Abstract
Autosomal-recessive congenital cerebellar ataxia was identified in Roma patients originating from a small subisolate with a known strong founder effect. Patients presented with global developmental delay, moderate to severe stance and gait ataxia, dysarthria, mild dysdiadochokinesia, dysmetria and tremors, intellectual deficit, and mild pyramidal signs. Brain imaging revealed progressive generalized cerebellar atrophy, and inferior vermian hypoplasia and/or a constitutionally small brain were observed in some patients. Exome sequencing, used for linkage analysis on extracted SNP genotypes and for mutation detection, identified two novel (i.e., not found in any database) variants located 7 bp apart within a unique 6q24 linkage region. Both mutations cosegregated with the disease in five affected families, in which all ten patients were homozygous. The mutated gene, GRM1, encodes metabotropic glutamate receptor mGluR1, which is highly expressed in cerebellar Purkinje cells and plays an important role in cerebellar development and synaptic plasticity. The two mutations affect a gene region critical for alternative splicing and the generation of receptor isoforms; they are a 3 bp exon 8 deletion and an intron 8 splicing mutation (c.2652_2654del and c.2660+2T>G, respectively [RefSeq accession number NM_000838.3]). The functional impact of the deletion is unclear and is overshadowed by the splicing defect. Although ataxia lymphoblastoid cell lines expressed GRM1 at levels comparable to those of control cells, the aberrant transcripts skipped exon 8 or ended in intron 8 and encoded various species of nonfunctional receptors either lacking the transmembrane domain and containing abnormal intracellular tails or completely missing the tail. The study implicates mGluR1 in human hereditary ataxia. It also illustrates the potential of the Roma founder populations for mutation identification by exome sequencing.
Main Text
Autosomal-recessive cerebellar ataxias (ARCAs) are a clinically and genetically heterogeneous group of disorders whose clinical and genetic classifications are still evolving as the field progresses and novel phenotypes and genes are described.1,2 Congenital cerebellar ataxias are a relatively small ARCA subset characterized by infantile onset of motor incoordination, developmental delay, and variable additional manifestations.3–10 Although a number of genes have already been implicated in ARCAs, many rare forms and their molecular basis remain to be discovered.
Here, we describe a form of congenital cerebellar ataxia identified in patients of Roma ethnicity in Bulgaria. The disorder was first encountered during our studies of congenital cataracts facial dysmorphism neuropathy11,12 (CCFDN [MIM 604168]) in patients M II-1 and M II-2, who belonged to an extended CCFDN-affected kindred. A similar phenotype was observed recently in two children (V III-1 and V III-2) from another Roma family, suggesting a common founder mutation.13 A search of old hospital records and field-trip notes identified three additional families for a total of ten living patients and 14 unaffected relatives participating in the study (Figure 1). All individuals belonged to the same Roma group, the Bowlmakers, a young population subisolate characterized by small founding size, historically stagnant demographic regime, and low genetic diversity.14,15 Parents were clinically healthy; those deceased or unavailable were described as symptom free, supporting autosomal-recessive inheritance. Written informed consent was provided by all participating subjects or guardians of minors. The study complied with the guidelines of the institutions involved.
Figure 1.
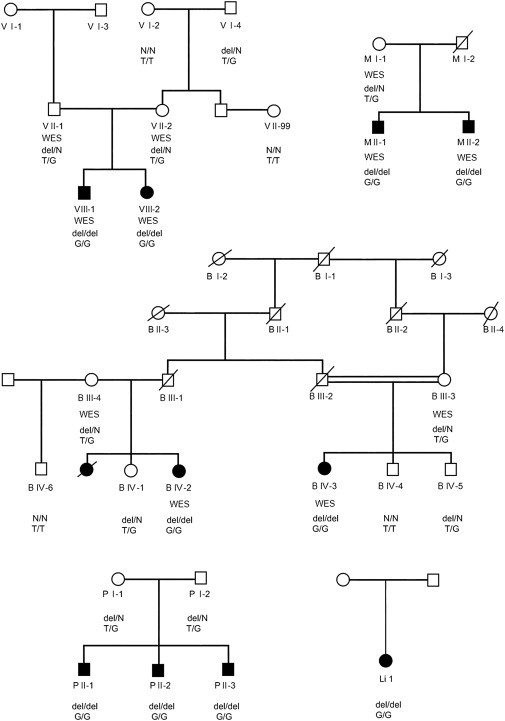
Pedigree Structure of the Ataxia-Affected Families Participating in the Study
The study included five unrelated Roma families, of which V, M, and B were recruited first and were included in the exome-sequencing analysis. “WES” designates individuals selected for exome sequencing. All participating subjects were genotyped for the GRM1 mutations. The following abbreviations are used: del, presence of c.2652_2654del; N, absence of c.2652_2654del; T, normal allele; and G, mutant allele of c.2660+2T>G.
Eight patients were admitted to university hospitals for detailed investigations, and two were examined during home visits. The clinical findings are summarized in Table 1.
Table 1.
Clinical Observations in the Ten Living Ataxia Patients
|
Patient |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Li 1 | V III-2 | V III-1 | B IV-2 | P II-2 | B IV-3 | P II-1 | P II-3 | M II-2 | M II-1 | |
| Age (years) at examination | 6 | 9 | 11 | 26 | 27 | 32 | 37 | 42 | 47 | 57 |
| Sex | female | female | male | female | male | female | male | male | male | male |
| Pregnancy and delivery | no data | normal | intranatal asphyxia | normal | lung atelectasis | normal | normal | normal | preterm | preterm |
| Age (years) at walking | 4 | 4 | 5 | childhood | never | 1 | never | never | 3 | no data |
| Age (years) at simple sentences | 4 | 2 | 3 | no data | never | no data | never | never | never | never |
| Height (cm) | 120 | 126 | 137 | 147 | 138 | 143 | 151 | 150 | 154 | 152 |
| Weight (kg) | 22 | 40 | 42 | 42.5 | 23 | 42.5 | 50 | 60 | 47.1 | 45.1 |
| Cerebellar Ataxia (SARA Scores) | ||||||||||
| Gait (0–8) | 4 | 5 | 5 | 5 | 8 | 4 | 7 | 8 | 6 | 7 |
| Stance (0–6) | 2 | 4 | 4 | 4 | 6 | 2 | 6 | 6 | 5 | 6 |
| Sitting (0–4) | 0 | 1 | 1 | 0 | 4 | 0 | 3 | 3 | 1 | 2 |
| Speech disturbance (0–6) | 2 | 2 | 2 | 2 | no speech | 2 | no speech | no speech | no speech | no speech |
| Finger chase (R + L)/2 (0–4) | 1 | 1 | 2 | 1 | not testeda | 1 | 1 | 1 | 2 | 1 |
| Nose-finger test (R + L)/2 (0–4) | 1 | 1 | 2 | 1 | not testeda | 1 | 1 | 2 | 1 | 0 |
| Fast alternating hand movements (R + L)/2 (0–4) | 1 | 2 | 2 | 1 | not testeda | 1 | 2 | 2 | 2 | 1 |
| Heel-shin slide (R + L)/2 (0–4) | 1 | 2 | 2 | 2 | not testeda | 1 | 2 | 3 | 2 | 2 |
| Total SARA score (0–40)b | 13/40 | 18/40 | 20/40 | 16/40 | 18/18 | 12/40 | 22/34 | 25/34 | 19/34 | 19/34 |
| Oculomotor Signs | ||||||||||
| Gaze-evoked horizontal nystagmus | no | no | no | yes | no | yes | yes | no | no | no |
| Hypometric saccades | no | yes | no | no | yes | no | yes | no | no | yes |
| Abduction deficit | yes | no | no | yes | yes | no | no | yes | no | no |
| Esotropia | no | yes | yes | no | yes | no | no | no | no | yes |
| Ptosis | no | no | no | yes | no | no | no | no | yes | yes |
| Additional | ||||||||||
| Intellectual deficit | mild | mild | mild | moderate | profound | moderate | moderate | severe | severe | severe |
| Hyperreflexia | yes | yes | yes | yes | no | yes | yes | yes | yes | yes |
| Spasticity | no | no | no | no | no | no | no | yes | no | no |
| Polyneuropathy | no | no | no | no | mild | no | no | no data | no data | no data |
| Seizures | no | at 2 years | at 5 months | no | one abnormal EEG | no | no | no | no | no |
The scoring of the eight SARA16 items is shown in the first column; 0 indicates a lack of impairment, and higher scores indicate increasing severity. The following abbreviations are used: R, right; L, left; SARA, Scale for the Assessment and Rating of Ataxia; and EEG, electroencephalography.
Profound intellectual deficit precluded these assessments in patient P II-2.
The second figure is the maximal possible total SARA score, which can be lower than the theoretical maximum of 40 if some assessments were not possible (e.g., a dysarthria score is missing in patients who never learned to speak).
Collected from previous hospital records and parental interviews, data on early manifestations and evolution of the disorder pointed to impaired psychomotor development. Three patients never walked, and five never learned to talk, whereas developmental milestones were markedly delayed in the remaining subjects (Table 1).
Upon current examination, height and weight were within normal limits in the three affected children but were markedly low in the adult patients.
Ataxia manifestations were evaluated with the Scale for the Assessment and Rating of Ataxia (SARA).16 Moderate to severe gait and stance ataxia were invariably present, had a mean score of 11.9 (SARA items 1–3; maximum score of 18), and tended to have higher values in older patients (Table 1). Mild dysarthria was present in all who were able to speak. Dysmetria, tremors, and dysdiadochokinesia were also mild and had a mean score of 5.2 for limb kinetic function (SARA items 5–8; maximum score of 16).
Pyramidal signs were relatively mild and localized mostly to the lower limbs.
Abnormalities revealed by ophthalmological examination included gaze-induced horizontal nystagmus, hypometric saccades, mild abduction deficits, strabismus, and ptosis (Table 1).
Cognitive function was assessed through interviews with the affected subjects and care providers with the use of the Diagnostic and Statistical Manual of Mental Disorders17 criteria targeting adaptive functioning. Intellectual deficit, ranging in severity from mild to profound, was present in all patients.
Brain-imaging data were collected on seven subjects, and spine imaging was performed on two subjects; magnetic resonance imaging (MRI [1.5T imagers, Signa Excite HDxt, GE Healthcare Milwaukee, USA]) was used for all of these patients, and additional computed tomography (CT) scans were used for two of the patients (Tables 2 and 3 and Figure 2). Generalized cerebellar atrophy was invariably present and was mostly classified as moderate to severe. A constitutional small inferior vermis was observed in three individuals, and a small brain was observed in five of the seven subjects. The middle cerebellar peduncle and brainstem were mostly normal. Isolated findings included atrophy of the right hippocampal body and bilateral-posterior-putamen signal change and atrophy associated with prominent perivascular spaces. The spine was normal.
Table 2.
Neuroimaging Findings in the Ataxia Patients
|
Patient |
|||||||
|---|---|---|---|---|---|---|---|
| Li 1 | V III-2a | V III-1a | B IV-2 | P II-2 | B IV-3 | P II-3 | |
| Age (years) at imaging | 6 | 6 | 11 | 24 | 27 | 32 | 37 |
| Modality | MRI | MRI | CT | MRI | MRI | MRI | MRI |
| Protocol | T1 FLAIR sagittal, T1 SPGR coronal, T2 FSE coronal, T2 FSE axial | PD and T2 axial | volumetric axial with orthogonal MPR | T1 sagittal, T2 propeller axial, T2 FLAIR coronal, DWI (b = 1,000) | T1 MEMP axial, T2 FSE coronal, DWI (b = 1,000), T2 GRE axial, T1 3D BRAVO with MPRs, T2 FSE axial, T2 FSEIR coronal, T2 FLAIR coronal | volumetric T1 MPRs—coronal and sagittal | T1 MEMP axial, T2 FSE coronal, DWI (b = 1,000), T2 GRE axial, T1 3D BRAVO with MPRs, T2 FSE axial, T2 FSEIR coronal, T2 FLAIR coronal |
| Global atrophy | no | no | no | no | no | no | no |
| Global small brain (score) | no (0) | no (0) | yes (1) | mild (1) | yes (1) | yes (1) | mild (1) |
| Ventricular system | fourth enlarged | fourth enlarged | mild to moderate generalized increase; no hydrocephaly | fourth enlarged | no | fourth enlarged | no |
| Selective cerebral atrophy | no | no | no | no | no | no | no |
| Selective cerebellar atrophy (score) | moderate generalized (2) | moderate to marked generalized (3) | moderate generalized (2) | mild hemispheric (1) | marked generalized (3) | moderate to severe generalized (3) | moderate generalized (2) |
| Cerebellar hypoplasia (score) | no (0) | no (0) | inferior vermian (1) | no (0) | inferior vermian (1) | inferior vermian (1) | no (0) |
| Posterior cranial fossa size (score) | normal (0) | normal (0) | small (1) | normal (0) | small (1) | normal (0) | normal (0) |
| Retrocerebellar cyst | no | yes | yes | no | yes | yes | yes |
| Middle cerebellar peduncle size (score) | normal (0) | normal (0) | mild decrease (1) | normal (0) | normal (0) | normal (0) | normal (0) |
| Brainstem size and signal | normal | normal | normal | normal | normal | normal | normal |
| Hippocampal size | normal | normal | normal | normal | normal | normal | right posterior body atrophic |
| Cerebral white matter | normal | normal | normal | normal | normal | normal | normal |
| Basal ganglia and thalami | normal | normal | normal | normal | posterior putaminal signal and atrophy | normal | normal |
| Total imaging scoreb | 2 | 3 | 6 | 2 | 6 | 5 | 3 |
| Total SARA scorec | 13/40 | 18/40 | 20/40 | 16/40 | 18/18 | 12/40 | 22/34 |
The following abbreviations are used: FLAIR, fluid attenuation inversion recovery; SPGR, spoiled gradient echo; FSE, fast spin echo; PD, proton density; MPR, multiplanar reformat; DWI, diffusion-weighted imaging; MEMP, multiecho multiplanar; GRE, gradient echo; BRAVO, brain volume; and FSEIR, fast spin-echo inversion recovery.
Longitudinal imaging data of V III-2 and V III-1 are shown in Table 3.
Scores for individual signs are 0 or 1 (absent or present, respectively). However, for cerebellar atrophy, 0, 1, 2, and 3 correspond to nil, mild, moderate, and marked, respectively.
Total SARA scores are as shown in Table 1.
Table 3.
Reassessment of CT and MRI Scans Performed at Different Ages in the Affected Siblings of Family V
|
Patient |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| V III-2 | V III-1 | ||||||||||
| Age (years) at imaging | 1 | 1 year, 8 months | 3 | 6 | 9 | 9 | 5 | 5 | 8 | 8 | 11 |
| Body region | head | head | head | head | head | T-spine | head | head | head | L-spine | head |
| Modality | CT | MRI | CT | MRI | CT | MRI | CT | MRI | CT | MRI | CT |
| nonvolumetric axial (8 mm slices) | T1 midline sagittal, PD, and T2 axial | nonvolumetric axial (5 mm slices) | PD and T2 axial | nonvolumetric axial (3 mm slices) | T2 frFSE sagittal, T1 FLAIR sagittal, T2 STIR sagittal, 2D MERGE axial | nonvolumetric axial (5 mm slices) | PD and T2 axial | nonvolumetric axial (3 mm slices) | T1 FSE sagittal, T2 IR-STIR sagittal, 2D MERGE axial, T1 FSE axial, T2 FSE coronal | volumetric axial with orthogonal MPR | |
| Ventricular system | no | no | fourth enlarged | fourth enlarged | fourth enlarged | − | mild generalized increase | mild to moderate generalized increase (no hydrocephalus) | mild to moderate generalized increase (no hydrocephalus) | − | mild to moderate generalized increase (no hydrocephalus) |
| Selective cerebellar atrophy | no | no | moderate generalized | moderate to marked generalized | moderate generalized | − | mild to moderate generalized | moderate generalized | moderate generalized | − | moderate generalized |
| Cerebellar hypoplasia | no | no | no | no | no | − | inferior vermian | inferior vermian | inferior vermian | − | inferior vermian |
| Posterior cranial fossa size | normal | normal | normal | normal | normal | − | small | small | small | − | small |
| Retrocerebellar cyst | no | no | yes | yes | yes | − | yes | yes | yes | − | yes |
| Middle cerebellar peduncle | normal | normal | normal | normal | normal | − | mild decrease | mild decrease | mild decrease | − | mild decrease |
| Comparison | new—cellebellar atrophy | slight increase in ventricular size and cerebellar atrophy | |||||||||
| Spine | normal | small lower spinal cord syrinx | |||||||||
The following abbreviations are used: CT, computed tomography; MRI, magnetic resonance image; PD, proton density; frFSE, fast-recovery fast spin echo; FLAIR, fluid attenuation inversion recovery; STIR, short-tau inversion recovery; MERGE, multiple-echo recombined gradient echo; FSE, fast spin echo; IR-STIR, inversion-recovery short-tau inversion recovery; and MPR, multiplanar reformat.
Figure 2.
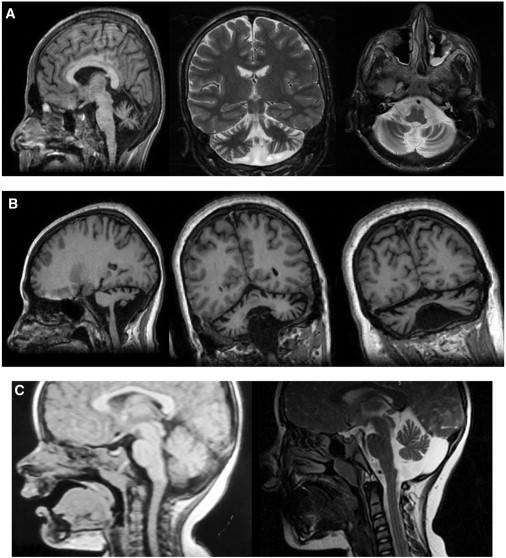
Brain Imaging of the Patients with Congenital Cerebellar Ataxia
The MRI data were obtained with 1.5T magnetic resonance imagers.
(A) Generalized cerebellar atrophy and inferior vermian hypoplasia in patient P II-2 (T1 3D brain volume [BRAVO] with multiplanar reformats [MPRs], coronal T2, and axial T2 sequences).
(B) Moderate to severe cerebellar atrophy and inferior vermian hypoplasia in patient B IV-3 (volumetric coronal and sagittal T1 MPR).
(C) Evolution of MRI changes in patient V III-2. The left panel shows normal findings at age 1 year and 8 months, and the right panel shows moderate to marked cerebellar atrophy (T1 midline sagittal, T2 sagittal) evident at 6 years of age.
Reassessment of CT and MRI scans performed at different ages in two patients (Table 3 and Figure 2) suggested a progressive nature of the structural brain changes: in V III-2, the findings were unremarkable at 1 year and 1 year and 8 months of age and became abnormal at the age of 3 (volume loss in the cerebellum), whereas in V III-1, the cerebellar atrophy became more prominent at 11 years of age compared to 8 years of age.
Nerve-conduction studies and needle electromyography (Dantec–Keypoint portable electromyograph [Natus, Copenhagen, Denmark]) were mostly normal.
Routine blood counts and biochemistry and blood and urine metabolites were within reference limits.
We sequenced the exomes of 11 individuals—six patients and five parents from families V, M, and B (Figure 1). Exome capture (Illumina TruSeq) and sequencing (Illumina HiSeq 2000) were performed by Axeq Technologies (Seoul, South Korea). The 101 bp paired-end reads were aligned to UCSC hg19 with Novoalign version 2.0.7. Reads mapping to multiple locations and presumed PCR duplicates were discarded with the Picard utility MarkDuplicates. Variants were detected with the mpileup and bcftools view commands from SAMtools 0.1.1818,19 with parameters -C50 and -q13. Variant filtering was performed with the vcfutils.pl varFilter script from the same program. Variants were annotated with the UCSC KnownGene annotation and ANNOVAR.20 The functional effects of coding nonsynonymous variants were assessed with SIFT21 and HumVar-trained PolyPhen2 version 2.1.0.22
We used SAMtools to infer from the exome sequence data genotypes at the location of HapMap Phase II SNPs.23 From 40,464 SNPs with inferred genotypes, we selected a subset of 6,855 that were in approximate linkage equilibrium, spaced at least 0.15 cM apart, and chosen for maximizing heterozygosity (average of 0.40) according to HapMap CEU (Utah residents with ancestry from northern and western Europe from the CEPH collection) genotypes. We used this subset to estimate relatedness between affected families and inbreeding within each family. The proportion of alleles shared identically by descent between every pair of the 11 individuals was assessed with PLINK.24 The findings (Table S1, available online) validated the known relationships within each of the three families. No sharing was detected between individuals from different families, indicating that although the affected families belonged to the same Roma group, they were not closely related. Inbreeding coefficients (F) were estimated with FEstim;25 we used CEU allele frequencies and starting values of 0.05 for the parameters F and A. All six patients were estimated to be inbred; F ranged from 0.023 to 0.113 (Table S2). Inbreeding was also detected in four of the five unaffected parents; the exception, mother V II-2, was known to have a non-Roma mother. Inbreeding and relatedness detection appeared to be robust to potential misspecification of allele frequencies as a result of the usage of CEU frequencies; known relationships were estimated accurately by both PLINK and FEstim.
Parametric multipoint linkage analysis was performed on the same subset of 6,855 SNPs with MERLIN26 under a fully penetrant autosomal-recessive model with a 0% phenocopy rate, a disease allele frequency of 0.00001, and CEU SNP allele frequencies. We added hypothetical consanguinity loops to the pedigrees to approximate the estimated inbreeding coefficients (Figure S1). Genome-wide linkage analysis identified in chromosomal region 6q24 a unique peak with a maximum heterogeneity LOD score of 6.009 (Figure S2). This was the only genomic region in which all three families showed linkage. The contribution of each family to the overall result is shown in Figure S3.
Examination of inferred 6q24 haplotypes with the use of HaploPainter27 showed a stretch of 11 SNPs (147.86– 151.16 cM) homozygous in all patients (Figure S4). Inspection of all polymorphic SNPs in the region showed homozygosity extending over 59 SNPs spanning 3.3 cM (3.8 Mb). The flanking (nonhomozygous) SNPs rs2073214 at 147.86 cM and rs2272998 at 152.38 cM defined an interval containing 32 genes (including nine pseudogenes), of which 12 have associated OMIM entries.
Of the total 565,618 sequence variants detected in at least one of the 11 exomes, 352,599 survived filtering. Of these, 294 (including 88 rare or novel [not found in any database] variants) were located within the linkage interval. Five were coding (one synonymous, two nonsynonymous, a nonframeshift deletion, and a stop-gain change), and one was within 2 bp of a predicted splice site. All six were within the shared homozygous haplotype. Only two of the six followed the expected segregation pattern, which is that all patients and parents are homozygous and heterozygous, respectively, for the mutant allele (illustrated for family V in Figure S5).
The two homozygous variants segregating with the disease are located in GRM1, the gene encoding metabotropic glutamate receptor 1 (mGluR1) and are c.2652_2654del and c.2660+2T>G (RefSeq accession number NM_000838.3). Database and literature searches identified multiple mutant animal models showing severe motor incoordination (Table S3), making GRM1 a highly plausible candidate in human ataxia. One mutation (c.2652_2654del [p.Asn885del]) (hg19, chr6: 146,720,827–146,720,829; RefSeq NM_000838.3) is a 3 bp deletion close to the 3′ end of exon 8. Seven base pairs downstream, the second mutation (c.2660+2T>G; hg19, chr6: 146,720,837) affects the second nucleotide of the canonical splice donor site of intron 8. The presence of the mutations was verified by Sanger sequencing (Figure S5); analysis of the 11 original samples, additional members of families V, M, and B, and two newly recruited families showed that genotypes for the two mutations predicted disease perfectly under the autosomal-recessive inheritance model (Figure 1).
Neither c.2652_2654del nor c.2660+2T>G is listed in the 1000 Genomes database release May 2011 (1,092 individuals) or the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project Exome Variant Server (5,379 individuals). Neither was detected in our sequencing analysis of 1,050 schizophrenia cases and controls28 or in a study of 1,300 schizophrenia and bipolar patients and controls.29 Thus, the overall number of control individuals negative for these mutations is 8,821.
Next, we examined carrier rates among 289 control individuals from three historically, linguistically, and genetically related Roma groups, which share ancestral mtDNA, Y chromosome lineages, and disease-causing mutations.13,30 The panel included 80 Bowlmakers, and the remaining individuals belonged to the Lom and Kalderash groups. In addition to assessing the spread of the GRM1 mutations, we aimed at understanding their simultaneous occurrence on the same chromosome and hypothesized that some control subjects might carry only one of the two variants. We used PCR-based restriction-fragment-length-polymorphism assays for the independent detection of the c.2652_2654del and c.2660+2T>G mutations (Figure S6) and used pyrosequencing as a confirmatory test (Figure S7). The screening identified two Bowlmakers heterozygous for both changes—a carrier rate of 2.5% (a 95% exact binomial confidence interval of 0.3%–8.7%) for this Roma group. No heterozygotes were detected among the Lom and Kalderash groups. Compared to the carrier rates of other disease-causing mutations in the Roma population,30 this carrier rate is relatively low. The fact that the mutations were confined to the Bowlmakers and were not found in the related groups (despite the fact that recent population fissions have given rise to the three subisolates) and the unusually large size of the conserved haplotype suggest a recent origin of the mutations. The events leading to the coexistence of these tightly linked variants on the same ancestral chromosome remain unexplained.
mGluR1, the protein product of GRM1, is a family C G protein-coupled receptor (GPCR). Class C GPCRs possess a large bilobed ligand-binding domain followed by a cysteine-rich domain, a highly conserved seven transmembrane (7TM) helical structure, and a large intracellular C-terminal tail.31 The 7TM domain, encoded by GRM1 exon 8, is essential for selective receptor coupling to G proteins and to allosteric modulators.32,33 The intracellular tail is encoded by the 3′ end of exon 8 and by exons 9 and 10, whose alternative splicing (Figure 3) generates mGluR1 isoforms with different lengths and amino acid compositions of the tail.34–36 Intracellular targeting, receptor activity, desensitization, and internalization of mGluR1 isoforms are largely dependent on protein-protein interactions involving the intracellular tail.34,36,37 A 4 aa sequence (Arg-Arg-Lys-Lys) at positions 876–879, exposed in the “short” isoforms, has been proposed to act as a signal for retention in the endoplasmic reticulum and to inhibit G protein coupling and agonist-independent receptor activity.38,39 Its masking by the long mGluR1α tail accounts for the membrane exposure, dendritic targeting, and higher activity of this isoform.34,38,39 The modulating effects on Ca2+ levels and neuronal activity require interactions of the tail with Homer proteins, ion channels, other receptors, protein kinases, and scaffolding and trafficking proteins.34,37
Figure 3.
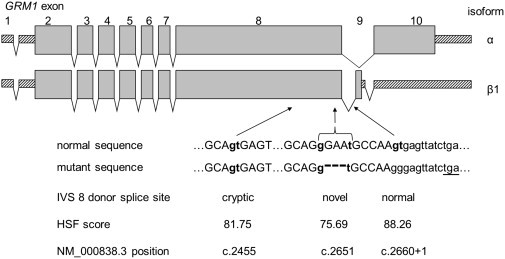
Genomic Organization of GRM1 and Location of the Ataxia Mutations
The upper panel (adapted from Ferraguti et al.34) shows alternative transcripts encoding the most abundant receptor isoforms, mGluR1α and mGluR1β1. Exons 1–8 are invariably included in all GRM1 transcripts, whereas downstream differences—inclusion or skipping of exon 9 and the usage of alternative splice acceptor sites in exon 10—are used for producing the receptor isoforms.34–36 The c.2660+2T>G mutation affects the intron 8 donor splice site, involved in generating all alternative transcripts. The lower panel shows potential consequences of the c.2660+2T>G mutation. In the surrounding sequence, alternative sites that are identified by the Human Splicing Finder tool as donor splice sites and that could be utilized in the absence of the normal signal are indicated in bold lowercase letters. The c.2652_2654del deletion and the novel splice site created as a result are shown with a bracket. In the absence of the normal splice signal, aberrant transcripts could also extend into intron 8 and use a downstream in-frame stop codon (underlined).
The mutations described here are located in the GRM1 region encoding the cytoplasmic tail (Figure 3). c.2652_2654del is predicted to result in the deletion of an asparagine residue at position 885, whereas c.2660+2T>G is expected to abolish the intron 8 donor splice site and have potential effects including exon 8 skipping, usage of alternative splice sites, and/or of an in-frame termination codon 10 nt into intron 8 (Figure 3).
We examined GRM1 transcripts in RNA extracted from lymphoblastoid cell lines (LCLs) of ataxia patients V III-2 and P II-2 and controls, which we selected to allow identification based on the presence of rare GRM1 SNPs. Brain RNA (Clontech, Mountain View, CA, USA) was used for comparison. RT-PCR reactions were performed as described,40 and product identity was verified by Sanger sequencing (primers are shown in Table S4). An initial RT-PCR experiment with primers targeting invariable upstream exons 3 and 4 confirmed GRM1 expression in LCLs from both controls and ataxia patients (Figure S8). Quantification of transcript abundance (Figure S8) indicated that, although its levels were lower in LCLs than in the brain, they were comparable in normal and mutant cells. Next, we used RT-PCR with primers in exons 7 and 10 to check the splicing patterns in normal LCLs and to examine the effects of the c.2660+2T>G mutation in ataxia cells. In normal LCLs, this primer pair generated the expected transcripts encoding the most abundant mGluR1α and mGluR1β1 isoforms (Figure 4). In ataxia cells, the normal products were replaced by much smaller fragments in which exon 8 was missing and the exon 7 sequence extended into exons 9 or 10 (Figure 4). Finally, RT-PCR with primers (in exon 7 and intron 8) targeting transcripts that continued from exon 8 into intron 8 generated a product in all cells tested (Figure 5). DNA contamination was ruled out in a parallel experiment with primers in exon 7 and intron 7 (Figure S9). The product in normal LCLs and the brain is likely to be identical to a previously described rare GRM1 transcript in cerebellar granule cells, confirmed at the protein level.41
Figure 4.
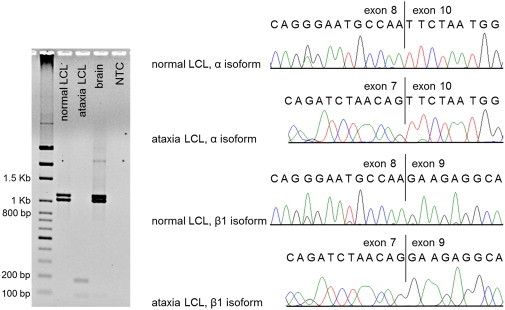
GRM1 Alternative Transcripts in Normal and Ataxia Cells
The left panel shows agarose gel electrophoresis of RT-PCR products. Primers in GRM1 exons 7 and 10 targeted the most abundant alternative transcripts encoding isoforms mGluR1α (skipping exon 9; expected fragment size is 1,030 bp) and mGluR1β (including exon 9; expected size is 1,115 bp). In ataxia cells, the expected fragments are replaced by products less than 200 bp in size. The right panel shows Sanger sequencing of the RT-PCR products in normal and ataxia LCLs. Sequencing was performed on purified products excised from the gel; the forward RT-PCR primer in exon 7 was used for ataxia LCLs, and a nested primer in exon 8 was used for controls. Transcript identity was confirmed in the control cells (derived from a subject heterozygous for the rare SNP rs362936 [G>A]). In ataxia cells, exon 8 was skipped and the transcripts extended from exon 7 into exons 9 and 10. Fragments resulting from the use of the cryptic splice site in exon 8 (expected sizes are 824 bp and 909 bp) or of the novel splice site created by the deletion (expected sizes are 1,020 bp and 1,105 bp) were not detected.
Figure 5.
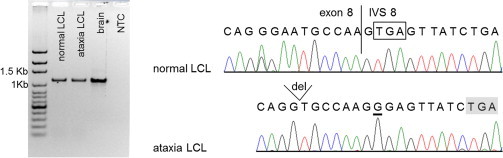
GRM1 Transcripts Extending from Exon 8 into Intron 8
The left panel shows gel electrophoresis of RT-PCR products with the use of primers in exon 7 and intron 8. Fragments of the expected size (1,144 bp) were observed in all cells. The right panel shows Sanger sequencing of the gel-purified fragments with a nested primer close to the normal junction of exon 8 and intron 8. The product observed in control LCLs and the brain could be identical to a previously described GRM1 transcript and its protein product,41 where the donor splice site of intron 8 is ignored and the immediately following termination codon (boxed) is utilized. In ataxia cells, a downstream in-frame termination codon (shaded) might be used for producing truncated receptor molecules.
GRM1 extraneural expression is limited by two conserved silencing elements, the neuronal restrictive silencing and the regulatory factor for X-box elements.42 Therefore, our LCL data, relying on low-level illegitimate transcription, should be interpreted as a general indication of the events potentially occurring in the affected brain. Our findings suggest that as a result of the c.2660+2T>G splicing mutation, ataxia cells produce two types of aberrant transcript that, if translated, would encode multiple species of abnormal, nonfunctional mGluR1 receptor molecules (Figure S10). Exon 8 skipping would lead to the absence of the 7TM domain; by itself, this absence would be equivalent to a lack of functional receptors. In addition, the out-of-frame 931 bp deletion caused by the skipping of exon 8 would produce a variety of C-terminal protein sequences, none of which correspond to the normal intracellular tails. In parallel, the transcripts ending in intron 8 would translate into truncated receptor molecules that have intact transmembrane domains but that are missing almost the entire intracellular tail. It is of interest to note that progressive cerebellar degeneration, a major feature of the human phenotype, is not observed in any of the animal models (Table S3). A degeneration mechanism that is worth exploring in future studies could involve impaired intracellular trafficking due to protein misfolding and/or exposure of the Arg-Arg-Lys-Lys sequence in the truncated mGluR1 receptors.
The mGluR1 receptor modulates intracellular Ca2+ levels and neuronal excitability through a signaling cascade involving phospholipase C (PLCβ4), second messengers inositol triphosphate (IP3) and diacylglycerol (DAG), and protein kinase C (PKCγ).34–36,43 It is most abundantly expressed in the cerebellar cortex and olfactory bulb, and it is also highly expressed in the hippocampus, lateral septum, globus pallidus, ventral pallidum, substantia nigra, and thalamic nuclei.34,36 Cerebellar expression of the “long” mGluR1 is localized to Purkinje cells, which integrate multiple input signals to provide the output of the entire vestibulocerebellum.34,36 The receptor is essential for the early postnatal development of the cerebellar cortex and plays a critical role in cerebellar and hippocampal synaptic plasticity, memory, and learning.43–45 Animal models with spontaneous or induced Grm1 mutations (Table S3) develop early-onset ataxia, impaired synaptic plasticity (cerebellar long-term depression and hippocampal long-term potentiation), and deficits in spatial and associative learning.34,43–45 In the Coton de Toulear dog model, which involves an insertion and possible frameshift in exon 8, saccadic dysmetria has been noted.46 Failure to establish the normal one-to-one relationship between Purkinje cells and climbing fibers leads to the persistent innervation of Purkinje cells by multiple climbing fibers; this abnormal innervation is an important neuropathological hallmark in Grm1-null mice.34,43
The clinical features in our patients can be summed up as global developmental delay, infantile onset of moderate to severe gait and stance ataxia, dysarthria, mild dysdiadochokinesia, dysmetria and tremor, intellectual deficit of variable severity, and mild pyramidal signs. The predominant brain-imaging findings were of moderate to marked generalized cerebellar atrophy with or without inferior vermian hypoplasia and/or a constitutionally small brain. In this small patient cohort, we found no clear-cut correlation between clinical severity and neuroimaging findings (Table 2). Although the disorder was perceived as nonprogressive by long-term care providers, our limited longitudinal imaging data pointed to a progressive nature of the cerebellar atrophy, and our clinical observations showed a trend to increasing severity of the ataxia (higher SARA scores) and of the intellectual deficit in older patients. A comparison of this phenotype to other forms of congenital ataxia (Table S5) highlighted the difficulties of clinical differential diagnosis. The same conclusion applied to the radiological findings: a spectrum of cerebellar hypoplasia and/or atrophy with selective involvement of the hemispheres and/or vermis are the most common ARCA findings and highlight the importance of genetic diagnosis.
Thus far, support for the contribution of mGluR1 to human cerebellar ataxia has come from rare patients with paraneoplastic or primary autoimmune reactions against the receptor protein.47,48 Sequencing GRM1 in 41 sporadic cases of early-onset cerebellar ataxia49 failed to identify any mutants. The present study is an example of a genetic GRM1 defect leading to human congenital ataxia. Identification of additional GRM1 mutations in human patients, as well as the development of a mouse model of the mutations we describe, will promote the understanding of pathogenetic mechanisms and interspecies differences. Our study also highlights the potential of linkage analysis, applied to genotypes inferred from exome-sequence data, to expedite mutation discovery. In addition, it illustrates the power of the Roma population, where the effects of long-term inbreeding in conjunction with close consanguinity are complemented by the sharing of founder mutations between distantly related affected families, for the identification of disease-associated genes.
Acknowledgments
We are grateful to the affected families participating in this study. D.N.A. is supported by National Health and Medical Research Council (NHMRC) Training Fellowship 634551; K.R.S. is supported by a PhD scholarship funded by the Pratt Foundation; and M.B. is supported by Australian Research Council Future Fellowship FT100100764 and NHMRC Program Grant 490037. The work of K.R.S. and M.B. is supported by Victorian State Government Operational Infrastructure Support and the Australian Government NHMRC Independent Research Institute Infrastructure Support Scheme. The Molecular Medicine Center is supported by grants from the National Science Fund; the Ministry of Education, Youth, and Science (DUNK01-2/2009); and the Science Fund of the Medical University of Sofia (grant 8I/2009). We thank Daniela Dacheva and Reni Tzveova for technical assistance.
Contributor Information
Melanie Bahlo, Email: bahlo@wehi.edu.au.
Luba Kalaydjieva, Email: luba.kalaydjieva@uwa.edu.au.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes Project, http://www.1000genomes.org/
ExPASy Translate Tool, http://web.expasy.org/translate/
Human Splicing Finder, http://www.umd.be/HSF/
Integrative Genomics Viewer, http://www.broadinstitute.org/igv/
Jackson Laboratory, http://www.jax.org/
NHLBI Exome Sequencing Project Exome Variant Server, http://evs.gs.washington.edu/EVS/
Novoalign, http://www.novocraft.com
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/
Picard, http://picard.sourceforge.net/
PolyPhen 2, http://genetics.bwh.harvard.edu/pph2/
SAMtools, http://samtools.sourceforge.net/
SIFT, http://sift.jcvi.org/
UniProt, http://www.uniprot.org/
Accession Numbers
The dbSNP accession numbers for the GRM1 sequences reported in this paper are ss534880213 and ss534880217.
References
- 1.Palau F., Espinós C. Autosomal recessive cerebellar ataxias. Orphanet J. Rare Dis. 2006;1:47. doi: 10.1186/1750-1172-1-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fogel B.L., Perlman S. Clinical features and molecular genetics of autosomal recessive cerebellar ataxias. Lancet Neurol. 2007;6:245–257. doi: 10.1016/S1474-4422(07)70054-6. [DOI] [PubMed] [Google Scholar]
- 3.Norman R.M. Primary degeneration of the granular layer of the cerebellum: An unusual form of familial cerebellar atrophy occurring in early life. Brain. 1940;63:365–379. [Google Scholar]
- 4.Steinlin M. Non-progressive congenital ataxias. Brain Dev. 1998;20:199–208. doi: 10.1016/s0387-7604(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 5.Mégarbané A., Delague V., Ruchoux M.M., Rizkallah E., Maurage C.A., Viollet L., Rouaix-Emery N., Urtizberea A. New autosomal recessive cerebellar ataxia disorder in a large inbred Lebanese family. Am. J. Med. Genet. 2001;101:135–141. doi: 10.1002/1096-8628(20010615)101:2<135::aid-ajmg1134>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 6.Bomar J.M., Benke P.J., Slattery E.L., Puttagunta R., Taylor L.P., Seong E., Nystuen A., Chen W., Albin R.L., Patel P.D. Mutations in a novel gene encoding a CRAL-TRIO domain cause human Cayman ataxia and ataxia/dystonia in the jittery mouse. Nat. Genet. 2003;35:264–269. doi: 10.1038/ng1255. [DOI] [PubMed] [Google Scholar]
- 7.Glass H.C., Boycott K.M., Adams C., Barlow K., Scott J.N., Chudley A.E., Fujiwara T.M., Morgan K., Wirrell E., McLeod D.R. Autosomal recessive cerebellar hypoplasia in the Hutterite population. Dev. Med. Child Neurol. 2005;47:691–695. doi: 10.1017/S0012162205001404. [DOI] [PubMed] [Google Scholar]
- 8.Lagier-Tourenne C., Tazir M., López L.C., Quinzii C.M., Assoum M., Drouot N., Busso C., Makri S., Ali-Pacha L., Benhassine T. ADCK3, an ancestral kinase, is mutated in a form of recessive ataxia associated with coenzyme Q10 deficiency. Am. J. Hum. Genet. 2008;82:661–672. doi: 10.1016/j.ajhg.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Türkmen S., Hoffmann K., Demirhan O., Aruoba D., Humphrey N., Mundlos S. Cerebellar hypoplasia, with quadrupedal locomotion, caused by mutations in the very low-density lipoprotein receptor gene. Eur. J. Hum. Genet. 2008;16:1070–1074. doi: 10.1038/ejhg.2008.73. [DOI] [PubMed] [Google Scholar]
- 10.Türkmen S., Guo G., Garshasbi M., Hoffmann K., Alshalah A.J., Mischung C., Kuss A., Humphrey N., Mundlos S., Robinson P.N. CA8 mutations cause a novel syndrome characterized by ataxia and mild mental retardation with predisposition to quadrupedal gait. PLoS Genet. 2009;5:e1000487. doi: 10.1371/journal.pgen.1000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angelicheva D., Turnev I., Dye D., Chandler D., Thomas P.K., Kalaydjieva L. Congenital cataracts facial dysmorphism neuropathy (CCFDN) syndrome: A novel developmental disorder in Gypsies maps to 18qter. Eur. J. Hum. Genet. 1999;7:560–566. doi: 10.1038/sj.ejhg.5200319. [DOI] [PubMed] [Google Scholar]
- 12.Varon R., Gooding R., Steglich C., Marns L., Tang H., Angelicheva D., Yong K.K., Ambrugger P., Reinhold A., Morar B. Partial deficiency of the C-terminal-domain phosphatase of RNA polymerase II is associated with congenital cataracts facial dysmorphism neuropathy syndrome. Nat. Genet. 2003;35:185–189. doi: 10.1038/ng1243. [DOI] [PubMed] [Google Scholar]
- 13.Kalaydjieva L., Morar B., Chaix R., Tang H. A newly discovered founder population: The Roma/Gypsies. Bioessays. 2005;27:1084–1094. doi: 10.1002/bies.20287. [DOI] [PubMed] [Google Scholar]
- 14.Kalaydjieva L., Calafell F., Jobling M.A., Angelicheva D., de Knijff P., Rosser Z.H., Hurles M.E., Underhill P., Tournev I., Marushiakova E., Popov V. Patterns of inter- and intra-group genetic diversity in the Vlax Roma as revealed by Y chromosome and mitochondrial DNA lineages. Eur. J. Hum. Genet. 2001;9:97–104. doi: 10.1038/sj.ejhg.5200597. [DOI] [PubMed] [Google Scholar]
- 15.Gresham D., Morar B., Underhill P.A., Passarino G., Lin A.A., Wise C., Angelicheva D., Calafell F., Oefner P.J., Shen P. Origins and divergence of the Roma (gypsies) Am. J. Hum. Genet. 2001;69:1314–1331. doi: 10.1086/324681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitz-Hübsch T., du Montcel S.T., Baliko L., Berciano J., Boesch S., Depondt C., Giunti P., Globas C., Infante J., Kang J.S. Scale for the assessment and rating of ataxia: Development of a new clinical scale. Neurology. 2006;66:1717–1720. doi: 10.1212/01.wnl.0000219042.60538.92. [DOI] [PubMed] [Google Scholar]
- 17.American Psychiatric Association . Fourth Edition. American Psychiatric Association; Washington, DC: 2000. Diagnostic and Statistical Manual of Mental Disorders. [Google Scholar]
- 18.Li H., Ruan J., Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H. Improving SNP discovery by base alignment quality. Bioinformatics. 2011;27:1157–1158. doi: 10.1093/bioinformatics/btr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K., Li M., Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 22.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith K.R., Bromhead C.J., Hildebrand M.S., Shearer A.E., Lockhart P.J., Najmabadi H., Leventer R.J., McGillivray G., Amor D.J., Smith R.J., Bahlo M. Reducing the exome search space for mendelian diseases using genetic linkage analysis of exome genotypes. Genome Biol. 2011;12:R85. doi: 10.1186/gb-2011-12-9-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leutenegger A.L., Prum B., Génin E., Verny C., Lemainque A., Clerget-Darpoux F., Thompson E.A. Estimation of the inbreeding coefficient through use of genomic data. Am. J. Hum. Genet. 2003;73:516–523. doi: 10.1086/378207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 27.Thiele H., Nürnberg P. HaploPainter: A tool for drawing pedigrees with complex haplotypes. Bioinformatics. 2005;21:1730–1732. doi: 10.1093/bioinformatics/bth488. [DOI] [PubMed] [Google Scholar]
- 28.Ayoub M.A., Angelicheva D., Vile D., Chandler D., Morar B., Cavanaugh J.A., Visscher P.M., Jablensky A., Pfleger K.D., Kalaydjieva L. Deleterious GRM1 mutations in schizophrenia. PLoS ONE. 2012;7:e32849. doi: 10.1371/journal.pone.0032849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frank R.A., McRae A.F., Pocklington A.J., van de Lagemaat L.N., Navarro P., Croning M.D., Komiyama N.H., Bradley S.J., Challiss R.A., Armstrong J.D. Clustered coding variants in the glutamate receptor complexes of individuals with schizophrenia and bipolar disorder. PLoS ONE. 2011;6:e19011. doi: 10.1371/journal.pone.0019011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morar B., Gresham D., Angelicheva D., Tournev I., Gooding R., Guergueltcheva V., Schmidt C., Abicht A., Lochmuller H., Tordai A. Mutation history of the roma/gypsies. Am. J. Hum. Genet. 2004;75:596–609. doi: 10.1086/424759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunishima N., Shimada Y., Tsuji Y., Sato T., Yamamoto M., Kumasaka T., Nakanishi S., Jingami H., Morikawa K. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 32.Pin J.P., Joly C., Heinemann S.F., Bockaert J. Domains involved in the specificity of G protein activation in phospholipase C-coupled metabotropic glutamate receptors. EMBO J. 1994;13:342–348. doi: 10.1002/j.1460-2075.1994.tb06267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knoflach F., Mutel V., Jolidon S., Kew J.N., Malherbe P., Vieira E., Wichmann J., Kemp J.A. Positive allosteric modulators of metabotropic glutamate 1 receptor: Characterization, mechanism of action, and binding site. Proc. Natl. Acad. Sci. USA. 2001;98:13402–13407. doi: 10.1073/pnas.231358298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferraguti F., Crepaldi L., Nicoletti F. Metabotropic glutamate 1 receptor: Current concepts and perspectives. Pharmacol. Rev. 2008;60:536–581. doi: 10.1124/pr.108.000166. [DOI] [PubMed] [Google Scholar]
- 35.Pin J.P., Waeber C., Prezeau L., Bockaert J., Heinemann S.F. Alternative splicing generates metabotropic glutamate receptors inducing different patterns of calcium release in Xenopus oocytes. Proc. Natl. Acad. Sci. USA. 1992;89:10331–10335. doi: 10.1073/pnas.89.21.10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicoletti F., Bockaert J., Collingridge G.L., Conn P.J., Ferraguti F., Schoepp D.D., Wroblewski J.T., Pin J.P. Metabotropic glutamate receptors: From the workbench to the bedside. Neuropharmacology. 2011;60:1017–1041. doi: 10.1016/j.neuropharm.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enz R. The trick of the tail: Protein-protein interactions of metabotropic glutamate receptors. Bioessays. 2007;29:60–73. doi: 10.1002/bies.20518. [DOI] [PubMed] [Google Scholar]
- 38.Mary S., Gomeza J., Prézeau L., Bockaert J., Pin J.P. A cluster of basic residues in the carboxyl-terminal tail of the short metabotropic glutamate receptor 1 variants impairs their coupling to phospholipase C. J. Biol. Chem. 1998;273:425–432. doi: 10.1074/jbc.273.1.425. [DOI] [PubMed] [Google Scholar]
- 39.Francesconi A., Duvoisin R.M. Alternative splicing unmasks dendritic and axonal targeting signals in metabotropic glutamate receptor 1. J. Neurosci. 2002;22:2196–2205. doi: 10.1523/JNEUROSCI.22-06-02196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pacheco R., Ciruela F., Casadó V., Mallol J., Gallart T., Lluis C., Franco R. Group I metabotropic glutamate receptors mediate a dual role of glutamate in T cell activation. J. Biol. Chem. 2004;279:33352–33358. doi: 10.1074/jbc.M401761200. [DOI] [PubMed] [Google Scholar]
- 41.Makoff A.J., Phillips T., Pilling C., Emson P. Expression of a novel splice variant of human mGluR1 in the cerebellum. Neuroreport. 1997;8:2943–2947. doi: 10.1097/00001756-199709080-00027. [DOI] [PubMed] [Google Scholar]
- 42.Crepaldi L., Lackner C., Corti C., Ferraguti F. Transcriptional activators and repressors for the neuron-specific expression of a metabotropic glutamate receptor. J. Biol. Chem. 2007;282:17877–17889. doi: 10.1074/jbc.M700149200. [DOI] [PubMed] [Google Scholar]
- 43.Kano M., Hashimoto K., Tabata T. Type-1 metabotropic glutamate receptor in cerebellar Purkinje cells: A key molecule responsible for long-term depression, endocannabinoid signalling and synapse elimination. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363:2173–2186. doi: 10.1098/rstb.2008.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lüscher C., Huber K.M. Group 1 mGluR-dependent synaptic long-term depression: Mechanisms and implications for circuitry and disease. Neuron. 2010;65:445–459. doi: 10.1016/j.neuron.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gil-Sanz C., Delgado-García J.M., Fairén A., Gruart A. Involvement of the mGluR1 receptor in hippocampal synaptic plasticity and associative learning in behaving mice. Cereb. Cortex. 2008;18:1653–1663. doi: 10.1093/cercor/bhm193. [DOI] [PubMed] [Google Scholar]
- 46.Coates J.R., O'Brien D.P., Kline K.L., Storts R.W., Johnson G.C., Shelton G.D., Patterson E.E., Abbott L.C. Neonatal cerebellar ataxia in Coton de Tulear dogs. J. Vet. Intern. Med. 2002;16:680–689. doi: 10.1892/0891-6640(2002)016<0680:ncaicd>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 47.Sillevis Smitt P., Kinoshita A., De Leeuw B., Moll W., Coesmans M., Jaarsma D., Henzen-Logmans S., Vecht C., De Zeeuw C., Sekiyama N. Paraneoplastic cerebellar ataxia due to autoantibodies against a glutamate receptor. N. Engl. J. Med. 2000;342:21–27. doi: 10.1056/NEJM200001063420104. [DOI] [PubMed] [Google Scholar]
- 48.Marignier R., Chenevier F., Rogemond V., Sillevis Smitt P., Renoux C., Cavillon G., Androdias G., Vukusic S., Graus F., Honnorat J., Confavreux C. Metabotropic glutamate receptor type 1 autoantibody-associated cerebellitis: a primary autoimmune disease? Arch. Neurol. 2010;67:627–630. doi: 10.1001/archneurol.2010.51. [DOI] [PubMed] [Google Scholar]
- 49.Rossi P.I., Vaccari C.M., Terracciano A., Doria-Lamba L., Facchinetti S., Priolo M., Ayuso C., De Jorge L., Gimelli S., Santorelli F.M. The metabotropic glutamate receptor 1, GRM1: Evaluation as a candidate gene for inherited forms of cerebellar ataxia. J. Neurol. 2010;257:598–602. doi: 10.1007/s00415-009-5380-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


