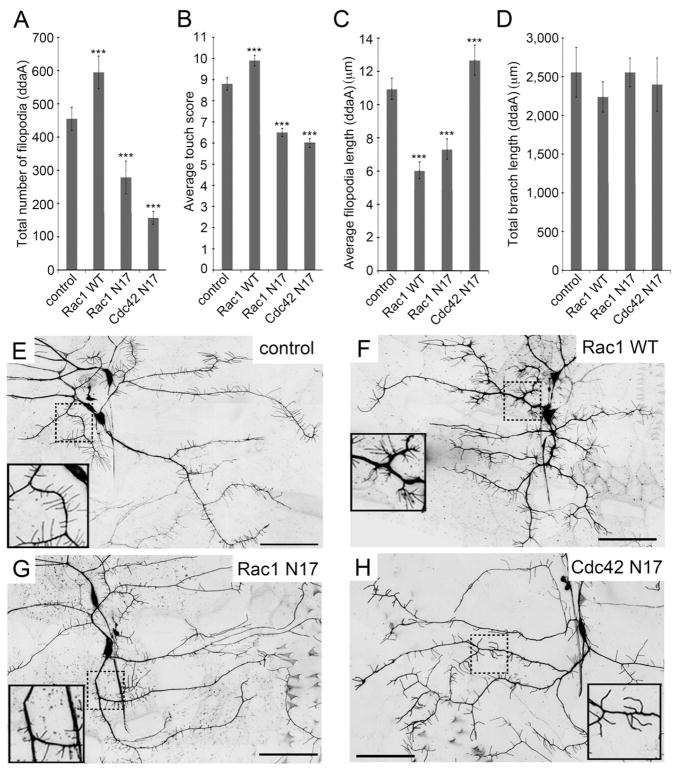
Figure 3. The number of dendritic sensory filopodia correlated with the strength of the gentle touch response.
(A) The number of sensory filopodia in class III ddaA neuron expressing Rho family GTPases. (B) Effect of Rho family GTPases on gentle touch responses. (N = 60 larvae of each genotype.) (C) Quantification of average length of sensory filopodia (D) Length of primary dendritic branches. (E–H) Dendritic morphology of representative class III ddaA neurons; (E) Neurons in control animals (w; 1003.3-GAL4 UAS-mCD8::GFP/+). (F) Neurons over-expressing Rac1 WT (w; UAS-Rac1 WT/+; 1003.3-GAL4 UAS-mCD8::GFP/+). (G) Neurons expressing dominant negative Rac1 (w; 1003.3-GAL4 UAS-mCD8::GFP/UAS-Rac1 N17). (H) Neurons expressing dominant negative Cdc42 (w; UAS-Cdc42 N17/+; 1003.3-GAL4 UAS-mCD8::GFP/+) Inset show higher magnification of the dashed boxes shown in the main panel. Scale bars = 50 μm. In panels A, C, and D data are means ± s.d. and panel B data are means ± s.e.m. ***P < 0.001 compared with the driver alone control (N = 10; N indicates number of larvae in each genotype). Statistical analyses were performed using one-way ANOVA with Tukey’s HSD post-hoc for pair-wise comparisons.
In contrast, when the dominant negative form of Rac1 or the dominant negative Cdc42 were expressed in the class III neurons, the number of sensory filopodia was decreased (Figure 3A, 3G, and 3H). These larvae also showed behavioral insensitivity to gentle touch stimuli (Figure 3B). Combined, these results suggested a correlation between the number of filopodia and behavioral responses to gentle touch. For each of these manipulations, we also quantified the length of the filopodia (Figure 3C) and the length of the primary dendritic branches (Figure 3D) in controls and in altered class III neurons but these features showed no obvious correlation with gentle touch behaviors.
Expression of Rac1 enhances force-triggered Ca2+ responses of class III neurons
To test whether actin manipulations produced measurable effects on the physiological responses of the class III neurons we investigated their effects on the force responses of these cells in our optical recording preparation. Intriguingly, relative to wild type controls, dramatically elevated Ca2+ responses to force were found in neurons overexpressing Rac1 (Figure 4A, B, D, E, Supplementary Figure 2A, B and Movie S3). These results suggest that the hypersensitive behavioral responses to touch seen in these animals is indeed a consequence of hypersensitive force sensitivity in these cells. Finally, expression of dominant negative Cdc42 in the class III neurons showed a significantly lower GCaMP3.0 change in response to force (Figure 4C, D, E, Supplementary Figure 2C and Movie S3). This result is consistent with the hypothesis that the impaired behavioral responses seen in Cdc42 N17 expressing animals is due to impaired sensory signaling as a consequence of a severe reduction in the number of sensory filopodia.
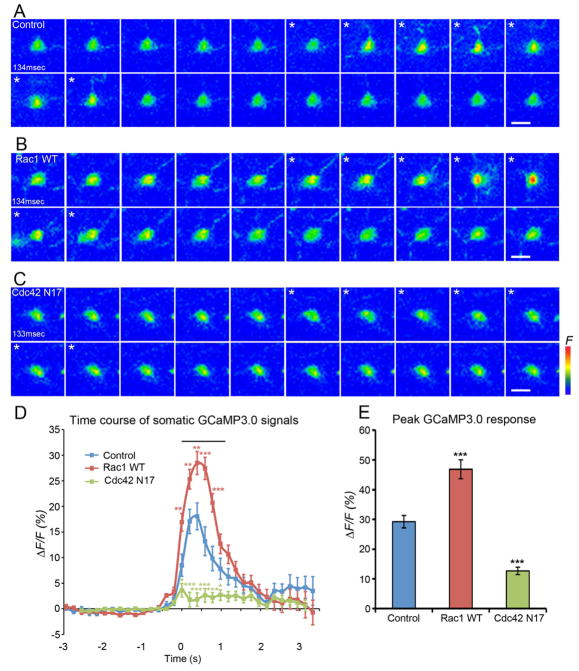
Figure 4. Expression of Rac1 enhances force triggered Ca2+ responses of class III neurons.
Panels A–C show maximum intensity projections from Z-stack time series. Panels D and E show quantification of data across trials. (A) Images of representative class III ddaA neuron expressing GCaMP3.0 (w; ppk-GAL4 UAS-GCaMP3.0/+) displaying increased GCaMP3.0 fluorescence during stimulation (asterisks) (1frame = 134 msec). (B) Images of representative Rac1 expressing ddaA neuron (w; ppk-GAL4 UAS-GCaMP3.0/UAS-Rac1) shows a higher increase in GCaMP3.0 fluorescence relative to controls during stimulation (asterisks) (1frame=134 msec). (C) Images of representative ddaA neuron expressing Cdc42 N17 (w;ppk-GAL4 UAS-GCaMP3.0/UAS-Cdc42N17) that did not show an increase in GCaMP fluorescence during stimulation (asterisks) (1frame=133 msec). Scale bars = 10 μm. (D) Traces showing average 3F/F0 % of ddaA neurons before, during, and after mechanical stimulation. The bar above the traces shows the average length of the mechanical stimulus (1.15 ± 0.02 sec, s.e.m). Statistical analyses was performed using MANOVA test P-value = 0.00074 (control v.s. Rac1 WT). P-value = 0.0000017 (control v.s. Cdc42 N17). Data are means ± s.e.m. ***P < 0.001, **P < 0.01, *P<0.05 compared with control in each time point by Student’s t-test. (N = 72 force stimuli) (E) Average peak ΔF/F0 % of ddaA neurons in response to stimulation of each genotype. Statistical analysis was performed using one-way ANOVA with Tukey’s HSD post-hoc for pair-wise comparisons. Data are means ± s.e.m. ***P < 0.001 compared with control. (N = 72 force stimuli). See also Figure S2 and Movie S3.
Genetic screen for ion channel genes required for the gentle touch response
To enhance our understanding of the molecular mechanisms generating the force responses of the class II and III neurons, we applied a tissue specific RNA interference (RNAi) approach. As a large body of evidence implicates ion channels in mechanosensation we focused our efforts on this class of genes. Using GAL4/UAS mediated expression of RNAi we searched for genes encoding ion channel subunits that were necessary for the response to gentle touch. Our initial screen was carried out using the painless-GAL4 driver to target the expression of RNAi [12]. This driver was chosen because it is broadly expressed in a variety of mechanosensory neurons including chordotonal neurons and all of the md neurons. We crossed the painless-GAL4 driver to 338 UAS-RNAi lines (Supplemental Table 1) targeting over 200 ion channel subunit genes and tested behavioral responses to gentle touch in the crossed progeny. Excluding the large iGluR olfactory ion channel gene family, the screened collection represented the entirety of the annotated ion channel genes of the Drosophila genome. We selected 39 candidate channel genes for further study as these showed potentially reduced gentle touch responses when knocked down by RNAi under control of pain-GAL4 (Supplementary Table 2).
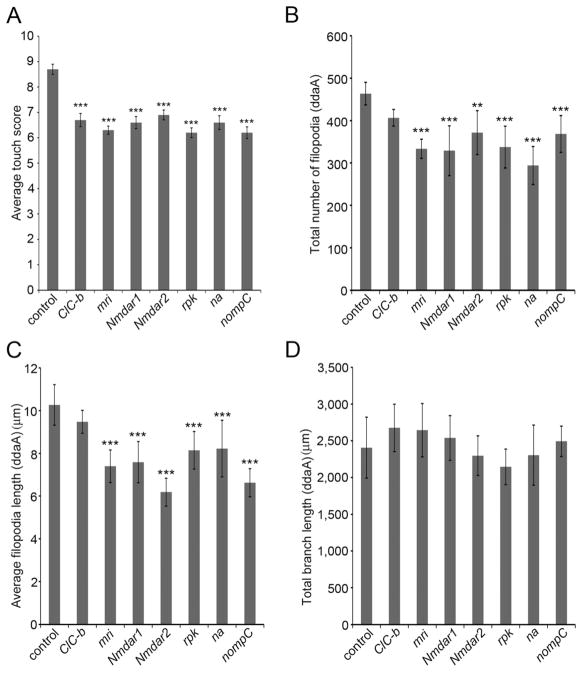
To determine which of the channel genes were likely to function in multidendritic neurons that were targeted by pain-GAL4 the UAS-RNAi lines were next crossed to md-GAL4;UAS-dicer-2. Ten of the candidate channels were found to be needed in all four classes of md-neurons (Supplementary Table 2) for normal touch insensitivity. Finally, the effects of knocking down each of these 10 candidate channel genes in the force responsive class II and class III neurons was tested by crossing the UAS-RNAi lines for these channels to the 1003.3-GAL4;UAS-dicer-2 driver (Figure 5A). The behavioral responses to touch in the crossed progeny showed that seven of the channel subunit genes (Chloride channel-b (ClC-b), mrityu (mri), narrow abdomen (na), no mechanoreceptor potential C (nompC), NMDA Receptor 1 (Nmdar1), NMDA Receptor 2 (Nmdar2), ripped pocket (rpk)) were required in the Class II and Class III neurons for normal responses (Figure 5A).
Figure 5. Ion channel genes are required for gentle touch response and dendritic morphology of class III neurons.
(A) Knockdown of the ion channel genes ClC-b, mri, Nmdar1, Nmdar2, rpk, na, and nompC caused an abnormal gentle touch response relative to the control animals (1003.3-GAL4;UAS-dicer-2 driver crossed to the isogenic w1118 strain of the VDRC). (Data are means ± s.e.m. N = 60 larvae per genotype.) No driver controls for all UAS-RNAi strains showed normal touch behaviors (data not shown). (B–D) Quantification dendrite morphology in ion channel RNAi strains (B) Number of sensory filopodia (C) Average length of sensory filopodia (D) Total length of primary dendrite branches. Data are means ± s.d. **P < 0.01, ***P < 0.001 compared with control. (N = 10 neurons per genotype.) Statistical analyses were performed using one-way analysis of variance (ANOVA) with Tukey’s HSD post-hoc test in pair-wise comparisons. See also Figure S3.
Based on sequence similarity to other known channels the molecular function of ClC-b is predicted to encode a chloride channel. The mri gene encodes a protein that is structurally similar to the tetramerization domain of voltage activated potassium channels [39]. NMDA receptors are well-known ionotropic glutamate receptors that form heteromeric channel complexes comprised of essential Nmdar1 subunits and Nmdar2 subunits [40]. Interestingly, mammalian NMDARs have been proposed to be force sensitive which suggests a potential role in force transduction in class III neurons [41]. The rpk gene encodes a DEG/ENaC channel subunit that is the closest homologue of the Pickpocket channel [42]. narrow abdomen (na) encodes a little studied cation channel unknown function [43]. Finally, nompC encodes a TRP channel with a well-known role in force transduction in the mechanosensory bristles of adult flies and a previously described role in larval touch sensing and proprioception as well as a role in adult hearing [11, 17, 44, 45].
Ion channel genes affect the formation of sensory filopodia
Given the evidence that the filopodia are required for the mechanosensory function of the class III neurons, we investigated whether the identified channel genes might play a role in the development of sensory filopodia. Interestingly, the number of filopodia, and the length of filopodia were significantly reduced by RNAi targeting mri, ripped pocket, nompC, Nmdar1 and Nmdar2 (Figure 5B–D, Supplementaly Figure 3A–3H). Thus, the reduced behavioral responses seen in these genotypes may be a consequence of a reduction in filopodia number. Interestingly, the morphology of Class III neurons (including the number of sensory filopodia) was normal in animals expressing RNAi targeting ClC-b.
ripped pocket is required for gentle touch sensation
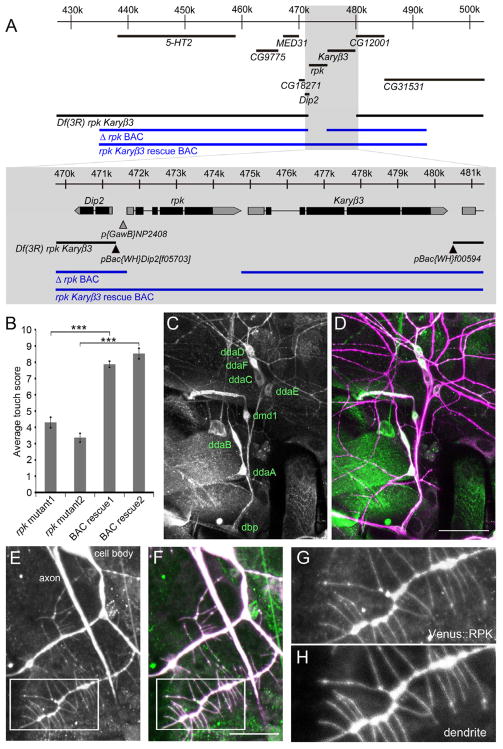
A growing body of evidence has implicated ion channels of the DEG/ENaC family in invertebrate mechanotransduction [3–6]. As the ripped pocket gene encodes a member of this gene family [42], we further investigated its role through the creation of a DNA null allele for the locus. We first constructed a deficiency line, Df(3R)rpk Karyβ3, using FLPase mediated recombination between the FRT site in the pBac{WH}Dip2f05703 line and pBac{WH}f00594 line (Figure 6A). This recombination resulted in the removal of both the rpk gene and the neighboring Karyopherin β-3 (Karyβ3) gene. Unfortunately, Df(3R)rpk Karyβ3 line was lethal at the early larval stage preventing its use in behavioral experiments.
Figure 6. The rpk locus is required for behavioral responses to gentle touch, rpk reporters are expressed in md neurons, and RPK localizes to primary dendrite branches and sensory filopodia.
(A) Schematic representation of the genomic interval surrounding the rpk locus. The regions covered by Δrpk BAC and the rpk Karyβ3 rescue BAC (CH321-22I15) are schematically shown in blue. Locations of transposable elements used to generate Df(3R)rpk Karyβ3 are shown as triangles. (B) Larval gentle touch responses of rpk null and BAC rescue animals. Genotypes were w; Δrpk BAC (22A3)/+; Df(3R)rpk Karyβ3/Df(3R)rpk Karyβ3 (rpk mutant1), w; Δrpk BAC (28E7)/+; Df(3R)rpk Karyβ3/Df(3R)rpk Karyβ3 (rpk mutant2), w; rpk Karyβ3 rescue BAC(22A3)/+; Df(3R)rpk Karyβ3/Df(3R)rpk Karyβ3 (BAC rescue1), w; rpk Karyβ3 rescue BAC (28E7)/+; Df(3R)rpk Karyβ3/Df(3R)rpk Karyβ3 (BAC rescue2). Data are means ± s.e.m. ***P < 0.001 for each value compared with rescue flies using Student t-test. (N = 30 larvae per each genotype.). (C and D) Expression pattern of an enhancer trap (w; p{GawB}NP2408/UAS-mCD8::GFP) at the rpk locus (green in D) was expressed in all md neurons. Neurons were immunolabeled with monoclonal antibody 22C10 (magenta in D). Scale bar = 50 μm. (E–H) Localization of Venus tagged RPK protein in UAS-Venus::RPK/UAS-mCD8::DsRed; 1003.3-GAL4/+ animals (detected with anti-GFP)(E, G, F(green)). Neuronal plasma membrane was labeled with mCD8::DsRed (detected with anti-CD8) (H,F(magenta)). G and H are high magnification images of structures highlighted by the white boxes in E and F (Scale bar = 20 μm).
To test whether the lethality of Df(3R)rpk Karyβ3 was due to removal of rpk or to the removal of Karyβ3, we generated transgenic flies in which we restored Karyβ3 to the Df(3R)rpk Karyβ3 deficiency background (Figure 6A). The restoration of Karyβ3 was achieved through the insertion of a Bacterial Artificial Chromosome (BAC) containing a genetically engineered deletion of rpk (Δrpk BAC). Two independent insertions of the Δrpk BAC were crossed into the Df(3R)rpk Karyβ3 genetic background and both transgenes resulted in deficiency flies that were viable at larval stages, but with somewhat reduced viability at the adult stage.
In addition, we generated transgenic flies with insertion of a BAC that completely restored both the rpk and the Karyβ3 loci ((CH321-22I15) rpk Karyβ3 rescue BAC) (Figure 6A). Two independent insertions of the rpk Karyβ3 rescue BAC were crossed into the Df(3R) rpk Karyβ3 genetic background and both of these were found to completely restore the viability of the deficiency strain.
To formally test the role of rpk in touch transduction we performed behavioral tests on animals of the genotype Δrpk BAC, Df(3R) rpk Karyβ3. These larvae (that were genetically null for rpk locus but rescued for Karyβ3) were strongly insensitive to gentle touch (Figure 6B). Two independent lines with the Δrpk BAC inserted at different insertion sites on the second chromosome (22A3 or 28E7) exhibited this touch insensitive phenotype (Figure 6B). The touch insensitive phenotype of the strain was due to loss of rpk because the presence of a BAC which contained the intact rpk locus completely rescued the touch insensitive phenotype of the rpk mutants. Combined, these data constitute formal genetic proof of a role for the rpk gene in the sensing of gentle touch in Drosophila larvae.
Expression pattern of rpk
A previous study described ripped pocket expression in early embryos but expression in neurons was not reported [42]. Given the effects of RNAi knockdown in class II and class III neurons expression in these cells seemed likely. Thus, to investigate the expression pattern of rpk in 3rd instar larvae, we examined a GAL4 enhancer trap line (p{GawB}NP2408) with an insertion approximately 200 base pairs upstream of the rpk transcriptional start site (Figure 6A). When crossed to UAS-mCD8::GFP, clear expression of GFP was detected in all md neurons (Figure 6C and 6D).
In order to investigate the subcellular localization of RPK in the md neurons we generated transgenic Drosophila that expressed RPK as a fusion with the Venus Fluorescent Protein in N-terminal region (1003.3-GAL4>UAS-Venus::RPK) (Figure 6E–6H). The Venus tagged RPK protein was detected throughout the primary dendritic arbor, cell body, and axons of md neurons (Figure 6E and 6F). Interestingly, Venus tagged RPK was detectable throughout the length of the filopodia in Class III neurons (Figure 6G and 6H).
Discussion
In this study, we have identified the first known function for the Class II and the Class III md neurons. These neurons showed physiological responses to force, their output was required for touch responses, and their optogenetic activation was sufficient to generate behavioral responses that resembled the behavioral responses to touch. We further focused our attention on the class III neurons and their actin-rich sensory filopodia. Our results indicated that the filopodia contribute to both physiological (Ca2+ responses) and behavioral responses to gentle touch.
Our time-lapse imaging indicated that sensory filopodia in class III neurons are very dynamic during the early third instar stage, which has interesting similarity to the actin mediated spine motility known to occur at immature synapses [46]. Interestingly, sensory filopodia are quite stable in late wandering third instar in comparison to the early third instar (AT and WDT unpublished observations), thus the dynamic nature of these filopodia may also be related to a developmental process in which the overall mechanical sensitivity of the neuron is tuned as the dendritic arbor scales during the profound growth period of the third instar.
An interesting possibility is that the actin-rich structures may hold mechanosensitive channels in a similar fashion to the stereovilli of hair cells in the vertebrate inner ear. If this is the case, growth and relaxation of the sensory filopoida may be important for fine-tuning the plasma membrane tension that is needed for proper mechanotransduction.
To begin to elucidate the molecular mechanisms by which class III neurons sense external signals, we performed a comprehensive RNAi screen for channel genes that were required for gentle touch responses. Ion channel gene families previously implicated in mechanotransduction (TRP, DEG/ENaC, and NMDARs) were identified in this screen. Although it is tempting to speculate from our results that one or more of the channel subunits isolated from our screen might be mechanically sensitive, further experimentation will be needed to adequately test this hypothesis.
We focused our efforts on the rpk gene, which is a member of the DEG/ENaC family [42]. We provide formal genetic proof that this gene is essential for gentle touch responses in the Drosophila larvae. Interestingly, the DEG/ENaC subunit encoded by the pickpocket locus is required for mechanical nociception [6, 22]. This function is consistent with the very specific expression pattern of the ppk gene in class IV neurons [6, 9]. Although ppk is not required for gentle touch responses, ppk-GAL4 is weakly expressed in the Class III neurons [6]. Thus, although not essential for gentle touch responses, PPK represents a potential heteromeric partner subunit for RPK in these cells [42].
Our results suggest that the identified ion channel subunits contribute to the stability of the sensory filopodia. Prior results implicated Calcium/calmodulin-dependent protein kinase II (CaMKII) in sensory filopodia growth [33], however the source of calcium ions to activate CaMKII in the Class III neurons is unknown. The NOMPC channel and the NMDARs identified here represent good candidates for providing mechanically activated Ca2+ entry that could drive the activation of CaMKII. Local activation of Rho family GTPases in post-synaptic dendritic spines can also be driven by CaMKII [47]. So the reduced filopodia formation seen in our ion channel-knockdown experiments may also involve pathways utilizing the same Rho family members that we have manipulated in this study. When we manipulated dendritic morphology by expressing wild type and dominant negative forms of Rho family GTPases in class III neurons, both filopodia number and behavioral sensitivity to gentle touch were dramatically changed.
In conclusion, actin-rich dendritic sensory filopodia on non-ciliated Drosophila class III neurons represent sensory organelles that play an important role in efficient somatosensory mechanosensation. An ensemble of ion channels interacts with these sensory filopodia in generating touch responses. Importantly, F-actin has been implicated in mechanosensory responses of mammalian somatosensory neurons [48, 49] and actin-rich protrusions have been observed on mechanosensory Merkel cells [50–52]. An interesting possibility is mammalian somatosensation relies upon structures that are evolutionarily homologous to the actin-rich sensory filopodia of the Drosophila Class III neurons. Thus, the further study of these structures may shed important light on the mechanisms of mammalian touch sensation.
Experimental Procedures
Fly maintenance and stocks
Drosophila stocks were raised on standard cornmeal molasses fly food medium at 25°C and 75% humidity on a 12/12 light/dark cycle. The following fly strains were used: w; GAL4 109(2)80 (md-GAL4), w; c161-GAL4, w; 1003.3-GAL4 [24], w; c755-GAL4, ppk1.9-GAL4, w; 2-21-GAL4, w; nompC-GAL4 [53], w; tutl-GAL4/Bc (Gift from Don van Meyel unpublished), w; 117y-GAL4, w; UAS-mCD8::GFP, w; UAS-moesin::GFP [34], w; UAS-mCD8::DsRed, w; UAS-TNT-E [26], w; UAS-ChR2-YFP (line C) [9], w; UAS-Rac1 WT, w; UAS-Rac1 N17, w; UAS-Cdc42 N17, tsh-GAL80, and UAS-Dicer2. Fly stocks with UAS marker, Rho family GTPases, and TRiP RNAi lines were provided by the Bloomington Stock Center. RNAi lines for screening were provided by the Vienna Drosophila RNAi Center [54]. The w; p{GawB}NP2408 strain was provided by the Kyoto Drosophila Genetic Resource Center, and w; pBac{WH}Dip2[f05703] and w; pBac{WH}f00594 strains were provided by the Exelixis collection at Harvard Medical School.
Gentle touch behavioral assay
The gentle touch behavioral tests were performed as described previously [6, 9, 12, 17, 25]. Each larva was tested 4 times and then discarded. The touch scores (0–4; Figure 1) were summed for obtain a score of 0–16.
Optogenetic activation
The behavioral tests were performed as described previously [6, 9, 31] stimulated with blue light (460 nm – 500 nm) for several seconds using the Hg light source of a Leica MZ16 FA stereomicroscope (14,500 lux). Behavioral responses were videotaped and analyzed offline.
Confocal imaging of dendrites
To observe dendritic morphology, early 3rd-instar larvae were anesthetizing larvae with ether mounted in Halocarbon oil 27 (Sigma). Images were taken every 30 seconds for 30 minutes using an Apochromat 40 × N/A 1.3 oil immersion lens.
Molecular cloning
We generated the Δrpk BAC through galK recombineering [55] of the P[acman] clone CH321-22I15 [56] in SW102 cells. The galK cassette was amplified from pGalK vector [55] using primers with 50 bp homology arms desigined to insert galK to the rpk locus ((forward primer 5′-CTAAATTGTCACCATCCGAACGTTTTTTTGCAAGTTCTCTCCAAGATCCTGCCTGTTGACAATTAATCATCGGCA-3′) (reverse primer 5′-CTTTTCTTGAACCGAAAAGCTTAAAAAGAAGCTGCACATGGGCTGCGATCTTCAGCACTGTCCTGCTCCTT-3′). The amplified galK cassette was transformed into SW102 cells harboring BAC CH321-22I15 followed by selection on M63 minimal media plates with galactose, leucine, biotin, and chloramphenicol. After incubation for 3 days at 32°C individual colonies were streaked on MacConkey chloramphenicol-indicator plates and Gal+ colonies were picked and tested for the desired galk insertion by PCR and by checking for diagnostic SpeI restriction fragments and agarose gel electrophoresis. The inserted galK cassette was then removed by negative selection against galK. A double-stranded oligo with the desired deficiency break point to remove the rpk gene (with the sequence: 5′-CTAAATTGTCACCATCCGAACGTTTTTTTGCAAGTTCTCTCCAAGATCCTGAGATCGCAGCCCATGTGCAGCTTCTTTTTAAGCTTTTCGGTTCAAGAAAAG-3′) was transformed into SW102 cells harboring the galK insertion BAC and the transformed cells were then plated onto M63 minimal media plates with glycerol, leucine, biotin, 2-deoxy-galactose (DOG), and chloramphenicol. After incubation for 3 days at 32°C, colonies that grew under the negative selection by DOG were further analyzed by digestion with SpeI and agarose gel electrophoresis. Colonies with the desired restriction pattern (that indicated removal of galK and the deletion of rpk) where further analyzed by PCR and sequencing across the deficiency breakpoint.
The pUAST-Venus::rpk plasmid was generated by PCR amplification from BAC DNA (CH321-22I15) (using the forward primer 5′-CACCATGACCATATCGGATTCGGAACTCGAC-3′ and the reverse primer 5′-TTATCCTTTAACCAGGCGCTTCAGATTGG-3′). The genomic rpk PCR product was then cloned into the pENTR/D-TOPO vector (Invitrogen) and fully sequenced. The desired clone was then used in the Gateway LR in vitro recombination reaction with the pTVW vector (Invitrogen).
Generation of Transgenic Flies
BAC DNA was purified by the PureLink HiPure Plasmid DNA Purification Kit (Invitrogen). Microinjections of the Δrpk BAC and the rpk Karyβ3 rescue BAC (CH321-22I15) were performed by BestGene Inc via PhiC31-mediated chromosome integration, with VK37(2L)22A3 or VK02(2L)28E7 as the docking sites [56]. The pUAST-Venus::rpk and pUAST-Venus::Nmdar1 plasmids were injected by GenetiVision for P-element-mediated transformation.
Immunostaining of larvae
Larvae were dissected and stained according to standard protocols (primary antibodies: rat anti-mouse CD8a antibody (1:100, Invitrogen), mouse GFP antibody (1:1000, Invitrogen), and mouse 22C10 antibody (1:300).
Calcium imaging
Wandering 3rd instar larvae expressing the genetically encoded Ca2+ sensor GCaMP3.0 [32] were filleted and immersed in a hemolymph like saline (HL-3), 70 mM NaCl, 5mM KCl, 1.5 mM CaCl2, 20 mM MgCl2, 10 mM NaCO3, 5 mM Trehalose, 115 mM Sucrose, and 5 mM HEPES (pH 7.2) [57]. The md neurons expressing GCaMP3.0 were then imaged using 20 × Plan-Apochromat lens N/A 0.8 and 488 nm laser through the transparent cuticle with the high-speed time-lapse ZEISS LSM 5 Live Confocal System. A piezo driven focus drive was used for fast acquisition of three-dimensional Z-stack time series. The dimensions in X-Y were 256 ×128 pixels. The pinhole size was 6.6 μm. The thickness of a single slice was 8μm and approximately 7 overlapping slices were acquired in each Z-stack image (total Z-stack depth ranged from_55 ± 1.8 μm s.e.m). The scan speed for acquisition of a single slice was approximately 15 msec which allowed us to acquire Z-stack images at an average rate of 8Hz. During the time series, force (~1mN) was applied to the imaged field of neurons using a nylon monofilament (diameter is approximately 200 μm) that was attached to a manually controlled micromanipulator. In each preparation, 10 force stimuli that each produced a visible 47 ± 2 μm s.e.m. displacement of the tissue in X-Y axes and ~25 μm in the Z axis, were consecutively applied to the filleted larva. The data were analyzed with the Zeiss LSM software physiology package. The cell body of individual cells was selected as regions of interest (ROI) in the maximum intensity projections from the confocal time series. To correct for movement of the cells in the XY axes, the position of the ROI was manually centered for each cell in each frame of the time series, and the measured fluorescence intensity was recorded. The average GCaMP3.0 intensity from the 15 frames prior to stimulus presentation was used as F0. 3F/F0 % was calculated with the formula 100% ×3F/F0 where F was the fluorescence intensity at each time point and F0 was the average baseline fluorescence intensity of the 15 frames preceding each stimulus. Average traces for each genotype were calculated by resampling of the raw data to 20 msec, decimating, and then down-sampling to 200 msec using the MATLAB software program (MathWorks Inc.).
Supplementary Material
Highlights.
Larval multidendritic neurons are required for sensing gentle touch in Drosophila.
Optogenetic activation of defined multidendritic neurons triggers touch behaviors.
Dendritic sensory filopodia are dynamic, F-actin-rich, organelles necessary for mechanosensing.
A high-throughput touch screen identifies NMDARs, NOMPC and Ripped Pocket.
Acknowledgments
We thank Dr. Don Van Meyel, Dr. Steven Stowers, the Vienna Drosophila RNAi Center and the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic RNAi fly stocks used in this study. We also thank the Bloomington Drosophila stock center for supplying other strains used in this study. Ken Honjo, Andrew Bellemer, Stephanie Mauthner, Kia Walcott, and Jessica Robertson made helpful suggestions on the manuscript. Jouchi Nakajima helped with the statistical analyses. This work was supported by grants to WDT from the National Institutes on Deafness and other Communication Disorders (R21DC010222) and the National Institute of Neurological Disorders and Stroke (5R01NS054899).
Footnotes
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Corey DP, Hudspeth AJ. Kinetics of the receptor current in bullfrog saccular hair cells. J Neurosci. 1983;3:962–976. doi: 10.1523/JNEUROSCI.03-05-00962.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Hagan R, Chalfie M, Goodman MB. The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals. Nat Neurosci. 2005;8:43–50. doi: 10.1038/nn1362. [DOI] [PubMed] [Google Scholar]
- 3.Geffeney SL, Cueva JG, Glauser DA, Doll JC, Lee TH, Montoya M, Karania S, Garakani AM, Pruitt BL, Goodman MB. DEG/ENaC but not TRP channels are the major mechanoelectrical transduction channels in a C. elegans nociceptor. Neuron. 2011;71:845–857. doi: 10.1016/j.neuron.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driscoll M, Chalfie M. The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration. Nature. 1991;349:588–593. doi: 10.1038/349588a0. [DOI] [PubMed] [Google Scholar]
- 5.Chatzigeorgiou M, Yoo S, Watson JD, Lee WH, Spencer WC, Kindt KS, Hwang SW, Miller DM, 3rd, Treinin M, Driscoll M, et al. Specific roles for DEG/ENaC and TRP channels in touch and thermosensation in C. elegans nociceptors. Nat Neurosci. 2010;13:861–868. doi: 10.1038/nn.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong L, Hwang RY, Tracey WD. Pickpocket is a DEG/ENaC protein required for mechanical nociception in Drosophila larvae. Curr Biol. 2010;20:429–434. doi: 10.1016/j.cub.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Price MP, Lewin GR, McIlwrath SL, Cheng C, Xie J, Heppenstall PA, Stucky CL, Mannsfeldt AG, Brennan TJ, Drummond HA, et al. The mammalian sodium channel BNC1 is required for normal touch sensation. Nature. 2000;407:1007–1011. doi: 10.1038/35039512. [DOI] [PubMed] [Google Scholar]
- 8.Price MP, McIlwrath SL, Xie J, Cheng C, Qiao J, Tarr DE, Sluka KA, Brennan TJ, Lewin GR, Welsh MJ. The DRASIC cation channel contributes to the detection of cutaneous touch and acid stimuli in mice. Neuron. 2001;32:1071–1083. doi: 10.1016/s0896-6273(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 9.Hwang RY, Zhong L, Xu Y, Johnson T, Zhang F, Deisseroth K, Tracey WD. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr Biol. 2007;17:2105–2116. doi: 10.1016/j.cub.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colbert HA, Smith TL, Bargmann CI. OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans. J Neurosci. 1997;17:8259–8269. doi: 10.1523/JNEUROSCI.17-21-08259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker RG, Willingham AT, Zuker CS. A Drosophila mechanosensory transduction channel. Science. 2000;287:2229–2234. doi: 10.1126/science.287.5461.2229. [DOI] [PubMed] [Google Scholar]
- 12.Tracey WD, Jr, Wilson RI, Laurent G, Benzer S. painless, a Drosophila gene essential for nociception. Cell. 2003;113:261–273. doi: 10.1016/s0092-8674(03)00272-1. [DOI] [PubMed] [Google Scholar]
- 13.Kwan KY, Allchorne AJ, Vollrath MA, Christensen AP, Zhang DS, Woolf CJ, Corey DP. TRPA1 contributes to cold, mechanical, and chemical nociception but is not essential for hair-cell transduction. Neuron. 2006;50:277–289. doi: 10.1016/j.neuron.2006.03.042. [DOI] [PubMed] [Google Scholar]
- 14.Kindt KS, Viswanath V, Macpherson L, Quast K, Hu H, Patapoutian A, Schafer WR. Caenorhabditis elegans TRPA-1 functions in mechanosensation. Nat Neurosci. 2007;10:568–577. doi: 10.1038/nn1886. [DOI] [PubMed] [Google Scholar]
- 15.Kang L, Gao J, Schafer WR, Xie Z, Xu XZ. C. elegans TRP family protein TRP-4 is a pore-forming subunit of a native mechanotransduction channel. Neuron. 2010;67:381–391. doi: 10.1016/j.neuron.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J, Chung YD, Park DY, Choi S, Shin DW, Soh H, Lee HW, Son W, Yim J, Park CS, et al. A TRPV family ion channel required for hearing in Drosophila. Nature. 2003;424:81–84. doi: 10.1038/nature01733. [DOI] [PubMed] [Google Scholar]
- 17.Kernan M, Cowan D, Zuker C. Genetic dissection of mechanosensory transduction: mechanoreception-defective mutations of Drosophila. Neuron. 1994;12:1195–1206. doi: 10.1016/0896-6273(94)90437-5. [DOI] [PubMed] [Google Scholar]
- 18.Zhong L, Bellemer A, Yan H, Honjo K, Robertson J, Hwang RY, Pitt GS, Tracey WD. Thermosensory and non-thermosensory isoforms of Drosophila melanogaster TRPA1 reveal heat sensor domains of a thermoTRP channel. Cell Rep. 2012;1:43–55. doi: 10.1016/j.celrep.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawashima Y, Geleoc GS, Kurima K, Labay V, Lelli A, Asai Y, Makishima T, Wu DK, Della Santina CC, Holt JR, et al. Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. J Clin Invest. 2011;121:4796–4809. doi: 10.1172/JCI60405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maingret F, Patel AJ, Lesage F, Lazdunski M, Honore E. Mechano- or acid stimulation, two interactive modes of activation of the TREK-1 potassium channel. J Biol Chem. 1999;274:26691–26696. doi: 10.1074/jbc.274.38.26691. [DOI] [PubMed] [Google Scholar]
- 21.Coste B, Xiao B, Santos JS, Syeda R, Grandl J, Spencer KS, Kim SE, Schmidt M, Mathur J, Dubin AE, et al. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature. 2012;483:176–181. doi: 10.1038/nature10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SE, Coste B, Chadha A, Cook B, Patapoutian A. The role of Drosophila Piezo in mechanical nociception. Nature. 2012;483:209–212. doi: 10.1038/nature10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coste B, Mathur J, Schmidt M, Earley TJ, Ranade S, Petrus MJ, Dubin AE, Patapoutian A. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330:55–60. doi: 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes CL, Thomas JB. A sensory feedback circuit coordinates muscle activity in Drosophila. Molecular and cellular neurosciences. 2007;35:383–396. doi: 10.1016/j.mcn.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caldwell JC, Miller MM, Wing S, Soll DR, Eberl DF. Dynamic analysis of larval locomotion in Drosophila chordotonal organ mutants. Proc Natl Acad Sci U S A. 2003;100:16053–16058. doi: 10.1073/pnas.2535546100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sweeney ST, Broadie K, Keane J, Niemann H, O’Kane CJ. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14:341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 27.Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- 28.Han C, Wang D, Soba P, Zhu S, Lin X, Jan LY, Jan YN. Integrins Regulate Repulsion-Mediated Dendritic Patterning of Drosophila Sensory Neurons by Restricting Dendrites in a 2D Space. Neuron. 2012;73:64–78. doi: 10.1016/j.neuron.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim ME, Shrestha BR, Blazeski R, Mason CA, Grueber WB. Integrins establish dendrite-substrate relationships that promote dendritic self-avoidance and patterning in drosophila sensory neurons. Neuron. 2012;73:79–91. doi: 10.1016/j.neuron.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honjo K, Hwang RY, Tracey WD., Jr Optogenetic manipulation of neural circuits and behavior in Drosophila larvae. Nat Protoc. 2012;7:1470–1478. doi: 10.1038/nprot.2012.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersen R, Li Y, Resseguie M, Brenman JE. Calcium/calmodulin-dependent protein kinase II alters structural plasticity and cytoskeletal dynamics in Drosophila. J Neurosci. 2005;25:8878–8888. doi: 10.1523/JNEUROSCI.2005-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dutta D, Bloor JW, Ruiz-Gomez M, VijayRaghavan K, Kiehart DP. Real-time imaging of morphogenetic movements in Drosophila using Gal4-UAS-driven expression of GFP fused to the actin-binding domain of moesin. Genesis. 2002;34:146–151. doi: 10.1002/gene.10113. [DOI] [PubMed] [Google Scholar]
- 35.Luo L. Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu Rev Cell Dev Biol. 2002;18:601–635. doi: 10.1146/annurev.cellbio.18.031802.150501. [DOI] [PubMed] [Google Scholar]
- 36.Jinushi-Nakao S, Arvind R, Amikura R, Kinameri E, Liu AW, Moore AW. Knot/Collier and cut control different aspects of dendrite cytoskeleton and synergize to define final arbor shape. Neuron. 2007;56:963–978. doi: 10.1016/j.neuron.2007.10.031. [DOI] [PubMed] [Google Scholar]
- 37.Emoto K, He Y, Ye B, Grueber WB, Adler PN, Jan LY, Jan YN. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 2004;119:245–256. doi: 10.1016/j.cell.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 38.Lee A, Li W, Xu K, Bogert BA, Su K, Gao FB. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development. 2003;130:5543–5552. doi: 10.1242/dev.00792. [DOI] [PubMed] [Google Scholar]
- 39.Rusconi JC, Challa U. Drosophila Mrityu encodes a BTB/POZ domain-containing protein and is expressed dynamically during development. Int J Dev Biol. 2007;51:259–263. doi: 10.1387/ijdb.062233jr. [DOI] [PubMed] [Google Scholar]
- 40.Ultsch A, Schuster CM, Laube B, Schloss P, Schmitt B, Betz H. Glutamate receptors of Drosophila melanogaster: cloning of a kainate-selective subunit expressed in the central nervous system. Proc Natl Acad Sci U S A. 1992;89:10484–10488. doi: 10.1073/pnas.89.21.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paoletti P, Ascher P. Mechanosensitivity of NMDA receptors in cultured mouse central neurons. Neuron. 1994;13:645–655. doi: 10.1016/0896-6273(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 42.Adams CM, Anderson MG, Motto DG, Price MP, Johnson WA, Welsh MJ. Ripped pocket and pickpocket, novel Drosophila DEG/ENaC subunits expressed in early development and in mechanosensory neurons. J Cell Biol. 1998;140:143–152. doi: 10.1083/jcb.140.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lear BC, Lin JM, Keath JR, McGill JJ, Raman IM, Allada R. The ion channel narrow abdomen is critical for neural output of the Drosophila circadian pacemaker. Neuron. 2005;48:965–976. doi: 10.1016/j.neuron.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 44.Kamikouchi A, Inagaki HK, Effertz T, Hendrich O, Fiala A, Gopfert MC, Ito K. The neural basis of Drosophila gravity-sensing and hearing. Nature. 2009;458:165–171. doi: 10.1038/nature07810. [DOI] [PubMed] [Google Scholar]
- 45.Gopfert MC, Albert JT, Nadrowski B, Kamikouchi A. Specification of auditory sensitivity by Drosophila TRP channels. Nat Neurosci. 2006;9:999–1000. doi: 10.1038/nn1735. [DOI] [PubMed] [Google Scholar]
- 46.Bonhoeffer T, Yuste R. Spine motility. Phenomenology, mechanisms, and function. Neuron. 2002;35:1019–1027. doi: 10.1016/s0896-6273(02)00906-6. [DOI] [PubMed] [Google Scholar]
- 47.Murakoshi H, Wang H, Yasuda R. Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature. 2011;472:100–104. doi: 10.1038/nature09823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drew LJ, Wood JN, Cesare P. Distinct mechanosensitive properties of capsaicin-sensitive and -insensitive sensory neurons. J Neurosci. 2002;22:RC228. doi: 10.1523/JNEUROSCI.22-12-j0001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piao L, Li HY, Park CK, Cho IH, Piao ZG, Jung SJ, Choi SY, Lee SJ, Park K, Kim JS, et al. Mechanosensitivity of voltage-gated K+ currents in rat trigeminal ganglion neurons. J Neurosci Res. 2006;83:1373–1380. doi: 10.1002/jnr.20810. [DOI] [PubMed] [Google Scholar]
- 50.Sekerkova G, Zheng L, Loomis PA, Changyaleket B, Whitlon DS, Mugnaini E, Bartles JR. Espins are multifunctional actin cytoskeletal regulatory proteins in the microvilli of chemosensory and mechanosensory cells. J Neurosci. 2004;24:5445–5456. doi: 10.1523/JNEUROSCI.1279-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haeberle H, Lumpkin EA. Merkel Cells in Somatosensation. Chemosens Percept. 2008;1:110–118. doi: 10.1007/s12078-008-9012-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toyoshima K, Seta Y, Takeda S, Harada H. Identification of Merkel cells by an antibody to villin. J Histochem Cytochem. 1998;46:1329–1334. doi: 10.1177/002215549804601113. [DOI] [PubMed] [Google Scholar]
- 53.Petersen LK, Stowers RS. A Gateway MultiSite recombination cloning toolkit. PLoS One. 2011;6:e24531. doi: 10.1371/journal.pone.0024531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 55.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Venken KJ, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stewart BA, Atwood HL, Renger JJ, Wang J, Wu CF. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J Comp Physiol A. 1994;175:179–191. doi: 10.1007/BF00215114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.