INTRODUCTION
The TP53 gene, which encodes the p53 protein, is the most frequent target for mutation in tumors, with over half of all human cancers exhibiting mutation at this locus (Vogelstein et al., 2000). Wild-type p53 functions primarily as a transcription factor and possesses an N-terminal transactivation domain, a centrally located sequence specific DNA binding domain, followed by a tetramerization domain and a C-terminal regulatory domain (Laptenko and Prives, 2006).
Unlike most tumor suppressor genes, which are predominantly inactivated as a result of deletion or truncation, the majority of mutations in TP53 are missense mutations, a few of which cluster at “hotspot” residues in the DNA binding core domain (Petitjean et al., 2007). In contrast to wild-type p53, which under unstressed conditions is a very short-lived protein, these missense mutations lead to the accumulation of full-length p53 protein with a prolonged half-life (Brosh and Rotter, 2009). While many tumor-derived mutant forms of p53 can exert a dominant-negative effect on the remaining wild-type allele, the end result in many forms of human cancer is frequently loss of heterozygosity, where only the mutant form is retained, suggesting that there is a selective advantage conferred by losing the remaining wild-type p53, even after one allele has been mutated (Brosh and Rotter, 2009).
Mutant forms of p53 can exert oncogenic, or gain-of-function, activities independent of their effects on wild-type p53. In vivo knock-in mice harboring two tumor-derived mutants of p53 (equivalent to R175H and R273H in humans) display an altered tumor spectrum as well as more metastatic tumors when compared to p53 null mice (Lang et al., 2004; Olive et al., 2004). The mutational status of p53 has been shown to predict poor outcomes in multiple types of human tumors, including breast cancer, and certain p53 mutants associate with an even worse prognosis (Olivier et al., 2006; Petitjean et al., 2007). Mutant p53 expression correlates with increased survival, invasion, migration and metastasis in preclinical breast cancer models (Adorno et al., 2009; Muller et al., 2009; Stambolsky et al., 2010). Nonetheless, mutant p53-induced phenotypic alterations in mammary tissue architecture have not been fully explored.
Breast cancer is thought to arise from mammary epithelial cells found in structures referred to as acini, which collectively form terminal ductal lobular units. Each acinus consists of a single layer of polarized luminal epithelial cells surrounding a hollow lumen (Bissell et al., 2002). While traditional two-dimensional (2D) cell culture has provided insight into the process of breast carcinogenesis, such in vitro culture conditions differ from the microenvironment that a cell would experience in vivo (Bissell et al., 2002). By contrast, a laminin-rich extracellular matrix allows normal mammary epithelial cells to form three-dimensional structures reminiscent of acinar structures found in vivo (Petersen et al., 1992). Since one of the hallmarks of breast tumorigenesis is the disruption of mammary tissue architecture, three-dimensional (3D) culture conditions allow one to readily distinguish normal and tumorigenic tissue by morphological phenotype (Petersen et al., 1992). In addition, inhibition of key oncogenic signaling pathways is sufficient to phenotypically revert breast cancer cells grown in 3D culture (Bissell et al., 2005). Here we implicate mutant p53 and the mevalonate pathway in the disruption of acinar morphology and our data have also revealed a potential mechanism by which mutant p53 increases expression of the genes in the mevalonate pathway.
RESULTS
Mutant p53 depletion in breast cancer cells leads to a phenotypic reversion in 3D culture
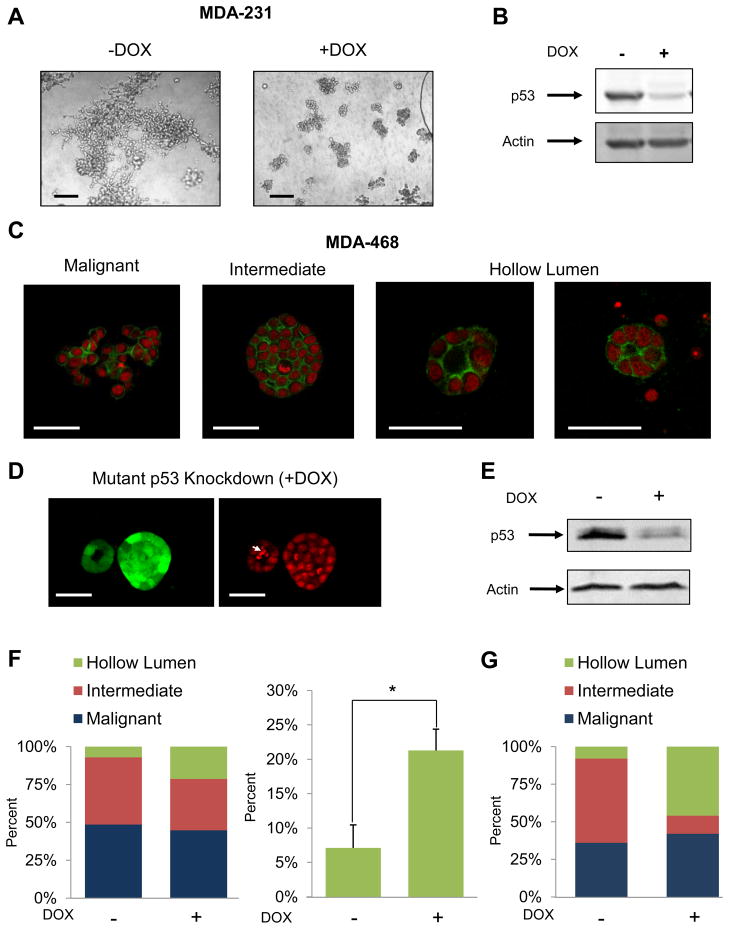
To investigate the role of mutant p53 in breast cancer, we employed the 3D culture protocol where mammary epithelial cells are grown in a laminin rich extracellular matrix. We examined the 3D morphologies of two cell lines derived from metastatic breast tumors that each expresses exclusively a single mutant p53 allele: MDA-231 (R280K) and MDA-468 (R273H). These cells were engineered to express miR30-based doxycycline-inducible shRNA targeting endogenous mutant p53 in the 3′ UTR (designated MDA-231.shp53 and MDA-468.shp53). In both cases mutant p53 reduction by shRNA led to dramatic changes in the behavior of the cells when cultured in a 3D microenvironment. MDA-231 cells, when grown in 3D culture, normally exhibit an extremely disordered and invasive morphology, which has been characterized as “stellate” (Kenny et al., 2007). Depleting these cells of mutant p53 in 3D culture conditions almost completely abrogated the stellate morphology of large, invasive structures with bridging projections (Figure 1A). Instead, MDA-231 cells with reduced p53 developed smaller, less invasive appearing cell clusters. By titrating doxycycline, we observed a progressive loss of malignant, invasive characteristics as a function of decreasing levels of mutant p53 (Figure S1A–B). Although this reduction in invasive behavior in 3D culture supports the recent findings that mutant p53 promotes the invasion of breast cancer cells (Adorno et al., 2009; Muller et al., 2009), MDA-231 cells with reduced p53 did not assume the ordered acinus-like morphology that is characteristic of non-malignant mammary epithelial cells.
Figure 1. Depletion of mutant p53 from breast cancer cells induces a phenotypic reversion in 3D culture.
(A) MDA-231.shp53 cells were grown under 3D conditions for 8 days in the absence or presence of DOX to induce an shRNA targeting p53. Representative differential interference contrast (DIC) images are shown. Scale Bar, 200 μm.
(B) MDA-231.shp53 cells grown as in (A) prior to immunoblotting analysis. p53 was detected using anti-p53 antibody (PAb1801).
(C) MDA-468.shp53 cells were grown in 3D cultures for 8 days and confocal microscopic structures were grouped into the three indicated categories. Actin (Green) and nuclear (Red) staining. Scale bar, 50 μm.
(D) MDA-468.shp53 cells were grown in 3D cultures for 8 days in the presence of DOX to deplete mutant p53. Left panel: GFP (Green) serves as a marker for shRNA induction. Right panel: Nuclei (Red). The larger structure is representative of intermediate colony morphologies, while the smaller structure is representative of acinus-like structures with hollow lumen morphology. White arrow indicates apoptotic cell debris within the luminal space. Scale bar, 50 μm.
(E) MDA-468.shp53 cells were grown and processed as in (B).
(F) Morphometry: A stable pool of MDA-468.shp53 cells was grown in 3D cultures for 8 days in the presence or absence of DOX and structures were analyzed by confocal microscopy and categorized as in (C). Left panel: population distribution. Right panel: percent structures with hollow lumens. Data presented as mean ± SD. *denotes p<0.01.
(G) Morphometry: A stable clone of MDA-468.shp53 cells was grown in 3D culture conditions and analyzed as in (F). An average of two experiments is shown.
MDA-468 cells exhibit a less invasive, but highly disorganized appearance, and have been classified as “grape-like” (Kenny et al., 2007). Under 3D culture conditions, MDA-468.shp53 cells displayed three types of cellular morphologies: (1) constellations of cells with a highly disordered “malignant” appearance that comprise about 30–40% of the population, (2) spherical cell clusters with an “intermediate” morphology that, while disordered, appear less malignant (about 55–65% of the population) and (3) a very small proportion (<5%) of structures that closely resemble small acini and contain a hollow lumen (examples of these categories are shown in Figure 1C). Strikingly, when mutant p53 was depleted from these cells, a significant proportion of the population underwent a full phenotypic reversion from highly disorganized structures to acinus-like structures with a hollow lumen (Figure 1D). These reverted structures also display proper localization of α6 integrin, suggesting that they have regained apicobasal polarization (Figure S1E). Using either a stable pool of MDA-468.shp53 cells (Figure 1F) or a stable clone derived from these cells (Figure 1G), we observed a significant increase in the hollow lumen population upon mutant p53 depletion, with nearly 50% of the population falling into this acinus-like morphology in the latter case. While in the experiment shown in Figure 1G, we observed a decrease in predominantly the intermediate population upon reversion to hollow lumen structures, in other cases we observed a decrease in both malignant and intermediate populations (e.g. Figure 4A below). Since the stable clone of MDA-468.shp53 cells exhibited a higher degree of reversion, all further experiments were carried out using these cells. Importantly, since both breast cancer cell lines express only mutant p53, these phenotypic changes may be attributed directly to the reduction in mutant p53 levels.
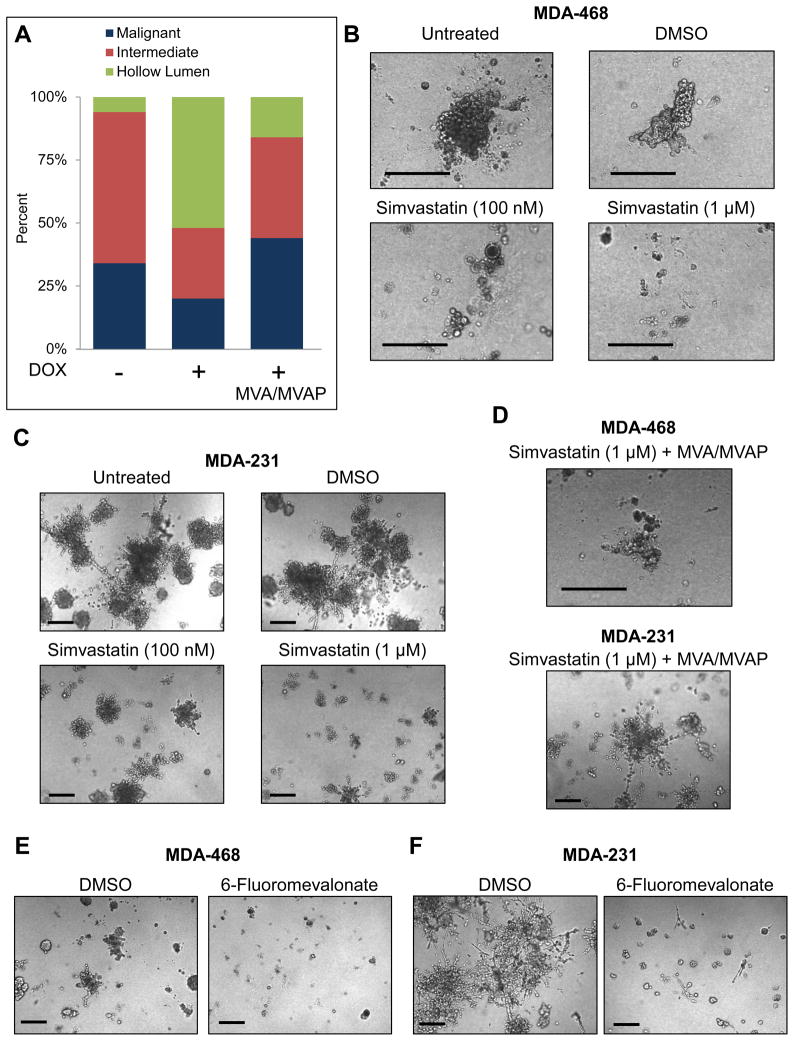
Figure 4. The mevalonate pathway is both necessary and sufficient to maintain the malignant state of breast cancer cells in 3D culture.
(A) MDA-468.shp53 cells were grown in 3D cultures for 8 days in the presence or absence of DOX to deplete mutant p53. Parallel cultures grown in the presence of DOX were supplemented with mevalonate pathway metabolites: mevalonic acid/mevalonic acid-phosphate (MVA/MVAP) beginning on Day 1. Morphological categories as indicated were determined using confocal microscopy and plotted as a percentage of the population. A representative experiment is shown here and a second representative experiment is shown in Figure S4.
(B) MDA-468 cells grown in 3D cultures for 13 days untreated or treated with DMSO, Simvastatin (100 nM) or (1 μM) as indicated. Representative DIC images are shown. Drugs were added on Day 4. Scale Bar, 200 μm.
(C) MDA-231 cells grown in 3D cultures and treated as in (B). Scale Bar, 200 μm.
(D) MDA-468 cells (top panel) or MDA-231 cells (bottom panel) were grown in 3D cultures for 13 days with Simvastatin (1 μM) as in (B) and (C), respectively, but were supplemented with MVA/MVAP. Scale Bar, 200 μm.
(E) MDA-468 cells were grown in 3D cultures for 8 days with DMSO or 6-Fluoromevalonate (200 μM), added on Day 1 of the experiment. Scale Bar, 200 μm.
(F) MDA-231 cells were grown as in (E). Scale Bar, 200 μm.
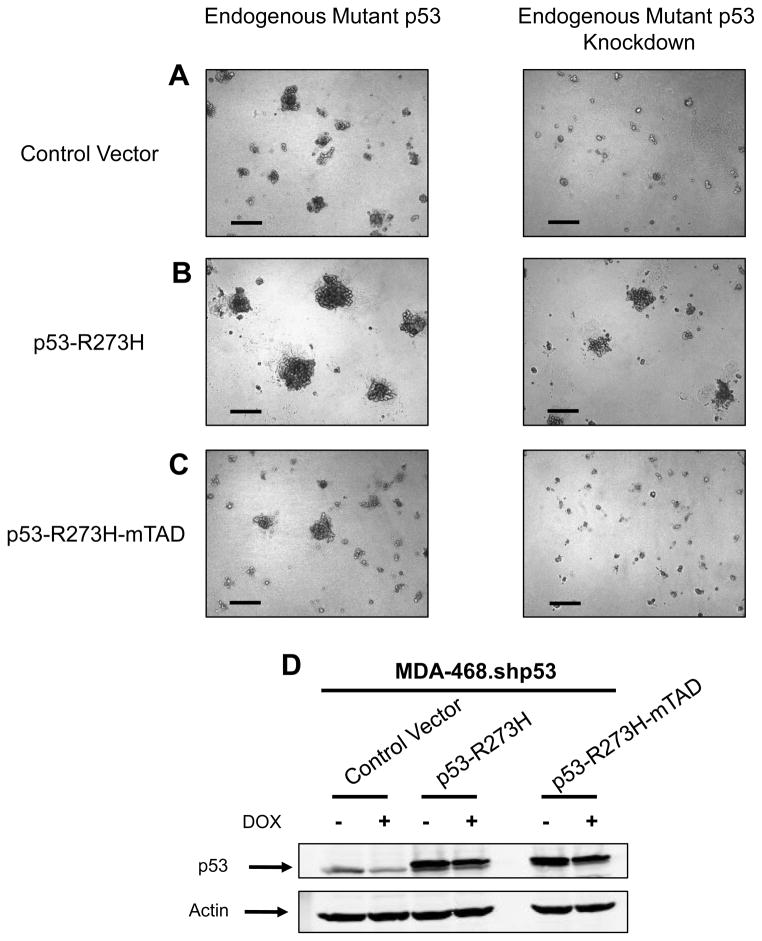
MDA-468.shp53 cells were then engineered to express an shRNA-resistant version of p53-R273H or a control vector (Figure 2A–B). Introducing this version of mutant p53 into MDA-468.shp53 cells prevented the phenotypic reversion that normally occurs after depleting cells of mutant p53 (Figure 2B) and the exogenous and endogenous p53 proteins together led to an even more highly disorganized and invasive phenotype than parental cells (compare left panels of Figure 2A and 2B).
Figure 2. Mutant p53 requires functional transactivation sub-domains to disrupt morphology of mammary cells in 3D culture.
(A) MDA-468.shp53 cells expressing a control vector were grown in 3D cultures for 5 days in the absence (left panel) or presence (right panel) of DOX. Representative DIC images are shown. Scale bar, 200 μm.
(B) MDA-468.shp53 cells expressing an shRNA-resistant Flag-tagged p53-R273H were grown in 3D cultures as in (A). Scale bar, 200 μm.
(C) MDA-468.shp53 cells expressing an shRNA-resistant Flag-tagged p53-R273H-mTAD (mutant p53 with non-functional transactivation region, p53-R273H-L22Q/W23S/W53Q/F54S) were grown in 3D cultures as in (A). Scale bar, 200 μm.
(D) Cell lines in (A–C) were grown in 3D cultures as in (A) and processed for immunoblotting for p53 (PAb240) or for actin. Flag-tagged mutant p53 variants migrate more slowly than endogenously expressed mutant p53.
Wild-type p53 primarily functions as a transcription factor and the transactivation domains of p53 have previously been implicated in oncogenic functions of mutant p53 (Lin et al., 1995; Matas et al., 2001; Yan and Chen, 2010). To interrogate the role of the transactivation domains in the effects of mutant p53 in 3D culture, we engineered MDA-468.shp53 cells to express an shRNA-resistant version of the endogenous mutant p53 that had been mutated at four key residues (L22Q/W23S/W53Q/F54S) shown previously to render its transactivation domains non-functional (Lin et al., 1994; Venot et al., 1999). As opposed to mutant p53 with a functional transactivation region (Figure 2B), the transactivation-dead version of mutant p53 failed to rescue the phenotypic reversion (Figure 2C), suggesting that the oncogenic effects in this system were due to transcriptional changes mediated by mutant p53.
To test whether the effects of mutant p53 on 3D morphology of breast cancer cells were generalizable between tumor-derived mutants of p53, we replaced the endogenous mutant p53 in MDA-231 cells (R280K) with an shRNA-resistant version of p53-R273H, the mutant that is endogenously expressed in MDA-468 cells. While control cells behaved like the cells with just the shRNA-targeting p53, expression of p53-R273H partially prevented the phenotypic changes of depleting endogenous p53-R280K (Figure S1F).
Non-malignant MCF10A mammary cells with wild-type p53 undergo a well-characterized progression of three-dimensional morphogenesis, which results in spherical acinus-like structures (Debnath et al., 2003; Petersen et al., 1992) (Figure S2A). We engineered the non-malignant human mammary epithelial cell line, MCF10A, to express Flag-tagged versions of the five most frequent p53 mutants found in breast tumors (p53-R175H, -R248Q, -R273H, -R248W, -G245S) (http://p53.free.fr). MCF10A cells infected with a control vector exhibited normal acinar morphogenesis. However, as recently reported (Zhang et al., 2011), expression of the four most frequent mutant p53 proteins led to an inhibition of luminal clearance, reminiscent of the filled lumen phenotype observed in ductal carcinoma in situ (DCIS), with R273H and R248W exhibiting the highest degree of luminal filling (Figure S2B–F). Further, transactivation-deficient versions (mTAD) of these same five p53 mutants were unable to block luminal clearance in MCF10A cells, with R273H and R248W displaying the highest dependence on their transactivation domains (Figure S2I–J). Thus, not only can depletion of mutant p53 from breast cancer cells lead to a phenotypic reversion in 3D culture, but also mutant p53 expression in non-malignant mammary epithelial cells is sufficient to disrupt their morphology in 3D culture and the transactivation domain of mutant p53 is required in both cases.
Tumor-derived mutants of p53 regulate the mevalonate pathway in breast cancer cells
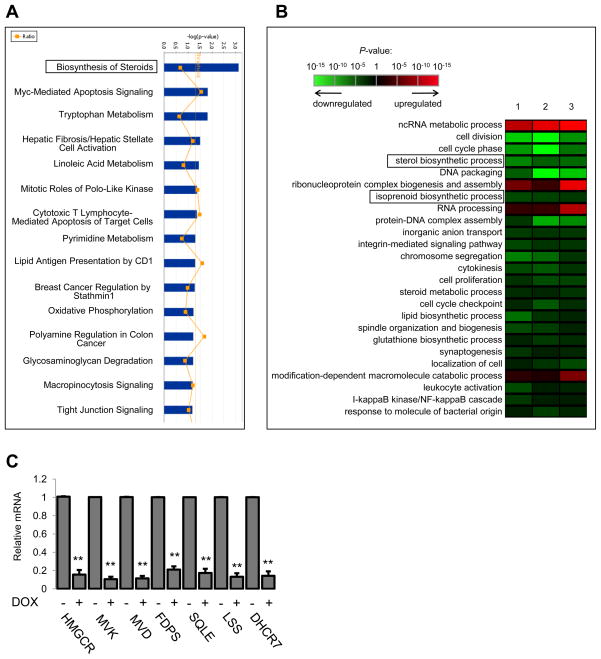
Based on the above findings we performed genome-wide expression profiling on MDA-468.shp53 cells grown in 3D culture, with or without full levels of mutant p53. We identified 989 genes as significantly altered (p<0.01) following shRNA-mediated downregulation of endogenous mutant p53, suggesting that mutant p53 acts promiscuously to affect many cellular processes. To guide our identification of those pathways/processes necessary for mutant p53 function in 3D culture, we employed two analysis methods, Ingenuity Pathway Analysis (IPA) and Gene Ontology (GO) Analysis. The mevalonate pathway was the most overrepresented pathway using IPA (labeled “Biosynthesis of Steroids” by Ingenuity); in fact, it was the only pathway detected with 99% confidence (p<0.01) following mutant p53 downregulation (Figure 3A). This pathway, along with the related isoprenoid biosynthetic process, was also detected using GO analysis as significantly downregulated upon mutant p53 ablation across three independent experiments (Figure 3B).
Figure 3. Knockdown of mutant p53 from breast cancer cells in 3D culture significantly downregulates the mevalonate pathway.
(A) Data were analyzed through the use of Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). Blue bars that cross the threshold line (p<0.05) represent pathways that are significantly changed following mutant p53 depletion from MDA-468 cells.
(B) Gene Ontology (GO) analysis. 1, 2, 3 represent three independent experiments. GO terms were sorted based on their significance and redundant terms were discarded.
(C) MDA-468.shp53 cells were grown in 3D cultures for 8 days in the presence or absence of DOX as indicated. qRT-PCR for the 7 sterol biosynthesis genes identified by IPA. Data represent mean ± SD of three independent experiments. **indicates p<0.005 by two-sided t-test.
The mevalonate pathway is responsible for de novo cholesterol synthesis as well as for many important non-sterol isoprenoid derivatives (Figure S3) (Goldstein and Brown, 1990). Of the many steps that convert acetyl-CoA to cholesterol, seven genes (HMGCR, MVK, MVD, FDPS, SQLE, LSS, DHCR7) encoding enzymes within the mevalonate pathway were found to be significantly reduced by mutant p53 depletion according to IPA. We confirmed in separate experiments that expression of all of these genes were markedly reduced (p<0.005) when mutant p53 was depleted by shRNA (Figure 3C). Using primers for intron containing transcripts we also showed that p53-mediated regulation of a subset of these genes in MDA-468 cells occurs as a result of RNA transcription (as opposed to mRNA stability or some later point of regulation) (data not shown).
The effects of mutant p53 on breast cancer morphology are mediated through the mevalonate pathway
Elevated or deregulated activity of the mevalonate pathway has been demonstrated in a number of different tumors, including breast cancer, and a number of studies have suggested that malignant cells are more highly dependent on the continuous availability of mevalonate pathway metabolites than their non-malignant counterparts (Buchwald, 1992; Larsson, 1996; Wong et al., 2002). While this pathway has been explored most extensively in the context of cholesterol production, which is necessary for membrane integrity and thus cell division, many of the intermediate metabolites and side products play key roles in other essential cellular processes. For example, farnesyl pyrophosphate (FPP) and geranylgeranyl pyrophosphate (GGPP) are critical for post-translational modifications of Ras and RhoA, respectively (Mo and Elson, 2004).
To test whether this pathway is necessary for the phenotypic effects of mutant p53, add-back experiments were performed in which breast cancer cells grown in 3D culture were depleted of mutant p53 and supplemented with intermediate metabolites produced by the mevalonate pathway. Addition of two upstream metabolites, mevalonic acid (MVA) and mevalonic acid phosphate (MVAP), dramatically inhibited the phenotypic reversion caused by mutant p53 knockdown in MDA-468 cells without affecting the amount of p53 depletion (Figure 4A and S4A–B). This confirms that activity of the mevalonate pathway is sufficient to compensate for the loss of mutant p53 and suggests that up-regulation of at least the initial steps of the mevalonate pathway is necessary for the effects of mutant p53 on tissue architecture.
HMG-CoA reductase, which catalyzes the formation of mevalonic acid, is the rate limiting step in cholesterol biosynthesis and is the target of numerous cholesterol reducing statins (Larsson, 1996). The use of statins is well established in the clinic to treat hypercholesterolemia and there have been multiple reports demonstrating that statins can exhibit anti-cancer activity; however, their anti-tumorigenic mechanism has not been firmly established.
We hypothesized that pharmacologic inhibition of the rate-limiting enzyme in the mevalonate pathway might be sufficient to mimic the effects of knocking down mutant p53. Strikingly, we found that treatment of breast cancer cells in 3D culture with Simvastatin, a lipophilic statin, used at clinically achievable concentrations (Wong et al., 2002), resulted in a reduction in growth in both cell lines, in addition to extensive cell death in MDA-468 cells (Figure 4B) and a significant reduction of the invasive morphology of MDA-231 cells (Figure 4C). In fact, in MDA-231 cells the morphological changes seen with either statin treatment or mutant p53 depletion were virtually the same. The consequence of inhibiting the mevalonate pathway in MDA-468 cells was even more dramatic than mutant p53 downregulation alone (cell death as opposed to formation of structures with a hollow lumen). On the other hand, inhibition of HMG-CoA reductase in wild-type p53 expressing MCF10A cells did not result in gross morphologic changes when used at clinically achievable concentrations (Figure S4C). This suggests that breast cancer cells bearing mutations in p53 upregulate the mevalonate pathway and eventually become dependent upon its activity for survival. Similar results were obtained with another lipophilic statin, Mevastatin (Figure S4D and E). Importantly, supplementation of MVA, the enzymatic product of HMG-CoA reductase, to either MDA-468 or MDA-231 cells treated with a statin blocked many of the phenotypic effects of statins (Compare Figure 4D to 4B or 4C). These results indicate that the effects of statins on breast cancer cells in 3D culture occur because of the function of HMG-CoA reductase to produce mevalonic acid, and further implicate the upregulated mevalonate pathway in the malignant 3D phenotype of these cells. In addition, we tested whether flux through the mevalonate pathway was sufficient to disrupt normal acinar morphogenesis and, similar to overexpression of tumor-derived p53 mutants, exogenous MVA was sufficient to block luminal clearance in MCF10A cells (Figure S4F).
We extended the effects of statins on breast cancer cells in three other assays. First, we observed that Simvastatin can significantly impair anchorage-independent growth in both MDA-468 and MDA-231 cells (Figure S5A). Second, in line with findings that HMG-CoA reductase inhibitors induce cell cycle arrest and/or apoptosis in 2D culture (Graaf et al., 2004), we noted a G1 cell cycle arrest, with a concomitant drop in S phase, in both breast cancer cell lines treated with 24 hours of Simvastatin at varying concentrations (Figure S5B–C). The phenotypic effects of statins in 3D culture are therefore likely due to a combination of factors (i.e. decreased growth, increased death and decreased invasion). Third, consistent with previous reports (Ghosh-Choudhury et al., 2010; Mori et al., 2009) using a xenograft model of MDA-231 breast cancer cells, Simvastatin significantly impaired tumor growth of these cells when implanted into immunocompromised mice (Figure S5D).
Geranylgeranylation is required for the phenotypic effects of mutant p53 in MDA-231 breast cancer cells grown in 3D culture
Next we tested whether inhibition of later enzymes within the mevalonate pathway would have similar phenotypic effects as mutant p53 depletion from breast cancer cells grown in 3D culture. An inhibitor of mevalonate decarboxylase, 6-Fluoromevalonate, had remarkably similar phenotypic effects on both MDA-468 and MDA-231 cells grown in 3D culture to that seen with statin treatment (Figure 4E and 4F). Thus, not only HMG-CoA reductase, but several downstream enzymatic steps in the mevalonate pathway are involved in the ability of mutant p53 to maintain malignant behavior of breast cancer cells in 3D culture conditions.
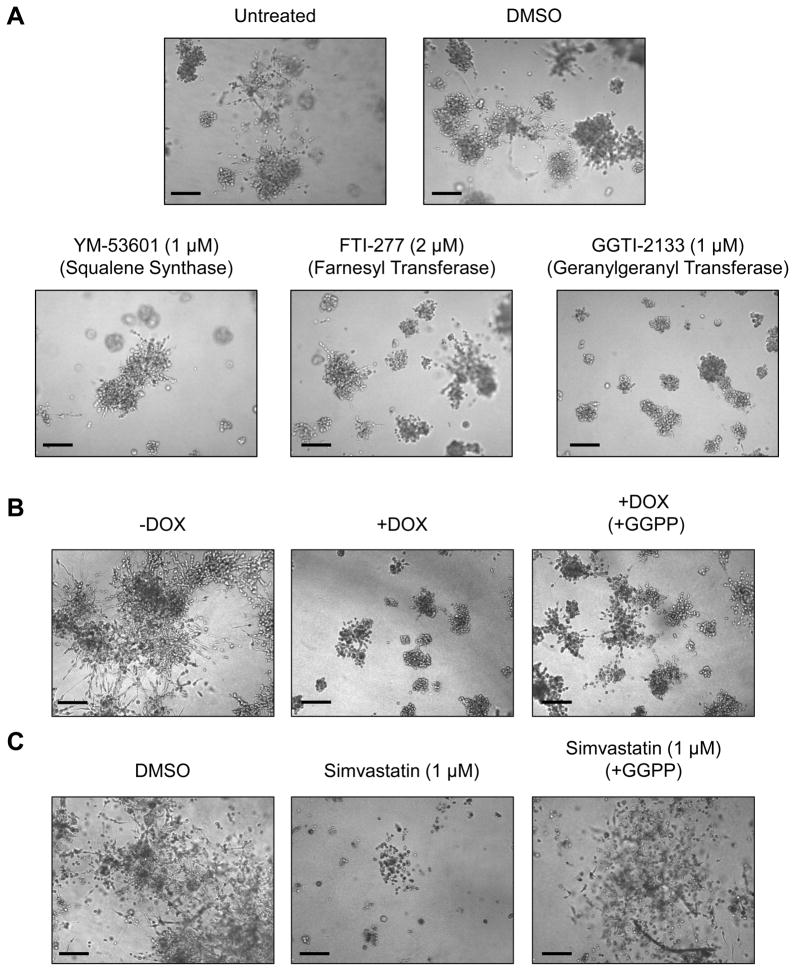
Because the mevalonate pathway is not only vital for producing cellular cholesterol, but also many other biologically active metabolites, we examined whether the phenotypic effects of mutant p53 knockdown were due to decreased cholesterol synthesis or the production of an earlier metabolite. To do this, we utilized three inhibitors that inhibit distinct actions of the mevalonate pathway (Figure S3). YM-53601 inhibits squalene synthase (and thus cholesterol production) at submicromolar concentrations (Ugawa et al., 2000), but spares all upstream intermediate metabolites. FTI-277 blocks farnesylation of proteins via inhibition of farnesyl transferase at nanomolar concentrations in whole cells, but has no effect geranylgeranyl transferase or squalene synthesis at low micromolar concentrations (Lerner et al., 1995). GGTI-2133 blocks geranylgeranylation of target proteins via inhibition of geranylgeranyl transferase, while sparing farnesylation and squalene synthesis (Vasudevan et al., 1999).
At low micromolar concentrations, inhibition of squalene synthase and farnesyl transferase had only a mild effect on the growth of MDA-231 cells in 3D culture, while inhibition of geranylgeranylation had a profound impact on both the growth and the invasive morphology of these cells in 3D culture (Figure 5A). To examine whether downregulation of geranylgeranylation is necessary for the phenotypic effects observed after mutant p53 depletion or HMG-CoA reductase inhibition, we performed add-back experiments using geranylgeranyl pyrophosphate (GGPP) to cells either depleted of mutant p53 or cells treated with Simvastatin (Figure 5B and C, respectively). Since supplementation with GGPP was sufficient to rescue the invasive phenotype in a portion of the population of MDA-231 cells in 3D culture, this suggests that geranylgeranylation is indeed a vital component of why mutant p53 upregulates the mevalonate pathway in cancer cells.
Figure 5. Modulation of geranylgeranylation mediates many of the phenotypic effects of mutant p53 depletion and HMG-CoA reductase inhibition in MDA-231 cells.
(A) MDA-231 cells were grown in 3D cultures for 8 days untreated or treated with DMSO, or inhibitors: YM-53601 (1 μM), FTI-277 (1 μM) or GGTI-2133 (1 μM) as indicated. Drugs were added on Day 1. Scale Bar, 200 μm.
(B) MDA-231.shp53 cells were grown in 3D culture conditions for 9 days in the absence or presence of DOX as indicated. Parallel wells of cells which were grown in the presence of DOX were supplemented with geranylgeranyl pyrophosphate (GGPP) (25 μM) beginning on Day 1. Scale Bar, 200 μm.
(C) MDA-231 cells were grown in 3D cultures for 13 days either treated with DMSO or Simvastatin (1 μM) as indicated. Parallel wells of cells which were grown in the presence of Simvastatin (1 μM) were supplemented with GGPP (25 μM) beginning on Day 1. Scale Bar, 200 μm.
Mutant p53 is recruited to the promoters of genes encoding mevalonate pathway enzymes
Much of the physiologic regulation of the sterol biosynthesis enzymes takes place at the transcriptional level in a manner that requires the sterol regulatory element binding proteins, SREBP-1 and SREBP-2 (Sato, 2010). SREBPs activate the genes encoding nearly every key enzyme in both the fatty acid and sterol biosynthetic pathways (Horton et al., 2002). Transcriptional regulation by mutant p53 may occur in many cases through interactions with other sequence-specific transcription factors binding to their cognate sites (Brosh and Rotter, 2009). We hypothesized that mutant p53 might serve as a co-activator with one or more of the SREBPs. The following experiments support this hypothesis.
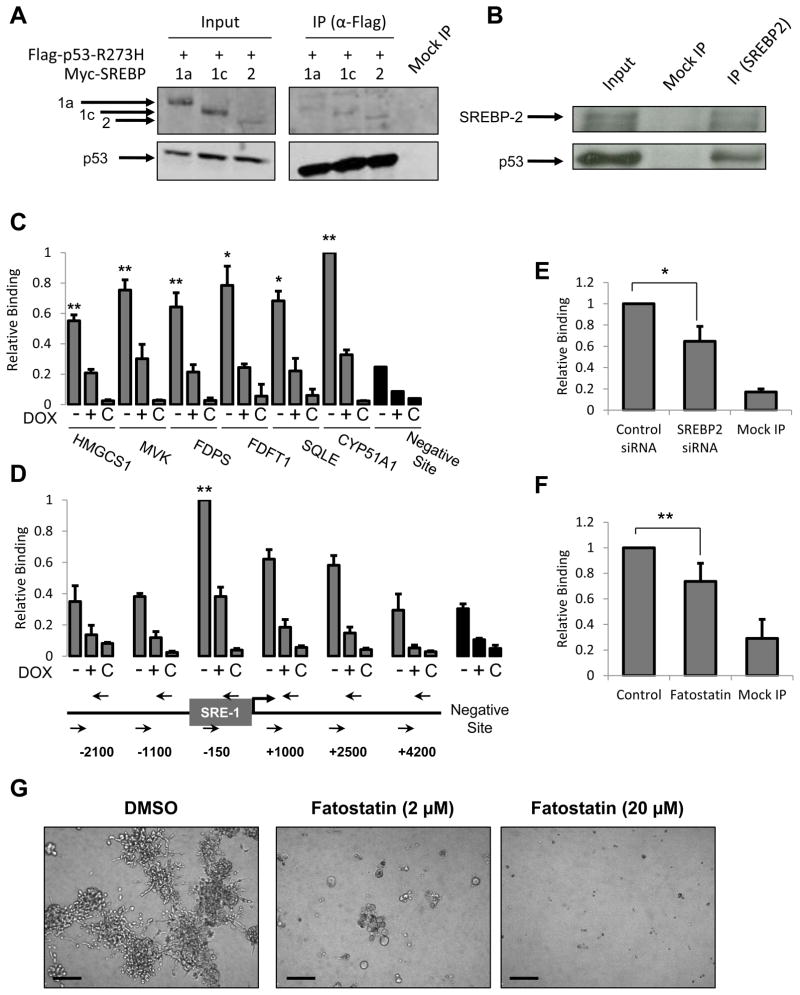
First, co-transiently expressed mutant p53 (p53-R273H) can interact with the mature forms of all three family members, SREBP-1a, SREBP-1c and SREBP-2 (Figure 6A) and endogenous mutant p53 can be co-immunoprecipitated with endogenous mature SREBP-2 in MDA-468 cells (Figure 6B) and MDA-231 cells (Figure S6A).
Figure 6. Mutant p53 is recruited to mevalonate pathway gene promoters and this recruitment is dependent on SREBP proteins.
(A) 293 cells were transfected with Flag-p53-R273H and either Myc-mSREBP-1a, -1c or -2. Cells were subjected to crosslinking with formaldehyde prior to lysis, sonication and immunoprecipitation as described in experimental procedures followed by SDS-PAGE and immunoblotting with anti-Myc (upper panel) and anti-Flag (lower panel). Input is 2.5% of IP sample.
(B) Nuclear lysates from serum-starved MDA-468.shp53 cells were immunoprecipitated with an anti-SREBP-2 antibody (1D2) or Mouse IgG (Mock IP) and then immunoblotted with anti-SREBP-2 (1D2) and anti-p53 antibodies (DO-1). Input is 10% of IP sample.
(C) MDA-468.shp53 cells were grown in 2D cultures for 8 days in the absence or presence of DOX and subjected to ChIP analysis as described in experimental procedures. Mock IP (C) serves as a negative control. Data is presented as mean ± SD of three independent experiments. Values were normalized to the highest immunoprecipitation signal. **indicates p<0.01 or *indicates p<0.05 compared to all of the following: negative site, +DOX and Mock IP.
(D) MDA-468.shp53 cells were grown and analyzed as in (C) to examine mutant p53 association with the HMGCR promoter. **indicates p<0.01 compared to all of the following: negative site, +DOX, Mock IP, upstream and downstream sites. Genomic locations of PCR primers are illustrated in the HMGCR promoter. SRE-1 denotes sterol regulatory element.
(E) MDA-468 cells were treated with siRNA targeting SREBP-2 and subjected to ChIP analysis as in (C) for mutant p53 recruitment to the vicinity of the SRE-1 site in the HMGCR promoter (−150 bp). Data presented as mean ± SD of three independent experiments. *designates p<0.05. See Figure S5C for extent of SREBP-2 knockdown.
(F) MDA-468.shp53 cells were treated with Fatostatin (20 μM) and subjected to ChIP analysis as in (E). Data is presented as mean ± SD of six independent experiments. **designates p<0.01.
(G) MDA-231.shp53 cells were grown in 3D cultures for 8 days treated with DMSO, Fatostatin (2 μM) or (20 μM). Drugs were added on Day 1. Representative DIC images are shown. Scale Bar, 200 μm.
Second, in addition to the seven sterol biosynthesis genes uncovered by our initial pathway analysis, we validated ten additional SREBP-regulated sterol biosynthesis genes that are regulated by mutant p53 in MDA-468 cells (Figure S6C), a subset of which are also regulated in MDA-231 cells (Figure S6D). Of note, DHCR24 was previously reported to be a transactivation target of another tumor-derived mutant, p53-R175H (Bossi et al., 2008). SREBP target genes that are not part of the mevalonate pathway are also regulated by mutant p53 in MDA-468 cells (Figure S6E). Three such genes (FASN, ELOVL6 and SCD) encode key enzymes within the fatty acid biosynthesis pathway, suggesting that this pathway may also be upregulated by mutant p53. Further, when a comprehensive list of SREBP target genes was queried (Reed et al., 2008) there was a marked enrichment of SREBP target genes in the set of genes which were affected after mutant p53 depletion from breast cancer cells (Figure S6B).
Third, mutant p53 is recruited to the promoter regions of genes encoding sterol biosynthesis enzymes. Using quantitative chromatin immunoprecipitation (ChIP) analysis of MDA-468 cells with or without full levels of mutant p53, we identified significant binding by mutant p53 in the vicinity of sterol regulatory elements (SRE-1), the cognate binding sites for the SREBPs, in the promoter regions of all seven genes tested (Figure 6C and 6D). This ChIP signal varied between 2- to 4-fold greater than the signal at a negative region within the CDKN1A gene locus, and was consistently reduced upon depletion of p53 (Figure 6C). ChIP analysis scanning regions upstream and downstream of the transcriptional start site (TSS) of the gene encoding HMG-CoA reductase (HMGCR) identified a region of peak binding by mutant p53 approximately 150 bp upstream of the TSS (Figure 6D), which corresponds to the known sterol regulatory element. The peak signal, which was on the order of 3-fold greater than the negative region, was again significantly reduced in cells with depleted p53 (Figure 6D).
Fourth, SREBPs are likely necessary for the full recruitment of mutant p53 to these gene promoters. Depletion of SREBP-2 substantially decreased the recruitment of mutant p53 to the HMG-CoA reductase gene promoter (Figure 6E), and depletion of SREBP-1 slightly, but significantly, reduced the level of binding to the same promoter (Figure S6).
Fifth, Fatostatin, a recently described inhibitor of SREBP activation (Kamisuki et al., 2009), significantly reduced the level of mutant p53 binding to the HMG-CoA reductase gene promoter (Figure 6F). Further, Fatostatin treatment had a dramatic effect on the 3D morphology of MDA-231 cells (Figure 6G) and MDA-468 cells (data not shown). Taken together, our results strongly implicate a functional interaction with SREBPs as being critical for mutant p53-mediated upregulation of the mevalonate pathway genes.
TP53 mutation correlates with elevated expression of mevalonate pathway genes in human breast cancer patients
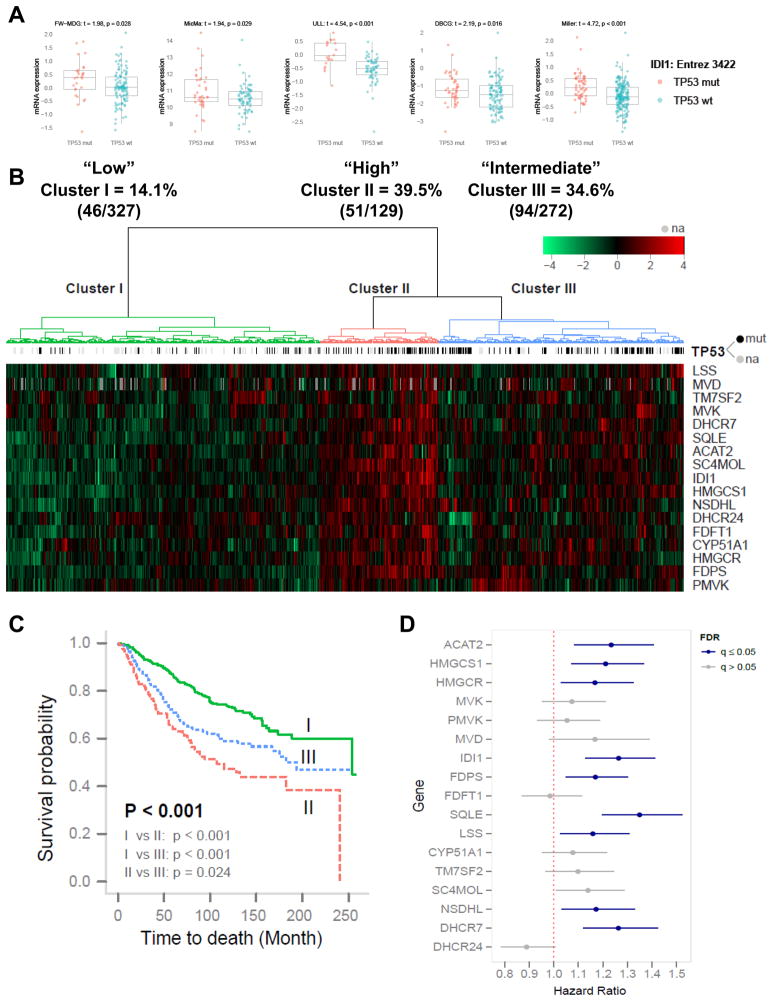
To investigate whether the regulation of the mevalonate pathway by mutant p53 is generalizable to human patients, we examined five datasets consisting of a total of 812 human breast cancer patient samples with expression data (728 had known TP53 mutational status). After stratifying patients based on the p53 mutational status of their tumors, we investigated the expression level of 17 mevalonate pathway genes. Remarkably, 11 of these exhibited significantly higher expression levels in mutant p53 breast tumors compared to those bearing wild-type p53 across multiple datasets (Figure 7A, S7 and Table S1).
Figure 7. Mutant p53 is correlated with higher expression of a subset of mevalonate pathway genes in human breast cancer patient datasets.
(A) Five human breast cancer patient datasets (see Supplemental Information for details) were analyzed to determine whether tumors bearing mutant p53 correlate with higher expression of sterol biosynthesis genes. Patients were stratified based on TP53 status (wild-type vs. mutant) and expression levels for sterol biosynthesis genes were analyzed. Isopentenyl Pyrophosphate Isomerase (IDI1), exhibited higher expression levels in mutant p53 tumors compared to wild-type p53 tumors (p<0.05) across all five datasets. p-value represents the result of a one-sided t-test. See Figure S7 and Table S1 for other genes.
(B) Unsupervised hierarchical clustering with Euclidean distance and ward linkage of expression matrix from the 17 sterol biosynthesis genes on 812 samples (728 of which have TP53 mutational status). MVD was not present in the DBCG dataset and its missing expression values were grayed out on the heatmap. Rows indicate the identity of the genes and columns indicate the identity of the patients. The TP53 mutational status for each tumor is depicted directly above each column. Cluster I exhibits the lowest expression of the mevalonate pathway genes, cluster III exhibits an intermediate expression level and cluster II exhibits the highest expression level of the mevalonate pathway genes.
(C) The Kaplan-Meier curves for the resulting clusters from the unsupervised hierarchical clustering in (B).
(D) Estimated hazard ratios (HRs; the relative risk for 1 unit increasing in the gene expression) with 95% confidence interval for risk of breast cancer specific death. Expression levels of following genes were positively associated with the risk of breast cancer specific death at False Discovery Rate (FDR) (q) 5%: ACAT2 (HR = 1.23, q = 0.0069), HMGCS1 (HR = 1.21, q = 0.007), HMGCR (HR = 1.17, q = 0.032), IDI1 (HR = 1.26, q < 0.001), FDPS (HR = 1.17, q = 0.012), SQLE (HR = 1.35, q < 0.001), LSS (HR = 1.16, q = 0.032), NSDHL (HR = 1.17, q = 0.032), DHCR7 (HR = 1.26, q < 0.001). Blue indicates q<0.05, grey indicates q>0.05.
We also performed a reciprocal analysis on these same patient datasets, stratifying tumors based on the expression of the mevalonate pathway genes and examining the mutation rate of TP53. Three main groups were observed from the hierarchical clustering of the expression matrix from 17 sterol biosynthesis genes on the 812 human breast cancer patient samples. Cluster I has the lowest sterol biosynthesis gene expression pattern and the lowest rate of TP53 mutations (14.1%). Cluster III exhibits an intermediate expression level and an intermediate rate of TP53 mutations (34.6%). Cluster II has the highest expression pattern and exhibits the highest rate of TP53 mutations (39.5%) (Figure 7B).
To test the biological significance of elevation of the mevalonate pathway in mutant p53 tumors, we examined whether upregulation of this pathway correlated with patient prognosis. It is striking that cluster I, which has the lowest expression level of the mevalonate pathway genes, is correlated with a more favorable prognosis, while cluster III, which has an intermediate expression pattern, correlates with an intermediate prognosis and cluster II, which has the highest expression of the mevalonate pathway genes, is associated with a significantly poorer survival probability (Figure 7C). Therefore, not only was elevation of the mevalonate pathway significantly correlated with a higher rate of p53 mutations, these breast cancer patients also had a significantly decreased survival. We then examined each sterol biosynthesis gene individually to investigate which genes contribute most to the prognostic value. Elevated expression of nine mevalonate pathway genes correlated with significantly poorer prognosis in these breast cancer patients (Figure 7D).
Since breast cancer cells bearing mutant p53 appear to be particularly sensitive to inhibition of the mevalonate pathway in the 3D culture system, the fact that multiple members of this pathway are upregulated in mutant p53 expressing human tumors and correlate with a poor prognosis may have important therapeutic implications.
DISCUSSION
Despite being one of the most well studied tumor suppressors, there is much evidence that once mutated, p53 can actively promote the progression of many cancers. With respect to breast cancer, tumor-derived mutants of p53 have been implicated in survival, chemoresistance, invasion, migration and metastasis (Brosh and Rotter, 2009). Since mammary tissue architecture is invariably disrupted during breast carcinogenesis, we sought to delineate the phenotypic effects of mutant p53 in breast cancer. This study describes a possible oncogenic role for certain missense mutants of p53 in disrupting acinar morphogenesis of breast cells, explored using a 3D culture system. In addition, we show that mutant p53 elevates expression of many mevalonate pathway genes and flux through the mevalonate pathway is both necessary and sufficient for the phenotypic effects of mutant p53 on breast cancer cell morphology in 3D culture.
The mevalonate pathway has recently been implicated in multiple aspects of tumorigenesis, including proliferation, survival, invasion and metastasis (Clendening et al., 2010; Kidera et al., 2010; Koyuturk et al., 2007). Competitive inhibitors of the rate-limiting enzyme in the mevalonate pathway, HMG-CoA reductase, collectively known as statins, have been reported to be cancer-protective for certain malignancies, including breast cancer (Ahern et al., 2011; Blais et al., 2000; Cauley et al., 2003). Yet the cancer-protective effects of statins are not without debate (Baigent et al., 2005; Browning and Martin, 2007). Statins have already been employed in multiple preclinical models of breast cancer (Kubatka et al., 2011; Shibata et al., 2004). In line with this, we were also able to confirm a significant impact of Simvastatin treatment on MDA-231 breast cancer cells in vivo.
It is interesting to note that at least one clinical study investigating the effect of statins in breast cancer noted a subgroup-specific protective effect: specifically, a significantly decreased incidence of hormone receptor-negative (ER−/PR−) tumors was documented in patients takings statins, while no such effect was observed for hormone receptor-positive tumors (Kumar et al., 2008). Preclinical models, employing either breast cancer cell lines or mouse models of breast cancer, also support a more dramatic role for statins in ER−/PR− breast cancers (Campbell et al., 2006; Garwood et al., 2010). Since the majority of breast tumors that bear p53 mutations most commonly are also immunohistochemically classified as ER−/PR− (Sorlie et al., 2001), it is tempting to speculate that the observed anti-tumorigenic effects of statins are a consequence of mutant p53’s upregulation of the mevalonate pathway.
Gene expression profiling of breast cancers has identified specific subtypes with important clinical, biologic and therapeutic implications (Perou et al., 2000). Using these expression signatures, most p53 mutations cluster in the basal-like subgroup of breast cancers, which has the poorest prognosis and is notoriously difficult to treat (Sorlie et al., 2001). Fascinatingly, using a combination of expression signatures and data from over 40,000 compounds screened in the NCI-60 cell lines, three FDA-approved drugs were predicted to be most effective for treating basal-like breast cancers, two of which, Simvastatin and Lovastatin, are inhibitors of HMG-CoA reductase (Mori et al., 2009). It will be exciting to examine whether stratifying breast cancer patients based on their p53 mutational status can resolve the apparent discrepancies within the rich body of literature linking statins and cancer.
Although we have implicated the mevalonate pathway in the phenotypic effects of mutant p53, it will be of great interest to further delineate the metabolite(s) as well as the downstream signaling pathways that are responsible for these phenotypic effects. While we have demonstrated that metabolic flux through the mevalonate pathway is necessary to maintain the malignant state, with a specific reliance on geranylgeranylation, we cannot rule out the possibility that one or more other metabolites are involved in the phenotypic effects that we observe in 3D culture. If geranylgeranyl pyrophosphate is in fact the key metabolite, it will be very interesting to delineate the geranylgeranylated protein target(s) that mediate the oncogenic effects of the mevalonate pathway in breast cancer cells in 3D culture.
It is interesting to note that, in addition to the mevalonate pathway, a number of fatty acid biosynthesis genes were also significantly affected by mutant p53 depletion from breast cancer cells in 3D culture (Figure 3B and S6E). Intriguingly, this is the other major pathway controlled by the SREBP family of transcription factors (Horton et al., 2002). While much of our data points to a role for SREBP proteins in the regulation of the mevalonate pathway by mutant p53, a direct link is yet to be established. This regulation is likely to occur through one or more of the SREBP proteins, but we cannot rule out the possibility that another factor or factors may also be involved. Mutant p53 may interact directly with elements in the promoters of the sterol biosynthesis genes or alternatively be recruited by a known mutant p53 interacting partner such as NF-Y, SP1, Ets-1 or VDR, which have been shown to recruit mutant p53 to their cognate binding sites (Brosh and Rotter, 2009; Stambolsky et al., 2010).
A number of scenarios have been proposed to explain why human tumors select for mutations in p53. First, mutant p53 may simply be selected for due to loss of wild-type p53 tumor suppressive activity. Second, mutant p53 may acquire neomorphic (i.e. novel gain-of-function) activities which promote tumor growth, many of which have actually been shown to be diametrically opposed to those performed by wild-type p53 (Peart and Prives, 2006; Stambolsky et al., 2010). In line with this hypothesis are the findings that Stearoyl-CoA desaturase (encoded by SCD) is a repression target of wild-type p53 (Mirza et al., 2003) and that wild-type p53 can suppress a subset of SREBP target genes in a mouse model of obesity (Yahagi et al., 2003). As the pro-survival roles of wild-type p53 are becoming more apparent (Kim et al., 2009), a third scenario can be envisaged in which mutant p53 may retain and exaggerate certain wild-type p53 functions, while selectively losing certain tumor suppressive mechanisms such as the ability to induce cell cycle arrest and apoptosis. Regarding the latter two hypotheses, it will be interesting for future studies to examine whether wild-type p53 and/or its family members (p63 and p73) serve to repress sterol biosynthesis genes. Alternatively, the maintenance of high levels of sterol biosynthesis genes by mutant p53 may be a remnant of an unrecognized wild-type p53 function.
These speculations raise another important consideration, that not all p53 mutations are equivalent. Genetic alterations in p53 are often grouped into two classes based on the type of mutant p53 that they produce. Contact mutants, exemplified by p53-R273H, involve mutation of residues that are directly involved in protein-DNA contacts. Conformational mutants, typified by p53-R175H, result in structural distortions in the p53 protein. Our findings that a subset of the sterol biosynthesis genes are significantly higher in large cohorts of human breast tumors bearing mutant p53 suggests that the ability of mutant p53 to upregulate the sterol biosynthesis genes is not constrained to a single class of mutations; however, it will be very interesting for follow-up studies to examine which tumor-derived mutants of p53 can regulate the levels of sterol biosynthesis genes.
In summary, our results demonstrate that mutant p53 can disrupt mammary acinar morphology and that downregulation of mutant p53 from malignant cells is sufficient to phenotypically revert these cells. Here we propose one mechanism, the upregulation of the mevalonate pathway, although one or more additional pathways may play a role. Specifically, we demonstrate that mutant p53 is recruited to the promoters of many sterol biosynthesis genes leading to their upregulation. We hypothesize that tumors bearing p53 mutations evolve to become highly reliant on metabolic flux through the mevalonate pathway, making them particularly sensitive to inhibition of this pathway. At a clinical level, inhibition of the mevalonate pathway, either alone or in combination with other therapies, may offer a novel, and much needed, therapeutic option for tumors bearing mutant p53.
EXPERIMENTAL PROCEDURES
Three Dimensional (3D) Culture
Three-dimensional culture was carried out as previously described (Debnath et al., 2003). Briefly, 8-well chamber slides were lined with growth factor reduced Matrigel and cells were seeded in Assay Medium containing 2% Matrigel.
Immunostaining and Microscopy
Cells were fixed using 2% formaldehyde. Confocal microscopy was conducted using an Olympus IX81 confocal microscope and analyzed using Fluoview software.
Microarray and Data analysis
MDA-468.shp53 cells cultured under 3D conditions were assayed for genome-wide expression changes by Affymetrix GeneChip array, analyzed with Ingenuity Pathway Analysis and Gene Ontology (GO) analysis.
Quantitative RT-PCR
As described in (Barsotti and Prives, 2009). Primer sequences are provided in Table S2.
Drug Treatments
Cells in 3D culture were treated on Day 1 or Day 4 of the 3D protocol (as described in the figure legends) and re-fed every 4 days with fresh drug.
Add-back experiments
On Day 1 of 3D culture cells cultured in the presence of doxycycline or Simvastatin were supplemented with MVA/MVAP or GGPP and re-fed every 4 days.
Quantitative Chromatin Immunoprecipitation
As previously described (Beckerman et al., 2009). Primer sequences are provided in Table S2.
Patient Data
Expression data for the sterol biosynthesis genes were extracted from individual cohorts (FW-MDG, MicMa, Ull, DBCG and Miller). Expression values of the 17 sterol biosynthesis genes on 812 samples were clustered using hierarchical clustering with Euclidean distance and ward linkage. The Kaplan-Meier survival curves were plotted for the resulting groups and the differences in clinical indications among the clusters were tested by a logrank test.
Supplementary Material
Acknowledgments
We would like to thank Prives laboratory members, Masha Poyurovsky in particular, for helpful discussions. Ella Freulich provided invaluable technical assistance. We would like to thank Carlos Cordon Cardo, Josep Maria Domingo-Domenech and Dennis Bonal for advice and help with xenograft models. This work was supported by CA87497, CA77742 and The Breast Cancer Research Foundation.
Footnotes
Accession Number
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, Solari A, Bobisse S, Rondina MB, Guzzardo V, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- Ahern TP, Pedersen L, Tarp M, Cronin-Fenton DP, Garne JP, Silliman RA, Sorensen HT, Lash TL. Statin Prescriptions and Breast Cancer Recurrence Risk: A Danish Nationwide Prospective Cohort Study. J Natl Cancer Inst. 2011 doi: 10.1093/jnci/djr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baigent C, Keech A, Kearney PM, Blackwell L, Buck G, Pollicino C, Kirby A, Sourjina T, Peto R, Collins R, et al. Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet. 2005;366:1267–1278. doi: 10.1016/S0140-6736(05)67394-1. [DOI] [PubMed] [Google Scholar]
- Barsotti AM, Prives C. Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene. 2009;28:4295–4305. doi: 10.1038/onc.2009.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckerman R, Donner AJ, Mattia M, Peart MJ, Manley JL, Espinosa JM, Prives C. A role for Chk1 in blocking transcriptional elongation of p21 RNA during the S-phase checkpoint. Genes Dev. 2009;23:1364–1377. doi: 10.1101/gad.1795709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Kenny PA, Radisky DC. Microenvironmental regulators of tissue structure and function also regulate tumor induction and progression: the role of extracellular matrix and its degrading enzymes. Cold Spring Harb Symp Quant Biol. 2005;70:343–356. doi: 10.1101/sqb.2005.70.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Radisky DC, Rizki A, Weaver VM, Petersen OW. The organizing principle: microenvironmental influences in the normal and malignant breast. Differentiation. 2002;70:537–546. doi: 10.1046/j.1432-0436.2002.700907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais L, Desgagne A, LeLorier J. 3-Hydroxy-3-methylglutaryl coenzyme A reductase inhibitors and the risk of cancer: a nested case-control study. Arch Intern Med. 2000;160:2363–2368. doi: 10.1001/archinte.160.15.2363. [DOI] [PubMed] [Google Scholar]
- Bossi G, Marampon F, Maor-Aloni R, Zani B, Rotter V, Oren M, Strano S, Blandino G, Sacchi A. Conditional RNA interference in vivo to study mutant p53 oncogenic gain of function on tumor malignancy. Cell Cycle. 2008;7:1870–1879. doi: 10.4161/cc.7.12.6161. [DOI] [PubMed] [Google Scholar]
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- Browning DR, Martin RM. Statins and risk of cancer: a systematic review and metaanalysis. Int J Cancer. 2007;120:833–843. doi: 10.1002/ijc.22366. [DOI] [PubMed] [Google Scholar]
- Buchwald H. Cholesterol inhibition, cancer, and chemotherapy. Lancet. 1992;339:1154–1156. doi: 10.1016/0140-6736(92)90744-n. [DOI] [PubMed] [Google Scholar]
- Campbell MJ, Esserman LJ, Zhou Y, Shoemaker M, Lobo M, Borman E, Baehner F, Kumar AS, Adduci K, Marx C, et al. Breast cancer growth prevention by statins. Cancer Res. 2006;66:8707–8714. doi: 10.1158/0008-5472.CAN-05-4061. [DOI] [PubMed] [Google Scholar]
- Cauley JA, Zmuda JM, Lui LY, Hillier TA, Ness RB, Stone KL, Cummings SR, Bauer DC. Lipid-lowering drug use and breast cancer in older women: a prospective study. J Womens Health (Larchmt) 2003;12:749–756. doi: 10.1089/154099903322447710. [DOI] [PubMed] [Google Scholar]
- Clendening JW, Pandyra A, Boutros PC, El Ghamrasni S, Khosravi F, Trentin GA, Martirosyan A, Hakem A, Hakem R, Jurisica I, et al. Dysregulation of the mevalonate pathway promotes transformation. Proc Natl Acad Sci U S A. 2010;107:15051–15056. doi: 10.1073/pnas.0910258107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Garwood ER, Kumar AS, Baehner FL, Moore DH, Au A, Hylton N, Flowers CI, Garber J, Lesnikoski BA, Hwang ES, et al. Fluvastatin reduces proliferation and increases apoptosis in women with high grade breast cancer. Breast Cancer Res Treat. 2010;119:137–144. doi: 10.1007/s10549-009-0507-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh-Choudhury N, Mandal CC, Ghosh Choudhury G. Simvastatin induces derepression of PTEN expression via NFkappaB to inhibit breast cancer cell growth. Cell Signal. 2010;22:749–758. doi: 10.1016/j.cellsig.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Graaf MR, Richel DJ, van Noorden CJ, Guchelaar HJ. Effects of statins and farnesyltransferase inhibitors on the development and progression of cancer. Cancer Treat Rev. 2004;30:609–641. doi: 10.1016/j.ctrv.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisuki S, Mao Q, Abu-Elheiga L, Gu Z, Kugimiya A, Kwon Y, Shinohara T, Kawazoe Y, Sato S, Asakura K, et al. A small molecule that blocks fat synthesis by inhibiting the activation of SREBP. Chem Biol. 2009;16:882–892. doi: 10.1016/j.chembiol.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidera Y, Tsubaki M, Yamazoe Y, Shoji K, Nakamura H, Ogaki M, Satou T, Itoh T, Isozaki M, Kaneko J, et al. Reduction of lung metastasis, cell invasion, and adhesion in mouse melanoma by statin-induced blockade of the Rho/Rho-associated coiled-coil-containing protein kinase pathway. J Exp Clin Cancer Res. 2010;29:127. doi: 10.1186/1756-9966-29-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Giese A, Deppert W. Wild-type p53 in cancer cells: when a guardian turns into a blackguard. Biochem Pharmacol. 2009;77:11–20. doi: 10.1016/j.bcp.2008.08.030. [DOI] [PubMed] [Google Scholar]
- Koyuturk M, Ersoz M, Altiok N. Simvastatin induces apoptosis in human breast cancer cells: p53 and estrogen receptor independent pathway requiring signalling through JNK. Cancer Lett. 2007;250:220–228. doi: 10.1016/j.canlet.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Kubatka P, Zihlavnikova K, Kajo K, Pec M, Stollarova N, Bojkova B, Kassayova M, Orendas P. Antineoplastic effects of simvastatin in experimental breast cancer. Klin Onkol. 2011;24:41–45. [PubMed] [Google Scholar]
- Kumar AS, Benz CC, Shim V, Minami CA, Moore DH, Esserman LJ. Estrogen receptor-negative breast cancer is less likely to arise among lipophilic statin users. Cancer Epidemiol Biomarkers Prev. 2008;17:1028–1033. doi: 10.1158/1055-9965.EPI-07-0726. [DOI] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- Larsson O. HMG-CoA reductase inhibitors: role in normal and malignant cells. Crit Rev Oncol Hematol. 1996;22:197–212. doi: 10.1016/1040-8428(96)00193-x. [DOI] [PubMed] [Google Scholar]
- Lerner EC, Qian Y, Blaskovich MA, Fossum RD, Vogt A, Sun J, Cox AD, Der CJ, Hamilton AD, Sebti SM. Ras CAAX peptidomimetic FTI-277 selectively blocks oncogenic Ras signaling by inducing cytoplasmic accumulation of inactive Ras-Raf complexes. J Biol Chem. 1995;270:26802–26806. doi: 10.1074/jbc.270.45.26802. [DOI] [PubMed] [Google Scholar]
- Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- Lin J, Teresky AK, Levine AJ. Two critical hydrophobic amino acids in the N-terminal domain of the p53 protein are required for the gain of function phenotypes of human p53 mutants. Oncogene. 1995;10:2387–2390. [PubMed] [Google Scholar]
- Matas D, Sigal A, Stambolsky P, Milyavsky M, Weisz L, Schwartz D, Goldfinger N, Rotter V. Integrity of the N-terminal transcription domain of p53 is required for mutant p53 interference with drug-induced apoptosis. EMBO J. 2001;20:4163–4172. doi: 10.1093/emboj/20.15.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza A, Wu Q, Wang L, McClanahan T, Bishop WR, Gheyas F, Ding W, Hutchins B, Hockenberry T, Kirschmeier P, et al. Global transcriptional program of p53 target genes during the process of apoptosis and cell cycle progression. Oncogene. 2003;22:3645–3654. doi: 10.1038/sj.onc.1206477. [DOI] [PubMed] [Google Scholar]
- Mo H, Elson CE. Studies of the isoprenoid-mediated inhibition of mevalonate synthesis applied to cancer chemotherapy and chemoprevention. Exp Biol Med (Maywood) 2004;229:567–585. doi: 10.1177/153537020422900701. [DOI] [PubMed] [Google Scholar]
- Mori S, Chang JT, Andrechek ER, Potti A, Nevins JR. Utilization of genomic signatures to identify phenotype-specific drugs. PLoS One. 2009;4:e6772. doi: 10.1371/journal.pone.0006772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, Lukashchuk N, Gillespie DA, Ludwig RL, Gosselin P, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, Theillet C, Rodriguez C, Lidereau R, Bieche I, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res. 2006;12:1157–1167. doi: 10.1158/1078-0432.CCR-05-1029. [DOI] [PubMed] [Google Scholar]
- Peart MJ, Prives C. Mutant p53 gain of function: the NF-Y connection. Cancer Cell. 2006;10:173–174. doi: 10.1016/j.ccr.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci U S A. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008;4:e1000133. doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato R. Sterol metabolism and SREBP activation. Arch Biochem Biophys. 2010;501:177–181. doi: 10.1016/j.abb.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Shibata MA, Ito Y, Morimoto J, Otsuki Y. Lovastatin inhibits tumor growth and lung metastasis in mouse mammary carcinoma model: a p53-independent mitochondrial-mediated apoptotic mechanism. Carcinogenesis. 2004;25:1887–1898. doi: 10.1093/carcin/bgh201. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambolsky P, Tabach Y, Fontemaggi G, Weisz L, Maor-Aloni R, Siegfried Z, Shiff I, Kogan I, Shay M, Kalo E, et al. Modulation of the vitamin D3 response by cancer-associated mutant p53. Cancer Cell. 2010;17:273–285. doi: 10.1016/j.ccr.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugawa T, Kakuta H, Moritani H, Matsuda K, Ishihara T, Yamaguchi M, Naganuma S, Iizumi Y, Shikama H. YM-53601, a novel squalene synthase inhibitor, reduces plasma cholesterol and triglyceride levels in several animal species. Br J Pharmacol. 2000;131:63–70. doi: 10.1038/sj.bjp.0703545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan A, Qian Y, Vogt A, Blaskovich MA, Ohkanda J, Sebti SM, Hamilton AD. Potent, highly selective, and non-thiol inhibitors of protein geranylgeranyltransferase-I. J Med Chem. 1999;42:1333–1340. doi: 10.1021/jm9900873. [DOI] [PubMed] [Google Scholar]
- Venot C, Maratrat M, Sierra V, Conseiller E, Debussche L. Definition of a p53 transactivation function-deficient mutant and characterization of two independent p53 transactivation subdomains. Oncogene. 1999;18:2405–2410. doi: 10.1038/sj.onc.1202539. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Wong WW, Dimitroulakos J, Minden MD, Penn LZ. HMG-CoA reductase inhibitors and the malignant cell: the statin family of drugs as triggers of tumor-specific apoptosis. Leukemia. 2002;16:508–519. doi: 10.1038/sj.leu.2402476. [DOI] [PubMed] [Google Scholar]
- Yahagi N, Shimano H, Matsuzaka T, Najima Y, Sekiya M, Nakagawa Y, Ide T, Tomita S, Okazaki H, Tamura Y, et al. p53 Activation in adipocytes of obese mice. J Biol Chem. 2003;278:25395–25400. doi: 10.1074/jbc.M302364200. [DOI] [PubMed] [Google Scholar]
- Yan W, Chen X. Characterization of functional domains necessary for mutant p53 gain of function. J Biol Chem. 2010;285:14229–14238. doi: 10.1074/jbc.M109.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yan W, Chen X. Mutant p53 disrupts MCF-10A cell polarity in 3-dimensional culture via epithelial-to-mesenchymal transitions. J Biol Chem. 2011 doi: 10.1074/jbc.M110.214585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.