Abstract
Objective
Multiple sclerosis (MS) is a multifactorial neurologic disease characterized by modest but tractable heritability. Genome Wide Association Studies (GWAS) have identified and/or validated multiple polymorphisms in approximately 16 genes associated with susceptibility. We aimed at investigating the aggregation of genetic MS-risk markers in individuals by comparing multi and single-case families.
Methods
A weighted log-additive integrative approach termed MS Genetic Burden (MSGB) was used to account for the well-established genetic variants from previous association studies and meta-analyses. The corresponding genetic burden and its transmission was analyzed in 1213 independent MS families (810 sporadic and 403 multi-case families).
Results
MSGB analysis demonstrated a higher aggregation of susceptibility variants in multi-case, compared to sporadic MS families. In addition, the aggregation of non-MHC SNPs depended neither on gender nor on the presence or absence of HLA-DRB1*15:01 alleles. Interestingly, while a greater MSGB in siblings of MS patients was associated with an increased risk of MS (OR=2.1, p=0.001), ROC curves of MSGB differences between probands and sibs (AUROC 0.57 [0.53; 0.61]) show that case-control status prediction of MS cannot be achieved with the currently available genetic data.
Interpretation
The primary interest in the MSGB concept resides in its capacity to integrate cumulative genetic contributions to MS risk. This analysis underlines the high variability of family load with known common variants. This novel approach can be extended to other genetically complex diseases. Despite the emphasis in assembling large case-control datasets, multigenerational, multi-affected families remain an invaluable resource for advancing the understanding of the genetic architecture of complex traits.
Introduction
Multiple sclerosis (MS) is a severe disease of the central nervous system and common cause of neurological disability in young adults (1, 2). MS displays several characteristics that are common to numerous autoimmune diseases including moderate polygenic heritability, evidence of environmental exposure, clinical and genetic heterogeneity, increased frequency in women, and susceptibility conferred primarily by an HLA-associated gene or genes 3–6. Recently completed genome wide association studies (GWAS) validated the predominant role of the MHC region in the genetic architecture of the disease, together with the identification of additional true susceptibility variants of modest effects (3–8).
The degree of familial aggregation of MS cases has been a subject of recent debate (9), with estimates ranging from 20 to less than 10 (10). In addition to potential biases in previous estimations, a decline in λs values compared with earlier estimates of 30 or higher could be explained by the increasing global prevalence of MS over the past century. Even at the lower threshold of these estimates, familial disease aggregation remains a pivotal element supporting the role of genetic influences on MS susceptibility. It is generally accepted that familial and sporadic MS are clinically indistinguishable. Given that environmental factors act most likely at the population level, it is conceivable that in addition to chance, an elevated genetic risk or burden may be operating in the families with first-degree co-affected relatives.
The role of genetic factors in the heightened susceptibility in females is unknown. Elegant experiments in MS animal models confirm the role of sex chromosomes in the female bias of autoimmune demyelination (11). Interestingly, the increase in the incidence of MS over the last century may have occurred primarily in women (12, 13). Since the distribution and frequency of genetic risk factors cannot have changed over such a short time, notwithstanding improved surveillance, this differential increasing frequency suggests the importance of non-genetic factors in the gender incidence bias. We address these questions by comparing the accumulated genetic risk in affected individuals from multi and single-case families, including gender in the modeling of the MS genetic “burden” (MSGB).
MATERIAL AND METHODS
In the present study, 1213 families (Table 1) are re-assessed in light of the contributions of recently identified genetic risk factors taken from various MS GWAS or meta analysis efforts (1, 4, 5, 14, 15) (Table 2). The UCSF institutional review board approved this study and written informed consent was obtained from all participants. All known ancestors were white and of European descent. The population studied here is comprised of two datasets of independent families: 403 “multi-case” families in which at least one first-degree relative of the affected proband also had clinically definite MS; and 810 “sporadic” (single-case) families in which the affected individual reported no known history of MS in any family member(16, 17). Families with ambiguous records of co-occurrence were omitted from the study. Diagnostic criteria and ascertainment protocols were identical for both datasets and are summarized elsewhere (16, 17). The patient characteristics are presented in Table 1. In this study, only parents of the proband and their offspring were studied. No significant differences were found between the 403 probands of multi-case families, the 412 affected relatives in the probands’ family, and the 810 probands of sporadic MS families in terms of gender, age of onset, disease duration, proportion with relapsing remitting MS, and proportion with secondary progressive MS (all p>0.05).
Table 1.
Clinical and demographic characteristics of study participants
| Multicase Families | Sporadic MS Families | ||
|---|---|---|---|
|
| |||
| Probands (n=403) | Affected relatives (N=394) | Probands (n=810) | |
|
| |||
| Gender Ratio (N female) | 305 | 276 | 612 |
| % [ 95% Confidence Interval] | 75.7% [71.5,79.9] | 70.1% [65.5,74.6] | 75.6% [72.6,78.5] |
|
| |||
| Average Age of Onset | 30 | 30 | 30 |
| Median( p25– p75) | (23–36) | (23–37) | (24–37) |
|
| |||
| Average Disease Duration | 10 | 13 | 8 |
| Median( p25– p75) | (5–18) | (6–22) | (4–15) |
|
| |||
| Relapsing Remitting and Secondary Progressive MS | 366 | 320 | 709 |
| % [ 95% Confidence Interval] | 90.8% [88,93.6] | 81.2% [77.4,85.1] | 87.5% [85.3,89.8] |
Table 2.
SNPs used for computing a Multiple Sclerosis Genetic Burden Score (MSGB)
| SNP rs# | “Risk” allele | Effect Size | Gene | Literature Reference |
|---|---|---|---|---|
| rs12708716 | A | 1.16 | CLEC16A | (Johnson et al. 2009) |
| rs10735781 | G | 1.14 | EVI5 | (Johnson et al. 2009) |
| rs2104286 | A | 1.15 | IL2RA | (De Jager et al. 2009) |
| rs2300747 | A | 1.30 | CD58 | (De Jager et al. 2009) |
| rs34536443 | G | 1.33 | TYK2 | (Johnson et al. 2009) |
| rs17445836 | G | 1.25 | IRF8 | (De Jager et al. 2009) |
| rs763361 | T | 1.13 | CD226 | (Hafler et al. 2009) |
| rs17824933 | G | 1.18 | CD6 | (De Jager et al. 2009) |
| rs727986 | C | 1.21 | GPC5 | (Baranzini et al. 2009) |
| rs6897932 | C | 1.12 | IL7R | (De Jager et al. 2009) |
| rs3135388 | T | 2.75 | HLA-DRB1 | (De Jager et al. 2009) |
| rs4149584 | T | 1.58 | TNFRSF1A | (De Jager et al. 2009) |
| rs4680534 | C | 1.12 | IL12A | (De Jager et al. 2009) |
| rs1790100 | G | 1.11 | MPHOSPH9 | (De Jager et al. 2009) |
| rs2760524 | G | 1.15 | RGS1 | (De Jager et al. 2009) |
| rs12122721 | G | 1.22 | KIF21B | IMSGC (IMSGC-consortium 2010) |
| rs1132200 | G | 1.24 | TMEM39A | IMSGC (IMSGC-consortium 2010) |
Table 2 present the rs# numbers for each SNP. A single SNP was taken into account for each genomic region showing evidence of association with MS susceptibility. The effect size corresponds to odds ratio in a dose dependent model. The references given for each SNP correspond to the study from which the effect size was estimated using subjects of European ancestry and not necessarily the original report of the association. rs3135388*T tags HLA-DRB1*15:01.
References:
International Multiple Sclerosis Genetics Consortium (IMSGC) Baranzini, S. E., Wang, J., Gibson, R. A., Galwey, N., Naegelin, Y., Barkhof, F., Radue, E. W., Lindberg, R. L., Uitdehaag, B. M., Johnson, M. R., Angelakopoulou, A., Hall, L., Richardson, J. C., Prinjha, R. K., Gass, A., Geurts, J. J., Kragt, J., Sombekke, M., Vrenken, H., Qualley, P., Lincoln, R. R., Gomez, R., Caillier, S. J., George, M. F., Mousavi, H., Guerrero, R., Okuda, D. T., Cree, B. A., Green, A. J., Waubant, E., Goodin, D. S., Pelletier, D., Matthews, P. M., Hauser, S. L., Kappos, L., Polman, C. H., and Oksenberg, J. R.: Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet18: 767–78, 2009
De Jager, P. L., Jia, X., Wang, J., de Bakker, P. I., Ottoboni, L., Aggarwal, N. T., Piccio, L., Raychaudhuri, S., Tran, D., Aubin, C., Briskin, R., Romano, S., Baranzini, S. E., McCauley, J. L., Pericak-Vance, M. A., Haines, J. L., Gibson, R. A., Naeglin, Y., Uitdehaag, B., Matthews, P. M., Kappos, L., Polman, C., McArdle, W. L., Strachan, D. P., Evans, D., Cross, A. H., Daly, M. J., Compston, A., Sawcer, S. J., Weiner, H. L., Hauser, S. L., Hafler, D. A., and Oksenberg, J. R.: Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet 41: 776–82, 2009
Hafler, J. P., Maier, L. M., Cooper, J. D., Plagnol, V., Hinks, A., Simmonds, M. J., Stevens, H. E., Walker, N. M., Healy, B., Howson, J. M., Maisuria, M., Duley, S., Coleman, G., Gough, S. C., Worthington, J., Kuchroo, V. K., Wicker, L. S., and Todd, J. A.: CD226 Gly307Ser association withmultiple autoimmune diseases. Genes Immun10: 5–10, 2009
IMSGC-consortium: Comprehensive follow-up of the first genome-wide association study of multiple sclerosis identifies KIF21B and TMEM39A as susceptibility loci. Hum Mol Genet19: 953–62, 2010
Johnson, B. A., Wang, J., Taylor, E. M., Caillier, S. J., Herbert, J., Khan, O. A., Cross, A. H., De Jager, P. L., Gourraud, P. A., Cree, B. C., Hauser, S. L., and Oksenberg, J. R.: Multiple sclerosis susceptibility alleles in African Americans. Genes Immun, 2009
The MSGB was computed based on a weighted scoring algorithm using one SNP per MS associated genomic region as found by trend-test association (meta-) analysis. This statistic is an extension of the log additive model, termed “Clinical Genetic Score (18), with weights given to each SNP based on its effect size as reported in the literature (Table 2). Homozygous individuals for the risk allele were consequently assigned twice the risk of heterozygous individuals. Gender was assigned an OR of 1.6 as a lower bound of the sex ratio observed in epidemiological longitudinal studies (13). The MHC component corresponding to HLA-DRB1*15:01 (see rs3135388 Supplementary Table 2) and the gender component were optionally implemented in the models to identify their specific effects on familial aggregation of MS.
SNP genotyping was completed using ABI custom TaqMan assays designed on File Builder 3.0 software and TaqMan predesigned SNP genotyping assays. TaqMan SNP genotyping assays were conducted in 384-well plates using TaqMan Universal PCR Master Mix on an ABI 7900HT Sequence Detection System using SDS 2.3 software. The overall genotype success rate was 99.28%. Sample sizes and statistical methods are specified along the analyses. Non parametric models were used in order to avoid assuming a normal distribution for the MSGB. All quality control and analyses were completed using R version 2.9 and STATA 10 (Stata Corp.). SNP names herein correspond to NCBI dbSNP build 130:human_9606.
RESULTS
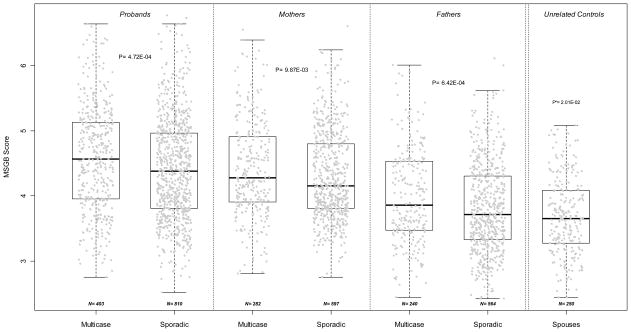
We found that the MSGB of probands is higher in multi-case families than in sporadic MS families (p=4.72 10−4, Figure 1) despite a large variability in both groups. The difference is still significant (p=1.46 10−4) when the gender term is excluded. In addition, the MSGB of both mothers and fathers are higher in multi-case families than in sporadic MS families (Figure 1, p=9.87 10−3 and p=6.42 10−4, respectively). The MSGB values of all patients and parents are significantly greater than genetically unrelated family controls, consisting of spouses of MS patients (taken as “genetically” unrelated controls). The MSGB differentiates multi-case families from sporadic MS families in both probands, mothers and fathers, suggesting a MSGB gradient from multi-case probands to unrelated controls through the parents of MS patients (Cuzick’s non parametric trend test across ordered groups (nptrend) p<10−3).
Figure 1. MSGB differentiates multi-case from sporadic MS families.
The distribution of Multiple Sclerosis Genetic Burden (MSGB) is presented using box-plots. MSGB is computed using components derived from gender, MHC and non MHC-SNPs. Gray dots represent the MSGB of an individual subject. Groups separated by dotted lines (probands, mothers of probands, fathers of probands, and unrelated controls) are divided into Multicase and Sporadic samples. Spouses of patients were considered genetically unrelated controls. Sample sizes are indicated at the bottom of each box-plot. P-values in each of the three left panels indicate the significance of Wilcoxon’s tests of the null hypothesis that MSGB of members of multi-case families are greater than those of members of sporadic MS families. The p-value in the right panel corresponds to the test that the MSGB of fathers of sporadic MS patients is different from unrelated controls (Wilcoxon’s test). 186/250 (74.8%) spouses are male; the comparisons remain significant when the MSGB is calculated without the gender component: p= 1.46 10−4 for multi-case probands vs. sporadic probands; p= 1.17 10−6 for sporadic fathers vs. controls.
When the gender component is removed from the MSGB, score differences between mothers and fathers are no longer significant (p=0.794 in multi-case families, p=0.402 in sporadic MS families). However, the MSGB of multi-case parents are still significantly greater than that of the sporadic MS parents (p= 5.52 10−05, p=1.53 10−3 after excluding the 32 mothers and 20 fathers who are also affected in the multi-case families). This observation leads us to conclude that there is no difference of MS genetic risk factors’ loading between unaffected fathers and unaffected mothers of probands. Without including gender in MSGB, the following hierarchy remains significant: probands of multi-case families > probands of sporadic MS families > parents of multi-case families > parents of sporadic MS families > spouses taken as unrelated controls (Table 3, nptrend p<10−3). When computed from non-MHC SNPs only, the same MSGB hierarchy is retained (Table 3, nptrend <10−3). When computed without the controls group, the same significant MSGB hierarchy is retained (data not shown, nptrend <10−3). Altogether, we demonstrate that in this dataset, members of MS families have a higher MSGB than controls and that individuals belonging to multi-case families, both probands and their parents, have higher genetic loads when compared to their counterparts in sporadic MS families. Gender and HLA are sufficient to explain the observed significant differences creating the MSGB gradient.
Table 3.
MSGB differentiates multicase families from sporadic families
| Components used in computing MSGB median (P25-P75) |
Multicase families probands n=403 |
Sporadic families probands n=810 |
Multicase families parents n=522 |
Sporadic families parents n=1161 |
Spouses as unrelated controls n=250 |
Cuzick’s Test P-value |
|---|---|---|---|---|---|---|
| Gender + HLA + non-MHC SNPs | 4.57 (3.95–5.13) | 4.38 (3.81–4.96) | 4.12 (3.63–4.65) | 3.96 (3.56–4.52) | 3.65 (3.28–4.08) | z=−20.56 p<10−16 |
| HLA + non-MHC SNPs | 4.28 (3.61–4.70) | 4.01 (3.46–4.59) | 3.78 (3.43–4.48) | 3.69 (3.34–4.31) | 3.49 (3.18–3.92) | z=−12.02 p<10−16 |
| Gender + non- MHC SNPs | 3.86 (3.55–4.15) | 3.83 (3.55–4.11) | 3.67 (3.41–3.98) | 3.66 (3.35–3.97) | 3.45 (3.17–3.76) | z=−20.30 p<10−16 |
| Non-MHC SNPs only | 3.48 (3.24–3.70) | 3.46 (3.20–3.73) | 3.47 (3.18–3.67) | 3.41 (3.16–3.66) | 3.35 (3.10–3.60) | z=−5.25 p= 1.5e-07 |
Table 3 presents medians and inter-quartile ranges (P25-P75) of the Multiple Sclerosis Genetic Burden (MSGB) in probands, parents, and controls. For each row, the MSGB is computed using the components in the first column. HLA stands for the HLA-DRB1*15:01 allele. The significance of the observed non parametric gradient from probands to controls through parents differentiating members of multi-case MS sporadic MS is assessed using a two-sided Cuzick’s test whose statistics; test’s statistics and p-values are given in the last column.
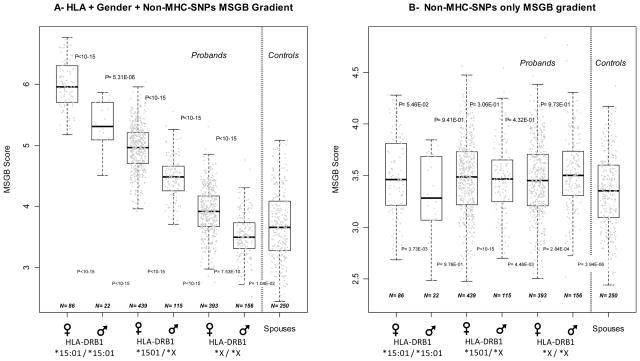
Figure 2 illustrates the separate contributions of the MHC, gender, and non-MHC SNPs to the MSGB. Figure 2A presents the “gender + MHC + non-MHC SNPs” MSGB gradient created in probands by stratification on the HLA-DRB1*15:01 tagging SNP (rs3135388) and further divided by gender stratification. As expected, two-by-two comparisons between probands’ subgroups are significant (all p<10−5). With the notable and interesting exception of HLA-DRB1*15:01 negative male probands (p=0.0104), all subgroups are significantly different from the unrelated controls. The expected HLA-DRB1*15:01 dose effect was present as well. Figure 2B presents the MSGB distribution for non-MHC SNPs only. The significant differences with controls are maintained (all p-value are below 5 10−3 except the one corresponding to the smallest sized patient group (n=22), which consists of male probands homozygous for HLA-DRB1*15:01 ( p=0.976)). The two-by-two comparisons between proband subgroups are no longer significant (all p>0.05) showing that virtually the entire observed signal is coming from gender and 1501. Table 4 shows the reduction of the MSGB gradient when HLA and gender are removed. Unexpectedly, the risk attributed to HLA and gender is not compensated by increased risk from non-MHC SNPs in the low risk HLA and gender groups (nptrend p=0.651). From Figure 2, two independent observations support this conclusion: (1 HLA negative male probands tend to have a lower MSGB than controls; (2) no compensatory gradient is observed when comparing sub groups of probands between Figures 2A and 2B. Similar observations are made in parents of patients (n=1712, data not shown). Altogether, the data presented in Figure 2 and Table 4 indicate that gender, HLA and non-MHC SNPs components independently contribute to the MSGB.
Figure 2. The contributions of HLA and gender (2A) to MSGB are not compensated by the contributions of non MHC SNPs (2B) in MS Patients.
The distribution of the Multiple Sclerosis Genetic Burden (MSGB) is presented using box-plots. The MSGB is computed using the gender, MHC and non-MHC SNPs components in Figure 2A, whereas only the non-MHC SNPs components are used in Figure 2B. Each gray dot corresponds to a given proband of MS families. Dotted lines separate probands from controls (spouses). In both panels, probands are organized according to the dose of HLA-DRB1*15:01 tagging allele (rs3135388*T; X stands for a non HLA-DRB1*15:01 alleles (rs3135388*C) ) and gender. Probands are members of both multi-case and sporadic MS families. Sample sizes are indicated at the bottom of each box-plot. P-values above the box-plots indicate the significance of two-by-two Wilcoxon’s tests of the null hypothesis that MSGB of the left sub-groups of probands are greater than those of the right sub-group of proband. P-values below the box-plots indicate the significance of two-by two Wilcoxon’s tests of the null hypothesis that MSGB of each of the sub-groups of probands are greater than the MSGB of the controls.
Table 4.
Gender and HLA-DRB1 contributes to the MSGB in Probands, absence of reverse aggregation of Non-MHC-SNPs
| MSGB Component Distribution | rs3135388 (T, T) HLA-DRB1*15:01 Double dose | rs3135388 (T, C) HLA-DRB1*15:01 Single dose | rs3135388 (C, C) HLA-DRB1*15:01 Negative | Cuzick’s Test P-value | Unrelated Controls (n=250) | |||
|---|---|---|---|---|---|---|---|---|
| P50 (P25-P75) | Female (n=86) | Male (n=22) | Female (n=439) | Male (n=115) | Female (n=393) | Male (n=156) | ||
|
| ||||||||
| Gender+ HLA+ non-MHC SNPs | 5.96 (5.71–6.29) | 5.31 (5.10–5.71) | 4.97 (4.70–5.21) | 4.48 (4.26–4.66) | 3.92 (3.68–4.18) | 3.50 (3.31–3.73) | z=30.2 p<10−16 |
3.65 (3.28–4.06) |
|
| ||||||||
| HLA + non-MHC SNPs | 5.49 (5.24–5.82) | 5.31 (5.10–5.71) | 4.50 (4.23–4.74) | 4.48 (4.26–4.66) | 3.45 (3.21–3.71) | 3.50 (3.31–3.73) | z=21.0 p<10−16 |
3.50 (3.18–3.89) |
|
| ||||||||
| Gender + non-MHC SNPs | 3.93 (3.68–4.27) | 3.28 (3.07–3.69) | 3.96 (3.69–4.20) | 3.47 (3.25–3.65) | 3.92 (3.68–4.18) | 3.50 (3.31–3.73) | z=6.0 p=2.4e-09 |
3.45 (3.18–3.76) |
|
| ||||||||
| Non-MHC SNPs only | 3.46 (3.21–3.80) | 3.28 (3.07–3.69) | 3.49 (3.22–3.73) | 3.47 (3.25–3.65) | 3.45 (3.21–3.71) | 3.50 (3.31–3.73) | z=0.45 p=0.65 |
3.36 (3.12–3.62) |
Table 4 presents medians and inter-quartile ranges (P25-P75) of the Multiple Sclerosis Genetic Burden (MSGB) in probands of multi-case and sporadic MS families grouped according to HLA-DRB1*15:01 dose (rs3135388*T) and gender, and controls. P-values correspond to two-sided Cuzick’s test of the MSGB gradient. MSGB is computed using the components indicated at the beginning of each row: gender, HLA (the HLA-DRB1*15:01 component in the MHC region), and non MHC-SNPs components.
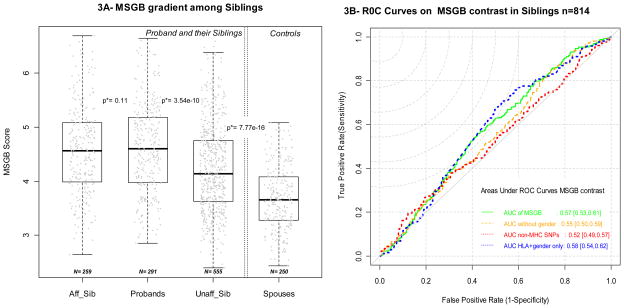
Figure 3A presents the distribution of the MSGB in multi-case families for the affected and non-affected siblings (brothers and sisters of the proband, when available). No difference in MSGB can be detected between the probands and the affected siblings of the same pedigree (matched Wilcoxon’s test p=0.11, n=259 pairs). Unaffected siblings, on the other hand, have lower MSGBs than the probands (matched Wilcoxon’s test p=3.54*10−10, n=555 pairs), but still carry greater MSGBs than controls (Wilcoxon’s test p=7.77*10-16). Consistent with the previous observations, these findings remain constant when removing the HLA and/or gender component of the MSGB (Table 5). Assuming that the genetic burden would be a good predictor of MS when individuals share similar environmental determinants, we asked whether the MSGB differences between siblings and probands can predict MS within families. In siblings of the same sibship, having a greater or equal MSGB than the proband is significantly associated with MS with an OR=2.1 [1.36, 3.2], (conditional logistic regression p=0.001, 164 informative pedigrees, 642 individuals). Figure 3B displays the Receiver Operating Characteristic (ROC) curves corresponding to the MSGB sib-proband contrast and demonstrates that contrasting MSGB in MS sibships is statistically significant but not fully informative. The AUROC (Area Under ROC) areas are close to 0.5: AUROC 0.57 [0.53; 0.61], AUROC for MSGB without gender 0.55 [0.51; 0.59], and AUROC for non-MHC SNPs only MSGB 0.53[0.49; 0.57]. The MSGB provides us with statistically significant differences between affected siblings and unaffected siblings but such differences are not predictive. With an AUROC of 0.58 [0.54; 0.62] HLA and gender provide more accurate information, suggesting that the non-MHC SNP component of the MSGB adds insufficient information while it contributes greatly to the variance of the MSGB, reducing then its informative content for predictive purposes.
Figure 3. Distribution of MSGB in siblings of MS multi-case families (3A) and informative potential of MSGB contrast (3B).
Figure 3A presents the distribution of Multiple Sclerosis Genetic Burden (MSGB) in siblings using box-plots. MSGB is computed with gender, MHC and non-MHC SNPs components. Gray dots correspond to the MSGB of an individual subject. The three left box-plots of panel A correspond to subject’s status in sibship (“Aff_Sib” = affected sibs, Probands in the middle, “Unaff_Sib” = unaffected siblings). The p-values correspond to Wilcoxon’s tests of the null hypothesis that the MSGB of side by side categories are the same. The box-plot of spouses of probands (controls) is given on the right. When several sibs are available within a group, one is randomly chosen. The p-value overlaying the dotted lines indicates the significance of (unmatched) Wilcoxon’s tests of the null hypothesis that MSGB of unaffected siblings of the probands are greater than the MSGB of the controls.
Figure 3B shows the Receiver Operating Curves (ROC) corresponding to the prediction of MS status of the sibs of the probands based on the contrast between the sib’s MSGB and proband MSGB. In red, MSGB contrasts are computed using the gender and the MHC components. In green, MSGB contrasts are computed using the gender, MHC and non MHC-SNPs components; in orange, MSGB contrasts are computed using the MHC and non MHC-SNPs components. In red, MSGB contrasts are computed using only the non MHC-SNPs components. In blue, MSGB contrasts are computed using only using the gender and MHC.
Table 5.
Contribution of the various MSGB components to the MSGB in siblings of multi-case MS families
| MSGB Distribution in Probands and their siblings | Patients | Unaffected | ||
|---|---|---|---|---|
| P50 (P25-P75) | Siblings (N=259 | Probands (N=291) | Siblings (N=555) | Unrelateds (N=250) |
|
| ||||
| Gender+ HLA + non-MHC SNPs | 4.57 (3.99–5.08) | 4.60 (3.98–5.18) | 4.14 (3.62–4.75) | 3.65 (3.28–4.06) |
|
| ||||
| HLA + non-MHC SNPs | 4.18 (3.65–4.72) | 4.35 (3.63–4.76) | 3.92 (3.40–4.48) | 3.50 (3.18–3.89) |
|
| ||||
| Gender + non-MHC SNPs | 3.80 (3.50–4.10) | 3.86 (3.50–4.15) | 3.67 (3.39–3.94) | 3.45 (3.18–3.76) |
|
| ||||
| Non-MHC SNPs only | 3.44 (3.17–3.71) | 3.48 (3.22–3.69) | 3.41 (3.19–3.66) | 3.36 (3.12–3.62) |
Table 5 presents medians and inter-quartile ranges (P25-P75) of Multiple Sclerosis Genetic Burden (MSGB) in sibship of multi-case families and controls. Patients are divided into two groups: probands and affected sibs of probands. Unaffected individuals are also presented into two groups: unaffected siblings of the probands, and unrelated unaffected individuals. MSGB is computed using the components indicated at the beginning of each row as in Table 4: gender, HLA (the HLA-DRB1*15:01 component of MS susceptibility in the MHC region), and non MHC-SNPs components.
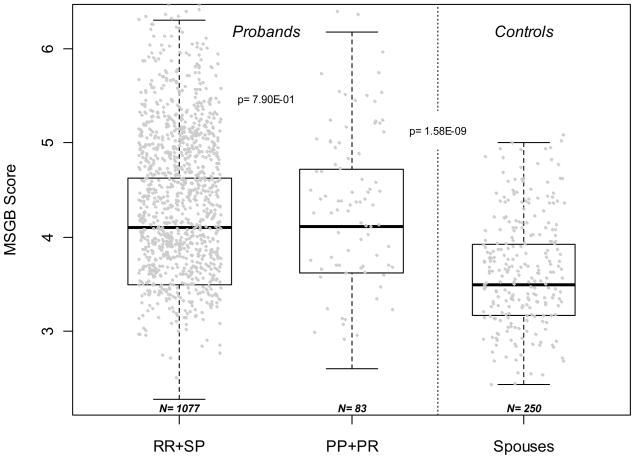
A limited set of gross clinical parameters (age of onset and severity as measured by EDSS and MSSS) failed to show significant associations with MSGB (data not shown). Interestingly, no differences in MSGB without gender component between primary progressive (PP) and relapsing-remitting (RR) MS was detected (Figure 4, p=0.79). MSGB is higher in PP MS (n=84) when compared to controls (n=254) (Figure 4, Wilcoxon’s test p=1.58 10−9). Taken together, it suggests that MS with Primary Progressive disease course shares the same common genetic variants associated with susceptibility as RR MS.
Figure 4. Absence of different MS Genetic Burden between patients with a Relapsing Remitting or Secondary Progressive form of MS vs. patients with Primary Progressive MS.
The distribution of Multiple Sclerosis Genetic Burden (MSGB) is presented using box-plots. MSGB is computed using components derived from MHC and non MHC-SNPs. Spouses of patients were considered genetically unrelated controls. Sample sizes are indicated at the bottom of each box-plot. The p-value in probands indicates the significance of Wilcoxon’s tests of the null hypothesis that MSGB of RR (Relapsing Remitting) + SP (Secondary Progressive) MS patients are different from those of PP (Primary Progressive) + PR (Progressive–Relapsing). The p-value in the right part of the figure corresponds to the test that the MSGB of PP (Primary Progressive) + PR (Progressive–Relapsing) MS patients are greater than those of from unrelated controls (Wilcoxon’s test).
DISCUSSION
In summary, using the most updated genetic information available for MS and a large and well-characterized familial dataset, we show the higher aggregation of susceptibility variants in multi-case families compared to sporadic MS. Using only external peer-reviewed publications to set the model, we avoided most of the over-fitting issue that can occur in the discovery-replication cohort estimates. In MS families, the aggregation of non-MHC SNPs depends neither on gender nor on presence or absence of HLA-DRB1*15:01 alleles. As suggested by the EVI5 association data (3, 15), other interactions may exist, but their computation into the MSGB is prevented by limited statistical power of the present study. Our results confirm and extend previous investigations of 5 SNPs aggregates in 43 multicase families (19). We also confirm the assumed (20), but never previously measured, higher genetic burden in parents of MS patients compared to controls. As a caveat, the distinction between multi-case and sporadic MS families should not be considered as definitive categories. For example, the sibship size is significantly lower in the so-called “sporadic” MS families compared to multi-case families (p=8.10−4). Thus, because of incomplete penetrance, bias may results in an underestimation of the difference between the sporadic and multi-case families. Although non-MHC SNPs add little additional information to HLA and gender for developing predictive tools for families affected by MS, the resulting AUROCs are lower or equal to those computed in unrelated individuals (18). Moreover, this study suggests that attempts to extend the paradigm of monogenic genetic counseling with models integrating GWAS identified SNPs is at least immature and possibly useless for MS families. It reinforces the need for caution about using genetics score to predict MS in general population and it suggests that family members and other “at risk” groups may be more appropriate targets for early implementations of genetic tests (18, 21). This paradigm may radically change if highly penetrant (rare) variants are identified (22) and/or high-impact environmental triggers are discovered.
The proposed MSGB model was parameterized following peer-reviewed publications with a large sample size. With the exception of HLA-centered publications, very few studies compared association models which could be implemented in the MSGB. In the same line, very few publications investigated genetic interactions. The next generation of MSGB models should account for various genetic models and genetic interactions. The absence of compensation by non-MHC SNPs for HLA alleles and gender may be due to insufficient modeling of the reported association by using only the trend model, as opposed to other genetic models to identify statistically associated SNPs. Hence, at a minimum six limitations of the study should be noted: 1) The MSGB would benefit from a better assessment of the genetic association of each SNP using recessive/dominant/dose-dependent models as commonly utilized in monogenic disease studies, 2) OR values taken from peer-review studies may inadequately estimate the association. 3) The MHC region needs more detailed modeling than a single SNP tagging HLA-DRB1*15:01. The contribution(s) of the MHC region to the MS genetic risk component cannot be efficiently summarized with a single SNP. Although HLA-DRB1*15:01 is the major HLA determinant of MS risk, there are additional, sometimes opposite, genetic contributions of class I and II regions (HLA-A, HLA-C, HLA-B, HLA-DRB5 (23–26)) to MS susceptibility that need to be considered. 4) Alternative SNP(s), or haplotype of SNPs, at a given locus might be more effective in capturing the contribution of a given gene/region to MS (5). Our model did not include any interactions between gender and SNPs, or SNPs and SNPs. Such interactions, between EVI5 and HLA-DRB1 (3, 15, 26), for example, are certainly relevant to the model estimations. 6) The effect sized of each SNP accounted for in the MSGB computation is not weighted for accuracy of the effect size estimations; effect sizes may vary also in different familial environmental background.
The primary interest in the MSGB concept resides in its capacity to integrate cumulative genetic contributions to MS risk and assessment of genetic heterogeneity across diverse phenotypes. This is reflected in the absence of compensatory aggregates of non-MHC SNPs in males and/or in the absence of HLA-DRB1*15:01 alleles. As a corollary, stratification or adjustment of GWAS data by gender and/or HLA-DRB1*15:01 may be minimally productive. MSGB as a cumulative score of common genetic variants captures part of the variance in case control design and may become a standard covariate in logistic regression. It would account for previous genetic evidence in a similar way that principal components account for population structure. The analyses also suggest that, in low MSGB families, the known genetic risks cannot explain the MS segregation patterns. The absence of difference in MSGB between MS with primary progression compared to classical RR and SP-MS further confirm that both course trajectories represent variants of the same disease rather than fundamentally different entities (23, 27, 28). Although determinants of disease course activity may not necessarily entail the same genetic factors as those involved in susceptibility, additional investigations of the genetic burden using more detailed phenotypic measures, including high-resolution MRI, are warranted. Together with the study of D’Netto et al.(19), the results suggest that recruiting patients from multi-case families may increase the power of case control association studies. MSGB metrics may also prove to be valuable in guiding re-sequencing or new linkage-based efforts in individuals or groups in the lower echelon of burden such as HLA-DRB1*15:01 males and their families. Finally, the analysis also suggests that MS patients carrying a low MSGB may have a larger environmental component in the cascade of events leading to noticeable clinical symptoms. In the last few years, credible environmental influences in MS were identified through epidemiological and laboratory approaches (vitamin D, Epstein-Barr virus infection, latitude, and smoking) (29, 30). The stringent prospective collection of this type of data in families may help, when combined with MSGB profiling, in developing strategies to reduce disease incidence. This conclusion can be easily extended to other etiologically complex autoimmune diseases. Fueled by the GWAS case-control design, identification, and ascertainment of multigenerational, multi-affected families has been neglected in recent years. We submit, however, that they constitute an invaluable resource for advancing the understanding of the architecture of complex disease genetics.
Acknowledgments
This study was supported by NIH grant RO1NS26799 and U19 AI067152, and National Multiple Sclerosis Society grant RG2901. We thank the MS patients, their relatives and healthy controls that participated in the original genetic studies. We thank Hourieh Mousavi, Rosa Guerrero and Robin Lincoln for sample management, as well as Refujia Gomez and Elise Taylor for management of the clinical data. We also thank Glenys Thomson for insightful comments of the manuscript.
ABBREVIATIONS
- AUROC
Area under Receiver Operating characteristic Curve
- GWAS
Genome Wide Association Studies
- HLA
Human Leucocyte Antigen
- MHC
Major Histocomaptibilty Complex
- MS
Multiple sclerosis
- MSGB
MS Genetic Burden
- nptrend
Cuzick’s non parametric trend test across ordered groups
- OR
Odds Ratio
- ROC
Receiver Operating characteristic Curve
- SNP
Single Nucleaotide Polymorphism
- PP
Primary Progressive MS
- RR
Relapsing-Remitting MS
Footnotes
CONFLICT OF INTEREST
The authors declare that none of the authors have a financial interest related to this work.
JO and PAG conceived the research and wrote the paper. PAG set up the analytical strategy. SC did the genotyping. AS was in charge of the database and formatted the datasets. JM, BJ, and SH contributed to the discussion and the manuscript.
References
- 1.De Jager PL, Jia X, Wang J, de Bakker PI, Ottoboni L, Aggarwal NT, Piccio L, Raychaudhuri S, Tran D, Aubin C, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oksenberg JR, Baranzini SE, Sawcer S, Hauser SL. The genetics of multiple sclerosis: SNPs to pathways to pathogenesis. Nat Rev Genet. 2008;9:516–526. doi: 10.1038/nrg2395. [DOI] [PubMed] [Google Scholar]
- 3.ANZGENE-consortium. Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009;41:824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 4.IMSGC-consortium. Comprehensive follow-up of the first genome-wide association study of multiple sclerosis identifies KIF21B and TMEM39A as susceptibility loci. Hum Mol Genet. 2010;19:953–962. doi: 10.1093/hmg/ddp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baranzini SE, Wang J, Gibson RA, Galwey N, Naegelin Y, Barkhof F, Radue EW, Lindberg RL, Uitdehaag BM, Johnson MR, et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet. 2009;18:767–778. doi: 10.1093/hmg/ddn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, Kwiatkowski DP, McCarthy MI, Ouwehand WH, Samani NJ, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comabella M, Craig DW, Camina-Tato M, Morcillo C, Lopez C, Navarro A, Rio J, Montalban X, Martin R. Identification of a novel risk locus for multiple sclerosis at 13q31.3 by a pooled genome-wide scan of 500,000 single nucleotide polymorphisms. PLoS One. 2008;3:e3490. doi: 10.1371/journal.pone.0003490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jakkula E, Leppa V, Sulonen AM, Varilo T, Kallio S, Kemppinen A, Purcell S, Koivisto K, Tienari P, Sumelahti ML, et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. Am J Hum Genet. 2010;86:285–291. doi: 10.1016/j.ajhg.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sawcer S, Ban M, Wason J, Dudbridge F. What role for genetics in the prediction of multiple sclerosis? Ann Neurol. 2010;67:3–10. doi: 10.1002/ana.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadovnick AD, Baird PA, Ward RH. Multiple sclerosis: updated risks for relatives. Am J Med Genet. 1988;29:533–541. doi: 10.1002/ajmg.1320290310. [DOI] [PubMed] [Google Scholar]
- 11.Smith-Bouvier DL, Divekar AA, Sasidhar M, Du S, Tiwari-Woodruff SK, King JK, Arnold AP, Singh RR, Voskuhl RR. A role for sex chromosome complement in the female bias in autoimmune disease. J Exp Med. 2008;205:1099–1108. doi: 10.1084/jem.20070850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentzen J, Flachs EM, Stenager E, Bronnum-Hansen H, Koch-Henriksen N. Prevalence of multiple sclerosis in Denmark 1950–2005. Mult Scler. 2010 doi: 10.1177/1352458510364197. [DOI] [PubMed] [Google Scholar]
- 13.Orton SM, Herrera BM, Yee IM, Valdar W, Ramagopalan SV, Sadovnick AD, Ebers GC. Sex ratio of multiple sclerosis in Canada: a longitudinal study. Lancet Neurol. 2006;5:932–936. doi: 10.1016/S1474-4422(06)70581-6. [DOI] [PubMed] [Google Scholar]
- 14.Hafler JP, Maier LM, Cooper JD, Plagnol V, Hinks A, Simmonds MJ, Stevens HE, Walker NM, Healy B, Howson JM, et al. CD226 Gly307Ser association with multiple autoimmune diseases. Genes Immun. 2009;10:5–10. doi: 10.1038/gene.2008.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson BA, Wang J, Taylor EM, Caillier SJ, Herbert J, Khan OA, Cross AH, De Jager PL, Gourraud PA, Cree BC, et al. Multiple sclerosis susceptibility alleles in African Americans. Genes Immun. 2009 doi: 10.1038/gene.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barcellos LF, Oksenberg JR, Green AJ, Bucher P, Rimmler JB, Schmidt S, Garcia ME, Lincoln RR, Pericak-Vance MA, Haines JL, et al. Genetic basis for clinical expression in multiple sclerosis. Brain. 2002;125:150–158. doi: 10.1093/brain/awf009. [DOI] [PubMed] [Google Scholar]
- 17.Goodkin DE, Doolittle TH, Hauser SS, Ransohoff RM, Roses AD, Rudick RA. Diagnostic criteria for multiple sclerosis research involving multiply affected families. Arch Neurol. 1991;48:805–807. doi: 10.1001/archneur.1991.00530200041016. [DOI] [PubMed] [Google Scholar]
- 18.De Jager PL, Chibnik LB, Cui J, Reischl J, Lehr S, Simon KC, Aubin C, Bauer D, Heubach JF, Sandbrink R, et al. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol. 2009;8:1111–1119. doi: 10.1016/S1474-4422(09)70275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Netto MJ, Ward H, Morrison KM, Ramagopalan SV, Dyment DA, DeLuca GC, Handunnetthi L, Sadovnick AD, Ebers GC. Risk alleles for multiple sclerosis in multiplex families. Neurology. 2009;72:1984–1988. doi: 10.1212/WNL.0b013e3181a92c25. [DOI] [PubMed] [Google Scholar]
- 20.Kantarci OH, Barcellos LF, Atkinson EJ, Ramsay PP, Lincoln R, Achenbach SJ, De Andrade M, Hauser SL, Weinshenker BG. Men transmit MS more often to their children vs women: the Carter effect. Neurology. 2006;67:305–310. doi: 10.1212/01.wnl.0000225070.13682.11. [DOI] [PubMed] [Google Scholar]
- 21.Ramagopalan SV, Giovannoni G. Can we predict multiple sclerosis? Lancet Neurol. 2009;8:1077–1079. doi: 10.1016/S1474-4422(09)70273-X. [DOI] [PubMed] [Google Scholar]
- 22.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barcellos LF, Sawcer S, Ramsay PP, Baranzini SE, Thomson G, Briggs F, Cree BC, Begovich AB, Villoslada P, Montalban X, et al. Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Hum Mol Genet. 2006;15:2813–2824. doi: 10.1093/hmg/ddl223. [DOI] [PubMed] [Google Scholar]
- 24.Bergamaschi L, Leone MA, Fasano ME, Guerini FR, Ferrante D, Bolognesi E, Barizzone N, Corrado L, Naldi P, Agliardi C, et al. HLA-class I markers and multiple sclerosis susceptibility in the Italian population. Genes Immun. 2010;11:173–180. doi: 10.1038/gene.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramagopalan SV, Knight JC, Ebers GC. Multiple sclerosis and the major histocompatibility complex. Curr Opin Neurol. 2009;22:219–225. doi: 10.1097/WCO.0b013e32832b5417. [DOI] [PubMed] [Google Scholar]
- 26.Chao MJ, Ramagopalan SV, Herrera BM, Lincoln MR, Dyment DA, Sadovnick AD, Ebers GC. Epigenetics in multiple sclerosis susceptibility: difference in transgenerational risk localizes to the major histocompatibility complex. Hum Mol Genet. 2009;18:261–266. doi: 10.1093/hmg/ddn353. [DOI] [PubMed] [Google Scholar]
- 27.Kremenchutzky M. Primary progressive MS. Int MS J. 2003;10:89–95. [PubMed] [Google Scholar]
- 28.Confavreux C, Vukusic S. Natural history of multiple sclerosis: a unifying concept. Brain. 2006;129:606–616. doi: 10.1093/brain/awl007. [DOI] [PubMed] [Google Scholar]
- 29.Ascherio A, Munger KL. Environmental risk factors for multiple sclerosis. Part II: Noninfectious factors. Ann Neurol. 2007;61:504–513. doi: 10.1002/ana.21141. [DOI] [PubMed] [Google Scholar]
- 30.Ascherio A, Munger KL. Environmental risk factors for multiple sclerosis. Part I: the role of infection. Ann Neurol. 2007;61:288–299. doi: 10.1002/ana.21117. [DOI] [PubMed] [Google Scholar]