Abstract
The gain, loss, and modification of gene regulatory elements may underlie a significant proportion of phenotypic changes on animal lineages. To investigate the gain of regulatory elements throughout vertebrate evolution we identified genome-wide sets of putative regulatory regions for five vertebrates, including human. These putative regulatory regions are conserved non-exonic elements (CNEEs), which are evolutionarily conserved yet do not overlap any, coding or noncoding, mature transcript. We then inferred the branch on which each CNEE came under selective constraint. This analysis identified three extended periods in the evolution of gene regulatory elements. Early vertebrate evolution was characterized by regulatory gains near transcription factors and developmental genes, but this trend was replaced by innovations near extra-cellular signaling genes, and then innovations near post-translational protein modifiers.
The gain, loss, and modification of gene regulatory elements has led to many phenotypic changes during animal evolution, including pigmentation changes in dogs, fish, and flies (1–3), bristle patterns on flies (4), and skeletal differences in fish (5, 6). A recent analysis of published genome-wide association studies also noted a strong enrichment for regulatory regions to be in linkage with trait/disease associated single nucleotide polymorphisms (7). Mutations in regulatory modules can avoid the pleiotropic effects that often result from protein-coding mutations, and hence provide an exceptionally flexible source of evolutionary change (8).
Computational methods can identify strong candidates for gene regulatory elements by detecting regions of the genome that show evolutionary conservation, yet do not appear in any coding or noncoding mature transcript. While many noncoding RNAs, or noncoding portions of protein-coding transcripts, may serve a regulatory purpose, we exclude these regions to focus on cis-regulatory elements that are functional at the DNA level. For this reason we use conserved non-exonic elements (CNEEs) to describe the set of putative regulatory regions. Strong conservation indicates a sustained purifying selection against mutations, suggesting the genomic region confers selective advantage and hence is functional. This procedure is independent of the tissue-type, developmental stage or environmental circumstance in which these elements become relevant. An experimental survey of CNEEs found that 50% of the 437 CNEEs tested in an in vivo mouse enhancer assay drove reproducible expression patterns during mouse development (9). Since only a single time-point during development was tested, many of the CNEEs that did not drive expression in the assay may act as transcriptional enhancers at other developmental stages. CNEEs have also been shown to act as repressors and insulators (10–12).
Early genome-wide studies of CNEEs not only noted their role in regulating the transcription of nearby genes, but also observed that CNEEs tend to be located near genes acting as developmental regulators, including many transcription factors, to finely control vertebrate development (13–16). Only a handful of vertebrate CNEEs date back to our chordate ancestor (17), suggesting that the vast majority of regulatory elements in the genomes of living vertebrates have arisen since their common ancestor 650 million years ago.
To better understand this gain of regulatory sequence on vertebrate lineages we identified genome-wide sets of CNEEs for the human genome as well as mouse, cow, medaka, and stickleback. Mouse and cow have well-assembled and annotated genomes that can leverage the densely sampled clade of placental mammals to identify and date CNEEs in their genomes. Ray-finned fish are the other clade of vertebrates with enough well-assembled genomes to identify and date CNEEs. Because the ray-finned fish diverged from placental mammals ~425 Mya their lineage offers a largely independent analysis of regulatory trends in vertebrate evolution.
Using multiple alignments of vertebrate genomes, a phylogenetic hidden Markov model was employed to determine CNEEs (18). For each CNEE we used genomic proximity to predict the gene being regulated. We assigned each CNEE to the gene with the closest transcription start site (18). We then inferred the evolutionary branch on which each CNEE came under selective constraint. To do this, for each CNEE we determined the most recent common ancestor of all species in the vertebrate-wide multiple-alignment that have an orthologous piece of DNA aligning to at least one-third of the CNEE (Fig. S1). For more than 99.97% of CNEEs we use this most recent common ancestor as the time of origin. In less than 1000 rare cases there is evidence that a significant onset of constraint occurred following the most recent common ancestor of all species in the alignment (19). In these rare cases we use the branch showing the onset of constraint as the origin of the CNEE (Fig. S1). The onset of constraint must be more significant than the rejection of the neutral model by the alignment as a whole (18). Finally, we examined the functional classes of genes that acquired the most regulatory innovations during each evolutionary epoch since the common ancestor of living vertebrates.
For our study of epoch-specific selection in the human lineage, we created alignments of 40 vertebrate genomes (including 31 mammals, 2 birds, 1 lizard, 1 amphibian, and 5 fish) projected onto the human genome (18). From this alignment we found 2,964,909 human CNEEs, averaging 28 bases in length and covering 2.9% of the genome.
To ensure that the CNEEs are enriched for regions that have been under selection, we examined the derived allele frequency spectrum of segregating single nucleotide polymorphisms found by the HapMap Consortium (20) in the Yoruban population (18). The frequency of derived alleles in the Yoruban population is shifted toward lower frequencies in the set of CNEEs compared to intronic regions (p<10−90) as is characteristic of sites under purifying selection, where the majority of mutations are deleterious and rarely progress to higher frequencies. Although the shift in derived allele frequencies for the set of CNEEs is not as strong as that for non-synonymous changes in coding regions, these results demonstrate that this set is strongly enriched for regions evolving under purifying selection in humans (Fig. S2).
Experimental methods such as ChIP-seq (21) and DNAse hypersensitivity (22), while still limited by the tissue-type and time point of the experiment, offer an alternative to in vivo mouse models, that allows the regulatory potential of CNEEs to be assessed on a much larger scale. We examined the overlap of the CNEEs with over 100,000 DNase hypersensitivity sites identified in human CD4+ cells, indicating regions of open chromatin (23). There is a 1.7-fold enrichment in these DNase hypersensitivity sites compared to a random control for overlap with the set of CNEEs (p<10−800) (18). More specific to being a regulatory region, although restricted to a single protein of interest, are locations of protein-DNA interactions discovered by ChIP-seq. We examined the intersection of CNEEs with ChIP-seq datasets for the factors SRF, GABP, and NRSF in human Jurkat cells (24). There is a 2.5-fold enrichment for SRF binding sites (p<10−142), a 3.5-fold enrichment for GABP binding sites (p<10−210), and a 4.7-fold enrichment for NRSF binding sites (p<10−220). This indicates that our set of CNEEs is strongly enriched for functional elements. Our set of CNEEs typically covers only 1 out of every 10 putative regulatory elements identified by these experimental approaches. However, evolutionary conservation will only detect those binding sites under considerable purifying selection in multiple species and not lineage-specific or recently created regulatory elements.
To assess the functions of genes putatively regulated by the CNEEs, we assigned each CNEE to the gene with the closest transcription start site and determined the Gene Ontology (GO) (25) terms associated with that gene. We tested for enrichment against the assumption that CNEEs are uniformly distributed throughout the genome, allowing for differences in genic and intergenic sizes among classes of genes (18).
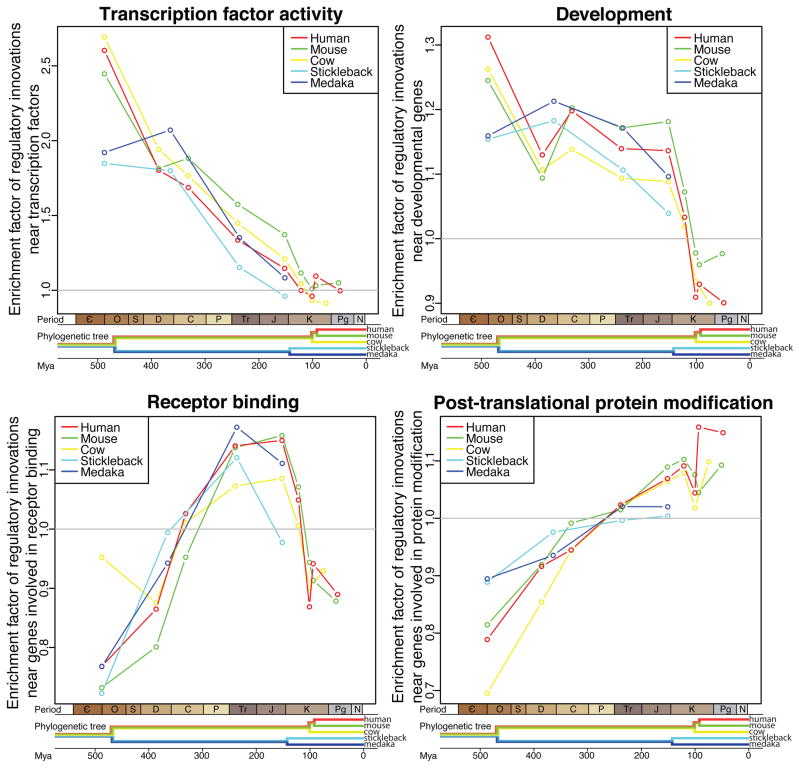
While the set of human CNEEs, when treated as a whole, is enriched in locations where the closest transcription start site is a “trans-dev” gene (“transcription factor activity” p<10−2000, “development” p<10−3000), these results showed that this enrichment is due to the subset of human CNEEs that came under selection prior to the boreoeutherian (human-cow) ancestor (Figs. 1 and S3 and Tables S1 and S2). Despite a dramatic enrichment for regulatory innovations near trans-dev genes in the earliest period of vertebrate evolution through the radiation of tetrapods onto land and to a lesser extent in our early mammalian ancestors, we found a sharp decrease from that enrichment to a rate expected by chance, or less, since the ancestor of placental mammals, approximately 100 Mya.
Fig. 1.
Regulatory innovation. Each panel shows data for the frequency of regulatory innovations near genes in a different GO category (panel title). Colors indicate the five lineages studied. Each data point (colored circle) represents the relative frequency of regulatory innovations on a specific lineage (color) as determined by analysis using the reference genome for that lineage. The relative frequency (enrichment factor) for a specific GO category is defined as the frequency of innovations in genes of this GO category as compared to what would be expected by selecting genomic regions at random (denoted by the horizontal line at the relative frequency of 1.0). Each data point is an estimate from at least 2800 putative regulatory innovations. The time associated with each data point, indicated on the horizontal axis, is the midpoint of the branch of the phylogenetic tree on which these innovations are inferred to have occurred by comparative genome analysis (Fig. S3 and Tables S1 and S2). The horizontal axis is annotated with both geologic time periods as well as speciation events for the lineages analyzed.
Separating developmental genes and transcription factors shows that developmental genes maintained a moderate enrichment for new regulatory elements until a sharp decline in our early placental ancestor. Transcription factors were dramatically enriched for regulatory elements in our early vertebrate ancestor, but this enrichment has consistently declined until reaching random expectation in our placental ancestor (Figs. 1 and S4).
We repeated the above study using the mouse, cow, medaka and stickleback species as reference species, each time starting with a new multiple alignment of other vertebrates to the chosen reference species. The analysis of each of these additional lineages gave similar results (Fig. 1).
Creating genome-wide alignments for vertebrate species is still an active area of research. We used alignments to infer the branch on which CNEEs came under selection, so inaccuracies in the alignment may result in inaccurate inferences as to when some CNEEs came under selection. False positive alignments to distantly related species may cause CNEEs to appear more ancient. False negatives in the alignment process, combined with CNEEs missing from assemblies due to low-coverage sequencing or deletions, may cause CNEEs to appear more recent. However, to explain the trends described there would need to be a systematic bias that treated the CNEEs associated with genes of one function differently than those associated with genes of other functions.
These findings are not due to biases in length, rate of evolution, or rate of turn-over between CNEEs near various functional classes of genes. We have greater statistical power to align both longer and slower evolving CNEEs over large evolutionary distances. We also have greater power to detect evolutionary conservation in longer and slower evolving CNEEs. Both of these factors contribute to our sets of very ancient and very recent CNEEs being enriched for longer and more slowly evolving elements. However, there is not a consistent trend for CNEEs associated with trans-dev genes or genes with any particular GO term to have different lengths or rates of evolution (Figs. S5 and S6). For this reason it is unlikely that our results are caused by either of these biases in creating alignments and detecting conservation. To show the results are not due to different rates of turn-over for CNEEs near trans-dev genes, we identified CNEEs in the human, mouse, and cow referenced alignments that have clear orthologs in other well-assembled mammalian genomes, but are not present in either the human and rhesus, mouse and rat, or dog and horse genome assemblies (18). These CNEEs are likely to have been lost in one of the mammalian lineages after having been present in the ancestor. We counted the number of these lost CNEEs near trans-dev genes versus other types of genes and found no consistent difference (Fig. S7). This indicates the results are unlikely to be the result of different rates of turn-over.
Neither are the results due to a bias in dating CNEEs whose time of origin is uncertain. The same trends seen in the entire set of CNEEs are present in the subset that have a clear point of origin, i.e. that exist precisely in all species descendant from a common ancestor, and in no additional species (Fig. S8).
To ensure that the changes we see in enrichments over time are robust against the alignment methods and against choices in what species are included in the analysis we have performed our analysis on a separate human-referenced alignment using only deeply-sequenced and well-assembled genomes along with stringent alignment parameters (Fig. S9). The results were similar. To ensure that our results are robust against the choice of gene set, CNEE to gene assignment algorithm, and GO term to gene mapping, we performed our enrichment analysis for the human lineage using a completely independent method (26) (Fig. S9). Again the results were similar. Hence, our conclusions are robust to a large number of variations in methodological approach.
Given the changes observed for trans-dev genes, we extended our analysis to each of the ~13,000 GO terms found in vertebrate genomes. We determined the approximate times of all regulatory innovations associated with each GO term and ranked all terms by their increase or decline over time, based on the slope of a linear model fit to time-vs-percent of CNEEs associated with the GO term. At one end of the spectrum, the top forty fastest-declining gene categories were “development,” “transcription factor activity,” and related GO terms, irrespective of the choice of human, mouse, cow, medaka, or stickleback as the reference genome (Tables S3 – S7).
At the other end of the spectrum, we found increases in the accumulation of regulatory innovations for several GO terms, suggesting that the decrease in regulatory innovations near trans-dev genes has been accompanied by an increase in innovations near genes of other functions. These include genes annotated with “post-translational protein modification,” “organelle membrane,” and other GO categories related to intra-cellular signaling (p<10−110, given the dating of CNEEs and the annotation of genes) (Tables S8 – S10).
This set of GO terms does not show a significant enrichment in the medaka or stickleback lineages; however, the fish lineages do not have the dense tree of closely related species that is required to identify recent regulatory innovations. Instead, the terms showing the sharpest increases on both fish lineages are “receptor binding,” “plasma membrane,” “signaling,” and other related terms (Tables S11 – S12). Upon analyzing inter-cellular membrane signaling genes in the mammalian lineages we found that mammalian ancestors too once had a high rate of regulatory innovations near this same set of genes, but such innovations have now returned to random expectation (Fig. 1). In fact, “receptor binding” genes show a peak of regulatory innovation between the time of our amniote ancestor and our placental mammalian ancestor, relative to the rate of innovation before and after this time period (p<10−250, given the dating of CNEEs and the annotation of genes).
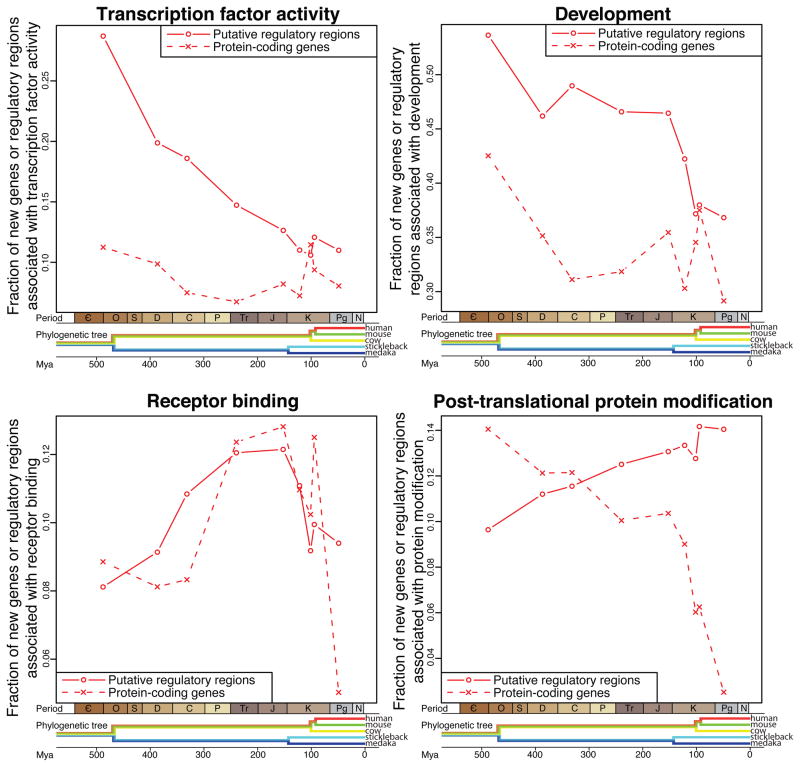
For some functional categories the trend in regulatory innovations appears to be correlated with an increased or decreased appearance of the genes themselves. The proportion of new genes that are associated with inter-cellular signaling closely mirrors the proportion of regulatory innovations near genes with this function (Fig. 2). During the time from the amniote ancestor to the placental mammalian ancestor there was an enrichment for both new genes and new regulatory elements associated with inter-cellular signaling. However, for transcription factors, which show the most dramatic change in their proportion of regulatory innovations, the proportion of gene births has stayed relatively constant. This hints that the selective pressures on novel regulatory elements and novel genes may be similar for some classes of genes and different for others. It could be that evolutionary advances in some cell functions may be realized both through novel genes and novel regulatory elements, while advances in other cell functions may preferentially happen through one of the two methods.
Fig. 2.
A comparative history of innovations in regulatory regions and genes. Each panel shows data for the frequency of regulatory innovations near genes in a different GO category (panel title) as well as the frequency of genic innovations for that GO category. Each data point for regulatory innovations (red circle) represents the percent of regulatory innovations appearing on a branch in the human lineage that are associated with a gene annotated with the given GO category. Each data point for protein-coding genes (red x) represents the percent of protein-coding genes appearing on a branch that are associated with the given GO category. The time associated with each data point is as described in Fig. 1.
As regulatory innovations near genes involved in post-translational protein modification have become increasingly common in placental mammals, the new appearance of these genes themselves has become increasingly rare. This emphasizes the fact that many new regulatory innovations are associated with ancient genes. For example, we have identified 10 regulatory innovations near protein kinase D1 (PRKD1, PKD1) since the human lineage split with cow. This is the most regulatory innovations of any post-translational protein modification gene during this recent time period, yet PRKD1 dates to at least our tetrapod ancestor. This enrichment for recent CNEEs near genes involved in post-translational protein modification is consistent across humans, cows, and to a lesser degree mouse, even when looking only at independent events after these species diverged (Tables S8 – S10).
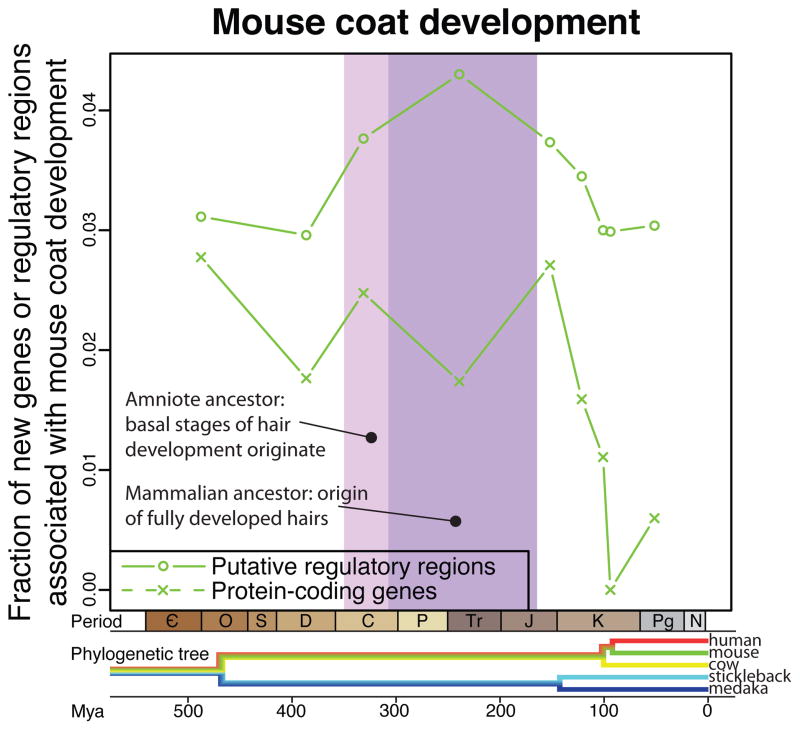
Finally, to further validate this approach, we looked outside of the Gene Ontology at a particularly well-studied gene set associated with the evolution of body hair, a phenotypic trait characteristic of mammals. There is a long history of genetic studies concerning mouse coat phenotypes that provides a set of almost 400 genes in the Mouse Genome Database for this process (27). Hair development begins as an area of epidermal thickening (epidermal placode) and shares the first developmental steps with avian feathers (28). The remaining stages in hair development do not appear until the mammalian ancestor, when body hair first originates. There is a slight enrichment for newly introduced regulatory regions to be associated with hair genes during the time of our amniote ancestor, when the basal stages of hair development originated (Fig. 3). Then the enrichment for putative novel regulatory regions near hair-associated genes peaks in the mammalian ancestor, at the time body hair first originates. We do not see a clear enrichment for new genes, only for new regulatory elements.
Fig. 3.
Enrichment for regulatory regions originating near genes involved in hair development during the time hair originated in evolution. For each branch we plot the percentage of genes or regulatory elements created on that branch that are associated with hair development.
It appears that at least three broad periods of regulatory innovation can be reliably detected in several vertebrate lineages. The first period, ranging from our vertebrate ancestor until about 300 Mya, when mammals split with birds and reptiles, is dominated by regulatory innovations near transcription factors and the key developmental genes they control. The second period, from about 300 Mya to 100 Mya, is characterized by a high frequency of regulatory innovations near receptors of extra-cellular signals, and a gradual decline in innovations near trans-dev genes. These two trends occurred independently in tetrapods and ray-finned fish. Finally, at least in placental mammals, we see a third period in which regulatory innovations for trans-dev and receptor genes have dropped to background frequencies, while regulatory innovations for genes involved in post-translational protein modification, including those in intra-cellular signaling pathways, are on the rise. Further sequencing of additional vertebrate species will make it possible to determine the pervasiveness of these trends and to look for additional functional categories that may be associated with epochs of evolutionary change in particular lineages.
Supplementary Material
Acknowledgments
This work was supported by the Howard Hughes Medical Institute (C.B.L., S.R.S., D.M.K., D.H.), the National Science Foundation (CAREER-0644282 to M.K., DBI-0644111 to A.S.), the National Institutes of Health (R01-HG004037 to M.K., P50-HG02568 to D.M.K., U54-HG003067 to K.L-T., 1U01-HG004695 C.B.L., 5P41-HG002371 B.J.R.), the Sloan Foundation (M.K.), and the European Science Foundation (EURYI to K.L-T.).
References
- 1.Karlsson EK, et al. Nat Genet. 2007;39:1321. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- 2.Miller CT, et al. Cell. 2007;131:1179. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB. Science. 2009;326:1663. doi: 10.1126/science.1178357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGregor AP, et al. Nature. 2007;448:587. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- 5.Colosimo PF, et al. Science. 2005;307:1928. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- 6.Chan YF, et al. Science. 2010;327:302. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hindorff LA, et al. Proc Natl Acad Sci USA. 2009;106:9362. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wray GA. Nat Rev Genet. 2007;8:206. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 9.Visel A, et al. Nat Genet. 2008;40:158. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee TI, et al. Cell. 2006;125:301. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim TH, et al. Cell. 2007;128:1231. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie X, et al. Proc Natl Acad Sci USA. 2007;104:7145. [Google Scholar]
- 13.Bejerano G, et al. Science. 2004;304:1321. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 14.Lindblad-Toh K, et al. Nature. 2005;438:803. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 15.Woolfe A, et al. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siepel A, et al. Genome Res. 2005;15:1034. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holland LZ, et al. Genome Res. 2008;18:1100. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Materials and methods are available as supporting material on Science online.
- 19.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Genome Res. 2010;20:110. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frazer KA, et al. Nature. 2007;449:851. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson G, et al. Nat Methods. 2007;4:651. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 22.Crawford GE, et al. Genome Res. 2006;16:123. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyle AP, et al. Cell. 2008;132:311. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valouev A, et al. Nat Methods. 2008;5:829. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashburner M, et al. Nat Genet. 2000;25:25. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLean CY, et al. Nat Biotechnol. 2010;28:495. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. Nucleic Acids Res. 2008;36:D724. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu P, et al. Int J Dev Biol. 2004;48:249. doi: 10.1387/ijdb.041825pw. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartz S, et al. Genome Res. 2003;13:103. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Proc Natl Acad Sci USA. 2003;100:11484. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanchette M, et al. Genome Res. 2004;14:708. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siepel A, Haussler D. Mol Biol Evol. 2004;21:468. doi: 10.1093/molbev/msh039. [DOI] [PubMed] [Google Scholar]
- 33.Pruitt KD, et al. Genome Res. 2009;19:1316. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hubbard T, et al. Nucleic Acids Res. 2002;30:38. doi: 10.1093/nar/30.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kent WJ. Genome Res. 2002;12:656. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu F, et al. Bioinformatics. 2006;22:1036. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 37.Pruitt KD, Tatusova T, Maglott DR. Nucleic Acids Res. 2005;33:D501. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu J, et al. PLoS Comput Biol. 2007;3:e247. doi: 10.1371/journal.pcbi.0030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherry ST, et al. Nucleic Acids Res. 2001;29:308. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Apweiler R, et al. Nucleic Acids Res. 2010;38:D142. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, et al. Nucleic Acids Res. 2006;34:D572. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lowe CB, Bejerano G, Haussler D. Proc Natl Acad Sci USA. 2007;104:8005. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green P. Genome Res. 2007;17:1547. doi: 10.1101/gr.7050807. [DOI] [PubMed] [Google Scholar]
- 44.Blair JE, Hedges SB. Mol Biol Evol. 2005;22:2275. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- 45.Benton MJ, Donoghue PC. Mol Biol Evol. 2007;24:26. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- 46.Kumar S, Hedges SB. Nature. 1998;392:917. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 47.Bininda-Emonds OR, et al. Nature. 2007;446:507. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 48.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Genome Res. 2007;17:413. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng Z, He S, Wang J, Wang W, Diogo R. Gene. 2006;370:113. doi: 10.1016/j.gene.2005.11.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.