Abstract
Systematically tunable in vitro platforms are invaluable in gaining insight to stem cell-microenvironment interactions in three-dimensional cultures. Utilizing recombinant protein technology, we independently tune hydrogel properties to systematically isolate the effects of matrix crosslinking density on cardiomyocyte differentiation, maturation, and function. We show that contracting human embryonic stem cell-derived cardiomyocytes (hESC-CMs) remain viable within four engineered elastin-like hydrogels of varying crosslinking densities with elastic moduli ranging from 0.45 to 2.4 kPa. Cardiomyocyte phenotype and function was maintained within hESC embryoid bodies for up to 2 weeks. Interestingly, increased crosslinking density was shown to transiently suspend spontaneous contractility. While encapsulated cells began spontaneous contractions at day 1 in hydrogels of the lowest crosslinking density, onset of contraction was increasingly delayed at higher crosslinking densities up to 6 days. However, once spontaneous contraction was restored, the rate of contraction was similar within all materials (71 ± 8 beats/min). Additionally, all groups successfully responded to electrical pacing at both 1 and 2 Hz. This study demonstrates that encapsulated hESC-CMs respond to 3D matrix crosslinking density within elastin-like hydrogels and stresses the importance of investigating temporal cellular responses in 3D cultures.
Introduction
Embryonic stem cells are versatile cells that hold great potential for regenerative medicine; but to harness their potential a greater understanding of how these cells proliferate, migrate, and differentiate in response to microenvironmental cues is needed. To date, stem cells are typically cultured on two-dimensional (2D), flat surfaces coated with extracellular matrix (ECM) proteins and exposed to soluble chemical cues to gain insight into stem cell-microenvironmental responses in vitro. While these 2D culture systems are advantageous for their simplified approach to identifying and assessing parameters that influence stem cell response, they fail to mimic most stem cell niches in the body that are comprised of three-dimensional (3D), hydrated networks of ECM proteins and sugars.
Native 3D environments provide a complex milieu of biochemical, biophysical, and biomechanical cues; therefore, reconstructing 3D environments in vitro is not trivial1. At the minimum, challenges in matrix design, successful cell encapsulation, and analysis of cell response need to be overcome before significant insight into stem cell response in 3D is gained. The development of highly tunable biomaterials has begun to address these challenges by providing in vitro platforms to systematically study stem cell responses to specific matrix parameters within a complex 3D culture environment.
Using recombinant protein technology, we have synthesized a family of recombinant elastin-like proteins that allow for the independent tuning of cell-adhesion ligand density, initial elastic modulus, and proteolytic degradation rate 2-3. In this system, we can tune hydrogel crosslinking density, thereby inherently modulating matrix stiffness, without altering cell-adhesion ligand density. This is achieved by varying the crosslinker to protein stoichiometry while maintaining a constant protein weight percent in the hydrogel. These modularly designed elastin-like biomaterials are attractive for their tunability, strength, and extensibility. Made entirely of amino acids, their proteolytic degradation profiles can be finely tuned, and protein degradation fragments are completely bioresorbable 3.
In this study, we utilize the elastin-like hydrogel platform to systematically explore the effects of matrix crosslinking density on the response of human embryonic stem cell-derived cardiomyocytes (hESC-CMs) within an embryoid body. In development, cardiomyocytes are derived from a soft mesoderm layer. With birth, the elastic modulus of the heart dramatically increases 4, and increased fibrillar collagen content with age further increases myocardium stiffness over time 5. Matrix properties of the myocardium can also be altered by injury, e.g., myocardial infarction, through the development of a fibrotic scar. The dynamic changes in myocardium stiffness throughout the course of development, maturation, aging, injury, and disease progression 6, suggest that stiffness may be a significant regulator of cardiomyocyte behavior.
Studies in 2D have shown that substrate stiffness can influence ESC pluripotency and differentiation7-8. In addition, 2D studies of neonatal cardiomyocytes cultured on matrices with elasticity matching that of native myocardium (~10-50 kPa) develop aligned sarcomeres, rhythmic contractility, and increased force generation 9-13. In contrast, cardiomyocytes cultured on more compliant or stiffer substrates show reduced cardiomyocyte function 9-13. As we move from 2D to 3D, fewer studies have attempted to investigate matrix stiffness and/or crosslinking density effects on cardiomyocyte response in more complex systems. In a study by Kraehenbuehl et al., matrix elasticity of matrix metalloproteinase-sensitive poly(ethylene glycol) (PEG)-based hydrogels was shown to affect cardiomyocyte commitment using P19 embryonal carcinoma cells 14. In addition, Shapira-Schweitzer and Seliktar encapsulated neonatal cardiomyocytes in PEGylated fibrinogen hydrogels of varying stiffness, by altering fibrinogen concentration, and demonstrated an inverse correlation between material stiffness and the amplitude of contraction 15. This inverse correlation was corroborated by Marsano et al. with rat cardiomyocytes in poly(glycerol sebacate) scaffolds 16.
To the best of our knowledge, separating out the effects of matrix crosslinking density from ligand density on cell response has not been examined for 3D cultures of hESC-CMs within biomaterials. Embryoid body cultures of hESCs are inherently 3D, and recent studies have demonstrated that physical forces act upon the embryoid body to alter cardiac differentiation potential. For example, Mohr et al. have demonstrated that controlling hESC embryoid body size through the use of microwells can regulate cardiac differentiation 17. The use of hydrodynamic forces was developed by Sargent et al. to influence both embryoid body size and to promote cardiac differentiation 18. Taken together, these previous studies suggest that 3D cultures of hESC-CMs are likely to be responsive to the mechanical properties of encapsulating biomaterials.
Materials and Methods
Protein Expression, Purification, and Crosslinking
Elastin-like protein (Figure 1) was expressed and purified using standard recombinant protein technology as previously reported 2. Briefly, protein sequences were cloned into pET15b plasmids using traditional recombinant techniques, expressed in Escherichia coli, BL21(DE3), for 3-5 hrs, and purified using an inverse temperature-cycling process. Typical protein yields were 50-100 mg/L. The modular protein design consists of four alternating repeats of an extended fibronectin-derived RGD sequence and a structural elastin-like domain. The recombinant protein chains are crosslinked through lysine residues using β[tris (hydroxymethyl) phosphino] propionic acid (THPP) (Pierce), where hydrogel crosslinking density is tuned by varying THPP hydroxy-methyl-phosphine:protein primary amine reactive group stoichiometry (4:1, 2:1, 1:1 and 0.5:1).
Fig. 1.
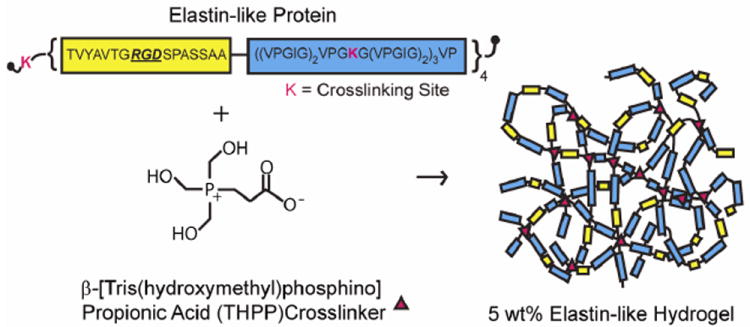
Schematic of elastin-like protein structure and hydrogel formation.
Mechanical Characterization
Acellular hydrogels were tested in unconfined compression on an ARG2 rheometer (TA Instruments). Hydrogels were compressed at 2 μm/s at 37°C in a custom-made phosphate buffered saline (PBS) bath. Elastic modulus in compression was determined by taking the slope of the stress-strain curve for small strains (<15%). Values are reported as mean ± SEM.
Embryonic Stem Cell Culture and Cardiomyocyte Differentiation
Human embryonic stem cells (H9, WiCell Research Institute) were cultured on mitotically-inactive primary mouse embryonic fibroblasts in DMEM:F12 knockout media (Invitrogen), supplemented with 20% knockout serum replacer (Invitrogen), 0.1 mM non-essential amino acids (Invitrogen), 2 mM L-glutamine (Invitrogen), 0.1 mM 2-mercaptoethanol (Gibco), and 8 ng/ml b-FGF (Peprotech). Human ESCs were spontaneously differentiated into cardiomyocytes through embryoid body (EB) formation in DMEM with 20% fetal bovine serum. Human ESCs were cultured in differentiation media for 8 days in suspension on ultra-low adhesion dishes to allow for EB formation, and then plated down on gelatin-coated 6-well plates for 8 days, prior to EB encapsulation. Whole EBs that exhibited spontaneous contractility were mechanically isolated and encapsulated in 5 wt% hydrogels (5 μl) with one EB per gel.
Cell Viability and Contractility Characterization
Cell viability was determined by metabolic activity via alamarBlue® cell proliferation assay and Live/Dead® viability/cytotoxicity kit (Invitrogen) over a 2 week period. Live/dead images were obtained on a Leica SPE confocal microscope and 3D visualizations were created using Volocity software. Contractility was characterized by the onset of spontaneous contractility, percentage of contracting hydrogels, beat rate, and electrical pacing. Spontaneous contraction and beat rate were determined visually using a Zeiss Axiovert 200M microscope equipped with a PECON incubation chamber (n=7-12, 2 independent trials). The percentage of the total population of hydrogels that exhibited spontaneous contractility within each group is reported as the number of spontaneously contractile hydrogels divided by total number of metabolically active hydrogels in each group. Beat rate is defined as the number of contractions observed per minute. Non-encapsulated EBs, plated in well plates, were used as comparative controls. EB-seeded hydrogels were also electrically paced after 14 days of culture using a commercial electrical pacer (MyoPacer, IonOptix LLC). A biphasic signal (+/-10 V, 10 ms) at varying frequencies (1 and 2 Hz) was applied, resulting in electrical pacing of cardiomyocytes within the hydrogel. EB response to electrical stimulation was characterized by pacing efficiency, which is defined as:
Immunohistochemistry
After 14 days of culture, hydrogels were immunostained for a mature cardiac marker, cardiac troponin T, and counterstained with 4’,6-diamidino-2-phenylindole (DAPI) for nuclei visualization. Samples were rinsed with PBS, fixed in 4% paraformaldehyde overnight, permeabilized using 0.2% Triton X-100 solution, blocked with 5% (w/v) non-fat milk, and incubated with primary mouse anti-cardiac troponin T (Isoform Ab-1, Thermo Scientific) at 1:100 dilution overnight, followed by secondary anti-mouse IgG-FITC (Santa Cruz Biotechnology, Inc.) at 1:200 dilution for 2 hrs. Images were taken with a Leica SPE confocal microscope, and 3D visualizations were created using Volocity software.
Hydrogel Degradation
Hydrogel degradation and remodeling was measured by protease activity in the culture medium. Proteolytic activity was assessed by zymography, as previously reported 19, with slight modifications. Samples were mixed with 2x Laemelli buffer without reducing agent and separated by 10% SDS-PAGE gels containing 0.1% gelatin or 0.2% elastin-like protein at room temperature. The gels were washed 2x with 2.5% Triton X-100 for 15 min, incubated in development buffer (0.05 M Tris-HCl, pH 8.8, and 5 mM CaCl2) overnight at 37°C, stained with 0.1% w/v Coomassie brilliant blue, and destained with destaining solution (45% methanol, 45% water, and 10% acetic acid). Collagenase type IV (Invitrogen) and porcine elastase (Sigma) were used as positive controls.
Results
Recombinant protein biomaterials can be used to systematically explore cellular responses to matrix cues. With molecular-level control over protein structure, several hydrogel properties can be independently tuned. Previously, we reported the design and synthesis of a family of elastin-like hydrogels 2, consisting of modular elastin-like and cell-adhesion domains. With the ability to deform and recoil, the elastin-like structural domains impart elasticity and resilience to the hydrogel, while serving as a relatively inert backbone. Cell-adhesion peptide sequences derived from the extracellular matrix (e.g., fibronectin) were designed into the recombinant elastin-like sequences to provide stable presentation of cell adhesion sites. Following recombinant protein synthesis and purification, the protein polymers were covalently crosslinked to form free-standing bulk hydrogels (Figure 1).
Hydrogel crosslinking density, and subsequently matrix modulus, was tuned systematically by varying the stoichiometry between crosslinker and protein reactive groups. The RGD cell-adhesion ligand density was maintained at 1.85 × 10-5 mol/cm3 for all hydrogels.
Using stoichiometric ratios of 0.5:1 to 4:1 crosslinker:protein reactive groups, the elastic modulus in compression could be tailored from 0.45 to 2.4 kPa without altering ligand density (Figure 2).
Fig. 2.
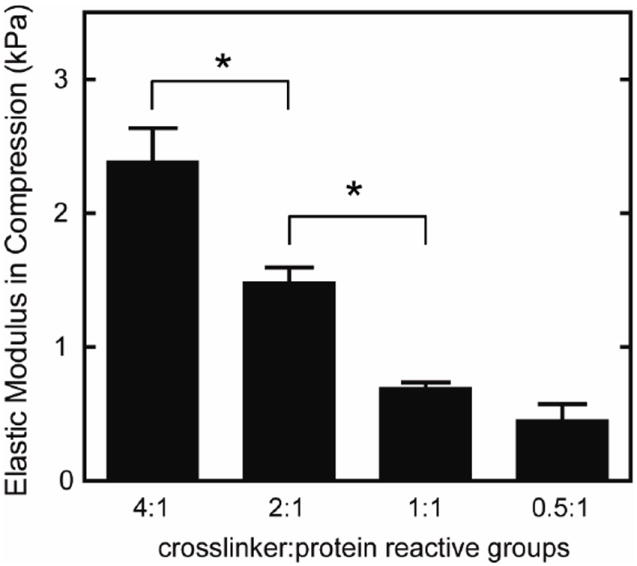
Elastic modulus in compression of elastin-like hydrogels with varying crosslinking densities, reported as mean ± SEM (n=4-6). Significant difference (p < 0.05) as calculated by Tukey’s post hoc test is denoted by *. Increasing the ratio of crosslinker to protein reactive groups increased the modulus of the elastin-like hydrogels.
Single human embryonic stem cell-derived embryoid bodies exhibiting spontaneous contractility were selected and encapsulated in individual elastin-like hydrogels. After encapsulation, embryoid bodies remained viable and metabolically active for up to 2 weeks (the latest experimental time point, Figures 3 and 4). As expected, individual samples showed varied metabolic activity, reflecting the heterogeneity of embryoid body populations (data not shown). When the activity of each embryoid body was individually normalized to its day 3 activity values, the data demonstrated similar levels or increased metabolic activity over time among all groups. No significant differences were detected among the groups at day 14. Verification of maintenance of cardiomyocyte differentiation within the encapsulated embryoid bodies was demonstrated by immunohistochemical staining of cardiac troponin T, a mature cardiac marker (Figure 5). Cardiac troponin T was present in embryoid bodies encapsulated within hydrogels of all crosslinking densities.
Fig. 3.
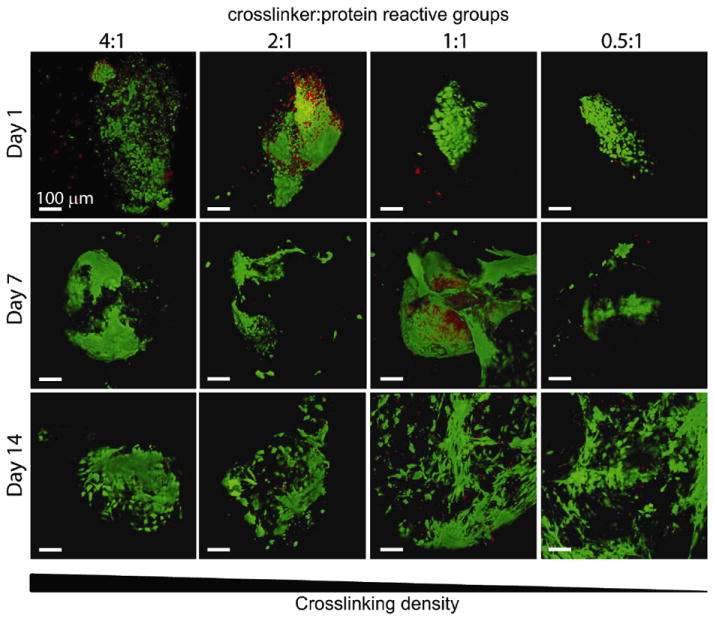
Representative 3D live (green) / dead (red) visualizations of encapsulated human embryoid bodies in elastin-like hydrogels of various crosslinking densities after 1, 7, and 14 days of culture, where each image is of a separate, representative EB. Scale bar = 100 μm.
Fig. 4.
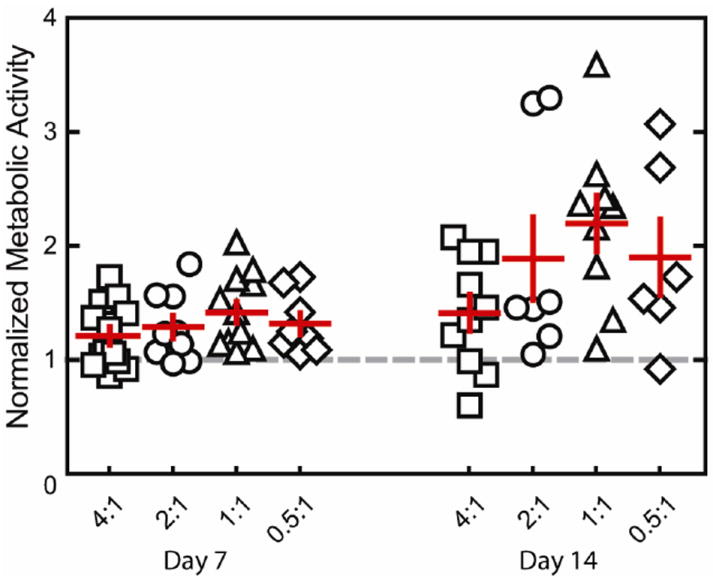
Metabolic activity normalized to day 3 (gray dashed line) for embryoid bodies encapsulated within hydrogels of various crosslinking densities, presented as mean ± SEM (n=6-13). Embryoid bodies remained metabolically active after encapsulation for 14 days of culture.
Fig. 5.
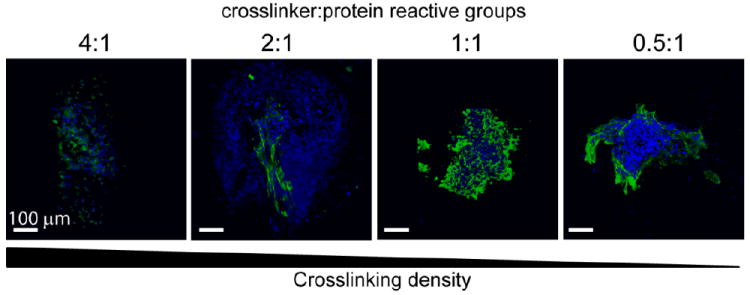
3D visualizations of representative immunohistochemical staining for cardiac troponin T (green) with nuclear stain (blue) for all groups after 14 days of culture. Scale bar = 100 μm. The presence of differentiated cardiomyocytes was confirmed by the presence of cardiac troponin T staining in all groups.
Contractility was measured visually, and the onset of spontaneous contraction in the hydrogels was recorded. The percentage of contracting hydrogels at each time point was quantified by dividing the number of contracting hydrogels by the total number of metabolically active hydrogels in each group. By analyzing metabolically active hydrogels, we ensure that only hydrogels with viable EBs are considered when characterizing the potential of each hydrogel group to regain contractility. If no metabolic activity was observed in the hydrogel, that hydrogel was eliminated from the data set, as the EB may have fallen out of the hydrogel during encapsulation. Encapsulation within a 3D hydrogel resulted in transient suspension of spontaneous contraction (Figure 6A,B). Across the four hydrogel formulations tested, increased crosslinking density resulted in longer delays before restored contraction (Figure 6A,B). While 100% of the embryoid bodies in the hydrogels with the lowest crosslinking density were contractile on day 1 (compared with only 7% in the highest crosslinking density), about 75% of all embryoid bodies had regained contractility by day 10. Once the cardiomyocytes regained contractility, similar beat rates among all groups and non-encapsulated EBs were observed (Figure 6C). Initial contractility rate for all groups was measured to be 50 ± 4 beats/min. Over time, beat rate began to diverge for all groups, but on average, a general increase with time was observed, where beat rate averaged 71 ± 8 beats/min by day 14. These beat rates fall slightly below the range of average human embryo heart rates, 80-196 beats/min,20 and within the range of the resting adult human, 60-80 beats/min.
Fig. 6.
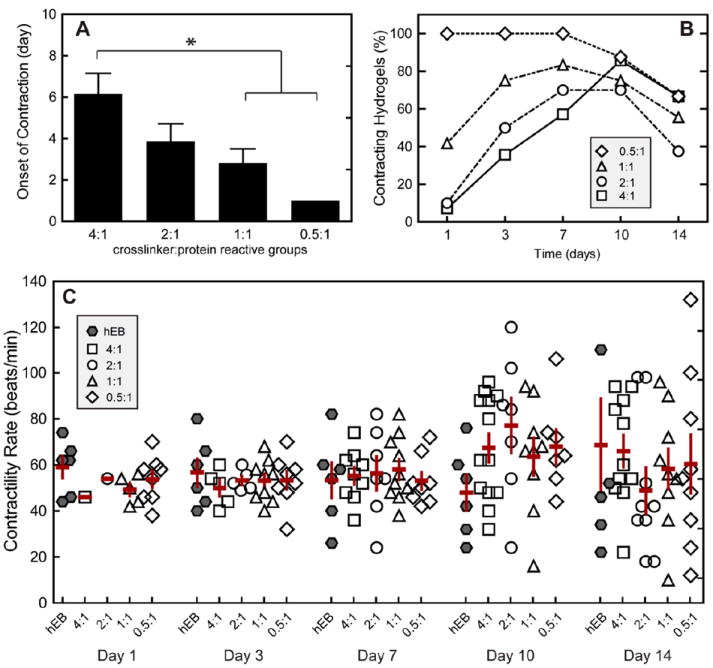
(A) Contractility of hESC-CMs within elastin-like hydrogels of various crosslinking densities characterized by onset of contraction (n = 7-12), (B) percent of contracting hydrogels of the total population of hydrogel in each group over time, and (C) contractility rate over time. Significant difference (p < 0.05) as calculated by Tukey’s post hoc test is denoted by *. Data are presented as mean ± SEM. Contractility of non-encapsulated human EBs (hEBs) is displayed for comparison. Increased crosslinking density resulted in a transient inhibition of cell contractility; however, after spontaneous contractility was regained, no significant differences were found for contractility rate among groups and non-encapsulated hEBs over the 14 days of culture.
After two weeks of culture, hydrogels were electrically paced with a commercial myopacer. The ability of the cells to respond to electrical stimulation was characterized by stimulated beat rate response and pacing efficiency (Figure 7). A pacing efficiency of 1 means that cells paced exactly at the stimulated rate, and a pacing efficiency of 0 means a lack of response to the electrical stimulation. If the pacing efficiency is greater than 1, then the observed contraction rate exceeded the stimulated rate; alternatively for efficiencies less than 1, the observed contraction rate did not meet the stimulated rate. Lastly, if the pacing efficiency is negative, this indicates that the cells responded in an inverse fashion, e.g., cells spontaneously beating faster than the pacing rate increased their contraction rate even further. In general, the majority of spontaneously contracting embryoid bodies in all groups was successfully electrically paced to 1 and 2 Hz. However, in some rare cases, where spontaneous contractility rate was faster than the electrically paced rate, irregular contractions were observed between stimulated contractions (Supplementary video A,B). Additionally, in a few exceptions where spontaneous contraction rates were much slower, these samples fell short of the stimulated frequency. Overall, these data demonstrate that hESC-CMs continue to respond to electrical stimulation while encapsulated within the 3D elastin-like matrix, and that cardiomyocyte response to electrical stimulation was not correlated to matrix crosslinking density.
Fig. 7.
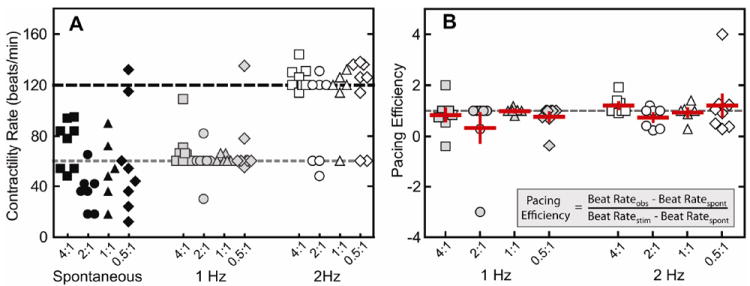
(A) Beat rates of embryoid bodies during spontaneous contraction (black) and electrical stimulation at 1 Hz (gray) and 2 Hz (open) (B), along with pacing efficiency at 1 Hz (gray) and 2 Hz (open). Data are presented as mean ± SEM
Macroscopic images of elastin-like hydrogels of the highest crosslinking density at day 0 and day 14 showed little to no evidence of bulk hydrogel degradation (Figure 8A). To further determine if embryoid body-induced hydrogel degradation and remodeling was occurring over the time course of the experiments, supernatants of the culture media were analyzed for protease activity. Media samples were separated by molecular weight using electrophoresis over polyacrylamide gels impregnated with gelatin (control) or the recombinant elastin-like protein. Zymography to detect elastin-like protein degradation revealed that protease activity was similar among all groups and was indistinctive from that of cell medium alone, i.e. medium that had never been exposed to cells (Figure 8B). Quantitative densitometry of the zymography gels displayed identical proteolytic activity profiles for samples with and without cells (Figure 8C). Collagenase type IV and porcine elastase were used as positive controls to confirm activity detection by both gelatin and elastin-like zymograms (Figure 8B,C).
Fig. 8.
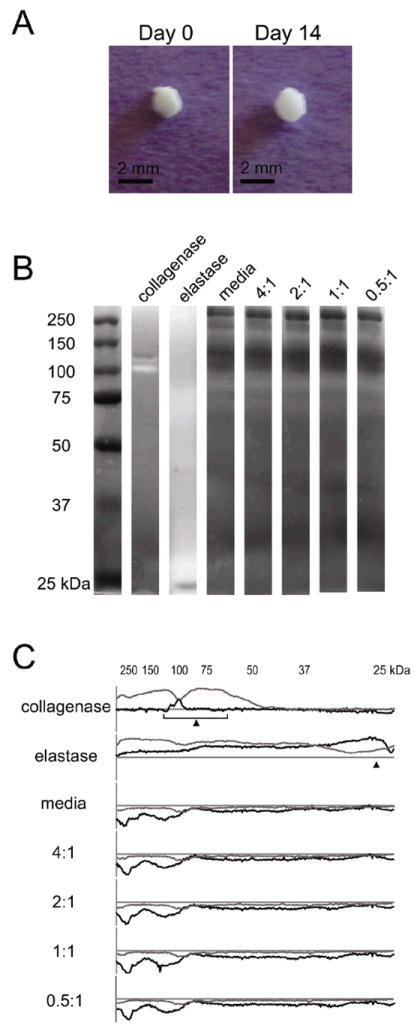
(A) Macroscopic images of a representative 4:1 hydrogel/embryoid body culture at days 0 and 14 displaying little to no visible degradation over the 2-week culture period. (B) Elastin zymogram showing no visible differences in proteolytic activity among unconditioned media and cell conditioned media from embryoid bodies encapsulated in hydrogels of varying crosslinking density. Collagenase type IV and elastase were used as positive controls. (C) Line profiles of gelatin (gray) and elastin (black) zymograms, where plots above the x-axis represent protein degradation and below the x-axis represent increased protein (e.g., proteins in the media). Expected molecular weights for collagenase type IV (68-130 kDa) and elastase (26 kDa) are indicated with arrows.
Discussion
The ECM evolves during development, aging, injury and disease. Changes to the content, quantity, and organization of the ECM can often direct stem cell fate and cell function. To dissect out the critical parameters that influence cell behavior in 3D environments, biomaterials with independently tunable properties are needed. Here we demonstrate the potential of elastin-like protein biomaterials to be used as an in vitro platform to isolate the effects of crosslinking density on encapsulated hESC-CMs.
Using recombinant protein engineering, we produced clinically relevant biomaterials with repeatable molecular precision and complete bioresorbability. The elastin-like protein provides both elasticity and the stable presentation of cell-adhesion ligands derived from fibronectin while allowing for the independent tuning of crosslinking density by varying crosslinker to protein stoichiometry (Figure 1). These unique biomaterials were found to support the viability and metabolism (Figure 3,4) of encapsulated human embryoid bodies, enabling sustained expression of a mature cardiomyocyte marker (Figure 5).
While most studies to date have focused on effects of substrate properties on cardiomyocyte function in 2D, less is known about matrix effects on hESC-CMs in 3D cultures. By changing the crosslinking density, we created a family of 3D matrices with moduli ranging from 0.45 to 2.4 kPa (Figure 2) without altering RGD ligand presentation (1.85 × 10-5 mol/cm3). Despite the small range of elastic moduli explored, we observed differences in transient contractility, suggesting that these cells are sensitive to small changes in matrix stiffness in 3D.
As with all hydrogel materials, changing crosslinking density also alters mesh size and hence the effective diffusion rate of nutrients, waste, and cell-secreted factors whose local accumulation or depletion in the matrix may lead to altered cell metabolism and growth. Confocal microscopic observations of embryoid bodies in hydrogels at all crosslinking densities confirmed the viability of cells throughout the entire gel during 14 days of in vitro culture (Figure 3). The isolated, random presence of dead cell clusters and the absence of cell death in the centers of the gels suggest that sufficient diffusion is occurring to support cell metabolism in all matrices. In addition to diffusion effects, changes in mesh size are also known to alter cell morphology and cellular processes affected by cell spreading such as proliferation and migration. For example, the small mesh size of highly crosslinked poly(ethylene glycol) networks physically inhibits cell growth if no proteolytic sites are present in the material 21-22. Results with other cell-laden 3D hydrogel systems typically observe increased cell proliferation and migration within hydrogels of lower crosslinking densities, although we observed no statistically significant changes in cell metabolism across the range of hydrogels tested here 14, 21, 23.
Encapsulated hESC-CMs exhibited bulk, synchronous contractions capable of deforming the bulk hydrogel. Interestingly, we observed a new phenomenon whereby spontaneous cellular contraction was transiently suspended after 3D encapsulation. The duration of this transient suspension was directly correlated with increased crosslinking density (Figure 6). In a hydrogel network, the work required to deform the material is directly correlated to crosslinking density; therefore, two distinct, yet not mutually exclusive, hypotheses may explain these results. First, the delay in contractility may reflect an adaptation period needed for cardiomyocytes to adjust to a stiffer microenvironment. Over time, the cardiomyocytes may mature and/or adapt to be capable of generating greater contraction forces on the surrounding matrix. Second, the cells may initiate remodeling of their local matrix to yield a more compliant material with lower crosslinking density on which they can then do work. These two hypotheses are not mutually exclusive, and based on previous work by others as described in more detail below, both processes are likely to be occurring simultaneously.
Determining cellular adaptation to the matrix environment is unfortunately harder to elucidate within a 3D matrix as compared to 2D substrates. Evidence of cell interaction and adaptation to the surrounding extracellular matrix has been shown in the form of integrin shedding 24 and cytoskeletal remodeling 25. Additionally, Jacot et al. demonstrated an immediate increase in elastic modulus in mouse myocardium after birth 12. This stiffening is hypothesized to aid in the functional maturation of cardiomyocytes. Uniaxial mechanical stretch has also been shown to increase cardiomyocyte alignment and sarcomeric banding 26. Thus, cardiomyocytes are capable of sensing and responding to external stimuli by altering their internal contractility machinery. As the field moves in the direction of 3D cultures, new non-destructive assays and technologies will be needed to assess cell adaptation to enable a better understanding of the mechanisms underlying cardiomyocyte maturation and function. Some steps have been made toward this direction with the introduction of 3D particle tracking 27-28 that is capable of visualizing 3D displacement vectors, but data processing can be computationally intensive and interpreting these results still remains a challenge, as matrix modulus is typically assumed to be constant and isotropic.
Rather than adapting to their surroundings, cells are also capable of degrading and remodeling the matrix around them. This ability to alter the surrounding extracellular matrix permits cells to migrate and proliferate, which is critical for developmental and regenerative processes 29, as well as disease progression 30. Unfortunately, cell-dictated matrix remodeling often results in local matrix inhomogeneities that cannot be assessed using bulk measurement techniques. Here, we used visual assessments of gels and detection of protease activity by zymography to assess cell-dictated matrix degradation. These results suggest that large-scale matrix remodeling is not occurring; however, this does not preclude local remodeling on the cellular length scale. To visualize matrix remodeling on a cellular length scale, others have utilized specialized techniques involving optical tweezers 31, microrheology 27, 32, Förster resonance energy transfer-based protease sensors 33-34, or non-linear microscopy like second harmonic generation 35-36. A more rigorous investigation of local cell-remodeling within the elastin-like hydrogels is outside the scope of this study, and left for future exploration.
With a growing interest in the use of embryonic stem cells, tunable systems that mimic nature’s 3D environments are needed to isolate principle parameters that regulate cell-matrix crosstalk. Here we demonstrate that recombinant elastin-like hydrogels are suitable biomaterials for systematic studies of 3D hESC-CM culture. By more fully understanding how hESC-derived cardiomyocytes respond to ECM properties, we may be able to predict how these cells will respond to and integrate with healthy (more compliant) or injured (more rigid) cardiac tissue in the body. In addition, our data demonstrate a new phenomenon of transient suspended contractility upon 3D hESC-CM encapsulation. The encapsulated embryoid bodies were able to overcome this suspended contractility after a time delay that was directly correlated to the matrix crosslinking density. Interestingly, the duration of time spent in the suspended contractility phase did not affect subsequent spontaneous beating frequency or the ability to respond to electrical stimulation. This period of suspended contractility is an intriguing observation, and future investigation into the mechanism of this delay may shed light on hESC-CM adaptability to mechanical properties of their surrounding environment.
Conclusions
Understanding cell-microenvironment interactions is important for harnessing the potential of embryonic stem cells. Using recombinant protein design, we systematically tailored elastin-like scaffolds to achieve a range of crosslinking densities with identical cell-adhesion ligand density. We have shown that hESC-CMs remain viable within elastin-like hydrogels, where the phenotype and function of these cells are maintained for up to 2 weeks. Increased crosslinking density was shown to transiently suspend contractility within these hydrogels. However, once contractility was restored, hESC-CMs in high crosslinking density hydrogels contracted with similar beat rates as their lower crosslinking density counterparts and were capable of responding to electrical stimulation demonstrating cardiomyocyte function. Taken together, these data demonstrate the suitability of these elastin-like hydrogels for in vitro systematic studies of hESC-CM responses to matrix cues.
Supplementary Material
Acknowledgments
The authors would like to thank Eric Chiao and Melanie Marchand from the Stanford Stem Cell Institute for Stem Cell Biology and Regenerative Medicine for hESC culture expertise. The authors acknowledge funding support from NSF EFRI-CBE-0735551, NSF DMR-084663, NIH 1DP2OD006477, NIH R33 HL089027, and CIRM RC1-00151-1.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
Notes and References
- 1.Lutolf MP, Gilbert PM, Blau HM. Nature. 2009;462:433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Straley KS, Heilshorn SC. Soft Matter. 2009;5:114–124. [Google Scholar]
- 3.Straley KS, Heilshorn SC. Adv Mater. 2009;21:4148. [Google Scholar]
- 4.Jacot JG, Martin JC, Hunt DL. J Biomech. 2010;43:93–98. doi: 10.1016/j.jbiomech.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradshaw AD, Baicu CF, Rentz TJ, Van Laer AO, Bonnema DD, Zile MR. Am J Physiol Heart Circ Physiol. 2010;298:H614–622. doi: 10.1152/ajpheart.00474.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaturvedi RR, Herron T, Simmons R, Shore D, Kumar P, Sethia B, Chua F, Vassiliadis E, Kentish JC. Circulation. 2010;121:979–988. doi: 10.1161/CIRCULATIONAHA.109.850677. [DOI] [PubMed] [Google Scholar]
- 7.Evans ND, Minelli C, Gentleman E, LaPointe V, Patankar SN, Kallivretaki M, Chen X, Roberts CJ, Stevens MM. Eur Cell Mater. 2009;18:1–13. doi: 10.22203/ecm.v018a01. discussion 13-14. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhury F, Li Y, Poh YC, Yokohama-Tamaki T, Wang N, Tanaka TS. PLoS One. 2010;5:e15655. doi: 10.1371/journal.pone.0015655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bajaj P, Tang X, Saif TA, Bashir R. J Biomed Mater Res A. 2010;95:1261–1269. doi: 10.1002/jbm.a.32951. [DOI] [PubMed] [Google Scholar]
- 10.Bhana B, Iyer RK, Chen WL, Zhao R, Sider KL, Likhitpanichkul M, Simmons CA, Radisic M. Biotechnol Bioeng. 2010;105:1148–1160. doi: 10.1002/bit.22647. [DOI] [PubMed] [Google Scholar]
- 11.Engler AJ, Carag-Krieger C, Johnson CP, Raab M, Tang HY, Speicher DW, Sanger JW, Sanger JM, Discher DE. J Cell Sci. 2008;121:3794–3802. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacot JG, McCulloch AD, Omens JH. Biophys J. 2008;95:3479–3487. doi: 10.1529/biophysj.107.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tracqui P, Ohayon J, Boudou T. J Theor Biol. 2008;255:92–105. doi: 10.1016/j.jtbi.2008.07.036. [DOI] [PubMed] [Google Scholar]
- 14.Kraehenbuehl TP, Zammaretti P, Van der Vlies AJ, Schoenmakers RG, Lutolf MP, Jaconi ME, Hubbell JA. Biomaterials. 2008;29:2757–2766. doi: 10.1016/j.biomaterials.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 15.Shapira-Schweitzer K, Seliktar D. Acta Biomater. 2007;3:33–41. doi: 10.1016/j.actbio.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Marsano A, Maidhof R, Wan LQ, Wang Y, Gao J, Tandon N, Vunjak-Novakovic G. Biotechnol Prog. 2010;26:1382–1390. doi: 10.1002/btpr.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohr JC, Zhang J, Azarin SM, Soerens AG, de Pablo JJ, Thomson JA, Lyons GE, Palecek SP, Kamp TJ. Biomaterials. 2010;31:1885–1893. doi: 10.1016/j.biomaterials.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sargent CY, Berguig GY, McDevitt TC. Tissue Eng Part A. 2009;15:331–342. doi: 10.1089/ten.tea.2008.0145. [DOI] [PubMed] [Google Scholar]
- 19.Hawkes SP, Li H, Taniguchi GT. Methods Mol Biol. 2010;622:257–269. doi: 10.1007/978-1-60327-299-5_16. [DOI] [PubMed] [Google Scholar]
- 20.Howe RS, Isaacson KJ, Albert JL, Coutifaris CB. J Ultrasound Med. 1991;10:367–371. doi: 10.7863/jum.1991.10.7.367. [DOI] [PubMed] [Google Scholar]
- 21.Bott K, Upton Z, Schrobback K, Ehrbar M, Hubbell JA, Lutolf MP, Rizzi SC. Biomaterials. 2010;31:8454–8464. doi: 10.1016/j.biomaterials.2010.07.046. [DOI] [PubMed] [Google Scholar]
- 22.Bryant SJ, Arthur JA, Anseth KS. Acta Biomater. 2005;1:243–252. doi: 10.1016/j.actbio.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Bryant SJ, Chowdhury TT, Lee DA, Bader DL, Anseth KS. Ann Biomed Eng. 2004;32:407–417. doi: 10.1023/b:abme.0000017535.00602.ca. [DOI] [PubMed] [Google Scholar]
- 24.Goldsmith EC, Carver W, McFadden A, Goldsmith JG, Price RL, Sussman M, Lorell BH, Cooper G, Borg TK. Am J Physiol Heart Circ Physiol. 2003;284:H2227–2234. doi: 10.1152/ajpheart.00920.2002. [DOI] [PubMed] [Google Scholar]
- 25.McCain ML, Parker KK. Pflugers Arch. 2011;462:89–104. doi: 10.1007/s00424-011-0951-4. [DOI] [PubMed] [Google Scholar]
- 26.Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD, Pabon L, Reinecke H, Murry CE. Circ Res. 2011;109:47–59. doi: 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bloom RJ, George JP, Celedon A, Sun SX, Wirtz D. Biophys J. 2008;95:4077–4088. doi: 10.1529/biophysj.108.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS. Nat Methods. 2010;7:969–971. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu P, Takai K, Weaver VM, Werb Z. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kandalam V, Basu R, Moore L, Fan D, Wang X, Jaworski DM, Oudit GY, Kassiri Z. Circulation. 2011;124:2094–2105. doi: 10.1161/CIRCULATIONAHA.111.030338. [DOI] [PubMed] [Google Scholar]
- 31.von Wichert G, Haimovich B, Feng GS, Sheetz MP. EMBO J. 2003;22:5023–5035. doi: 10.1093/emboj/cdg492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotlarchyk MA, Shreim SG, Alvarez-Elizondo MB, Estrada LC, Singh R, Valdevit L, Kniazeva E, Gratton E, Putnam AJ, Botvinick EL. PLoS One. 2011;6:e20201. doi: 10.1371/journal.pone.0020201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller MA, Barkal L, Jeng K, Herrlich A, Moss M, Griffith LG, Lauffenburger DA. Integr Biol (Camb) 2011;3:422–438. doi: 10.1039/c0ib00083c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi L, De Paoli V, Rosenzweig N, Rosenzweig Z. J Am Chem Soc. 2006;128:10378–10379. doi: 10.1021/ja063509o. [DOI] [PubMed] [Google Scholar]
- 35.Brown E, McKee T, diTomaso E, Pluen A, Seed B, Boucher Y, Jain RK. Nat Med. 2003;9:796–800. doi: 10.1038/nm879. [DOI] [PubMed] [Google Scholar]
- 36.Lee PF, Yeh AT, Bayless KJ. Exp Cell Res. 2009;315:396–410. doi: 10.1016/j.yexcr.2008.10.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


