Abstract
Folding studies have been focused mainly on small, single-domain proteins or isolated single domains of larger proteins. However, most of the proteins present in biological systems are composed of multiple domains, and to date, the principles that underlie its folding remain elusive. The unfolding of Pfk-2 induced by GdnHCl has been described by highly cooperative three-state equilibrium (N2↔2I↔2U). This is characterized by a strong coupling between the subunits’ tertiary structure and the integrity of the dimer interface because “I” represents an unstructured and expanded monomeric intermediate. Here we report that cold and heat unfolding of Pfk-2 resembles the N2↔2I step of chemically induced unfolding. Moreover, cold unfolding appears to be as cooperative as that induced chemically and even more so than its heat-unfolding counterpart. Because Pfk-2 is a large homodimer of 66 kDa with a complex topology consisting of well-defined domains, these results are somewhat unexpected considering that cold unfolding has been described as a special kind of perturbation that decouples the cooperative unfolding of several proteins.
Introduction
The folding of small proteins has proven to be very informative for understanding the folding code of single-domain proteins. However, large and multidomain proteins with molecular masses >20 kDa are much more representative in typical biological systems (1). Phosphofructokinase-2 (Pfk-2) from Escherichia coli, a member of the ribokinase family of enzymes (2), is a medium-sized homodimer, composed of 33-kDa subunits, which show a complex topology. Each subunit is composed of a large domain based on the architecture of a α/β/α sandwich and a four-stranded β-sheet that covers the active site as a lid. The two lids of the dimer are packed together to form an orthogonal β-barrel—an intertwined structure, stabilized by a single hydrophobic core that includes residues from both subunits, called a “β-clasp” (3). Thus, the overall architecture of Pfk-2 and its close homologs can best be described as a homodimer composed of two α/β/α domains and one bimolecular domain (the β-clasp) that unites them (see Fig. 1). The complexity of the fold is further enhanced by the fact that the large domain is discontinuous, i.e., it is not formed by a contiguous stretch of polypeptide chain, but instead has a small four-stranded β-sheet inserted within it.
Figure 1.
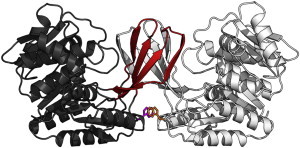
(Ribbon representation) Homodimeric structure of Pfk-2 (PDB:3CQD). For a single subunit, the five β-strands are located in the β-clasp interface (red); large domain (gray). (Sticks) Side chain of a single tryptophan residue (W88) on the interface between subunits, used for intrinsic fluorescence measurements. Figure was prepared with the software PyMol (www.pymol.org).
We have studied how the folding and subunit association of Pfk-2 occurs, looking for equilibrium or transient intermediates accumulated during guanidine hydrochloride (GdnHCl)-induced unfolding or folding experiments. No compact monomers were detected under equilibrium conditions (4) or during folding (5), supporting the fact that the β-clasp and an important portion of the α/β/α domain do not seem to behave independently with respect to their conformational stability and structural integrity. However, global unfolding transitions induced by chaotropic agents can hide intermediates or low-energy structures populated under native conditions, as has been shown by hydrogen-exchange experiments (6).
Given a large and positive ΔCp value, usually observed for the overall unfolding of a polypeptide chain (7,8), the thermal stability of proteins is dominated by concave free-energy dependence with temperature. Accordingly, the Gibbs-Helmholtz equation for a two-state system predicts two thermal unfolding transitions for a protein—cold unfolding and heat unfolding (9). Nevertheless, protein unfolding induced by cold has been documented for only a few proteins because nonnative species induced by cold are predicted to be populated below the freezing point of water.
Interestingly, some studies have shown that cold and heat unfolding might occur by different mechanisms because equilibrium intermediates are frequently found along the cold- but not the heat-induced transition pathway (10–15), although some proteins seem to unfold by a two-state mechanism at both temperature extremes (16–18). Freire et al. (19) have rationalized the different mechanism for both events as a natural consequence of the thermodynamics associated with exposing hydrophobic surfaces upon partial unfolding of a given structure. Within this framework, cold unfolding is proposed to be a special kind of perturbation able to probe the presence of low-energy substructures populated under native conditions but hidden upon heat or chemical perturbation.
In this work, the reversible thermal unfolding of Pfk-2 was characterized under equilibrium conditions. Strikingly, in addition to the typical heat-induced transition, Pfk-2 shows a clear cold-induced transition which is completed at ∼3°C. Different from several other proteins, the direct observation of a cold-induced transition in Pfk-2 did not require such additional perturbations as chaotropic agents (11,20), high pressure (21), extreme pH (22), or point mutations (16), typically used to decrease the protein stability to allow us to observe cold unfolding transitions above the freezing point of water. Cold-induced unfolding of Pfk-2 seems to be a highly cooperative process significantly disrupting the dimer structure without generating additional intermediates, as was also found to be the case in chemical unfolding.
Materials and Methods
Purification and storage of Pfk-2
Pfk-2 was purified and stored as described by Babul (23). At least four independent preparations of Pfk-2 were used in this work. The specific activity of Pfk-2 (80–100 U/mg) was used to check the quality of the protein preparations. Previous to the unfolding or refolding experiments, the storage buffer was changed to standard buffer (50 mM Tris pH 8.2, 5 mM MgCl2, and 2 mM dithreitol) by using a HiTrap desalting column (Amersham Biosciences, Uppsala, Sweden). The enzyme was concentrated using a Centricon-60 concentrator (Amicon, Beverly, MA) and further concentration was achieved using a Microcon Ultracel YM-10 unit (Millipore, Billerica, MA). Protein concentration was determined by the Bradford assay (24) using bovine serum albumin as the standard and is expressed in terms of the monomer concentration. Pipette tips, cuvettes, and all material necessary for the manipulation of the enzyme were equilibrated at the working temperature before withdrawing the protein from the Eppendorf tube.
Circular dichroism measurements
Circular dichroism (CD) spectroscopy was performed using a model No. J-810 instrument (JASCO, Essex, UK). The spectra were measured in the far ultraviolet region, from 260 to 190 nm. Cuvettes with optical paths of 0.1, 1, and 10 mm were employed depending on the protein concentration used. Each spectrum was obtained from the accumulation of at least three scans at the working temperature.
Intrinsic and extrinsic fluorescence measurements
Measurements were made using a model No. PC-5031 spectrofluorimeter (Shimadzu, Kyoto, Japan). Protein samples at several different GdnHCl concentrations or temperatures were excited at 295 nm and emission spectra were recorded from 300 to 480 nm. The concentration of the stock 8-anilino-1-naphthalene sulfonic acid (ANS) solution (Molecular Probes, Eugene, OR) was determined using a ε = 7800 M−1 cm−1 at 372 nm in methanol. Pfk-2 samples unfolded at different GdnHCl concentrations were incubated in 100 μM ANS for 3 h in the dark at 3 or 20°C. The mixture was excited at 380 nm and the emission spectra recorded from 400 to 580 nm.
Sedimentation experiments
Sedimentation velocity experiments were conducted on an Optima XL-I analytical ultracentrifuge (Beckman-Coulter, Palo Alto, CA) equipped with an An-60 Ti rotor. Experiments were performed at 25 and 4°C using a rotor speed of 41,000 rpm and a protein concentration of ∼40 μM. Pfk-2 was incubated at 3°C for 24 h in standard buffer (using tris(2-carboxyethyl)phosphine instead of dithreitol as the reducing agent) and the centrifuge, rotor, and centerpieces were cooled down to 4°C before centrifugation. The moving boundary was monitored by absorbance optics at 280 nm. Sedimentation velocity data were analyzed using the software program SEDFIT (25) and the continuous size-distribution model was chosen to generate the sedimentation coefficient distribution plots (25). It is worth noting that this model can be applied to interacting molecules if the reaction is slow on the timescale of sedimentation and if the species are stable during sedimentation.
Dynamic light scattering measurements
Dynamic light scattering (DLS) measurements were performed on a DynaPro-MS800 instrument (Protein Solutions, Chicago, IL). Before starting the experiment, the solution was centrifuged for 10 min at 14,000 rpm in an Eppendorf microfuge to remove any particulate matter. Cold-induced transitions were obtained by increasing the temperature of the sample, previously unfolded at 3°C, using a protein concentration of 30 μM. The hydrodynamic radius was calculated via the Stokes-Einstein equation from the diffusion coefficient, which was obtained from the measured autocorrelation function, using the DYNAMICS software supplied with the instrument (Protein Solutions).
Unfolding induced by GdnHCl and low temperatures
Cold unfolded Pfk-2 was diluted at various GdnHCl concentrations and left for 24 h to reach equilibrium at 3°C. Each property, intrinsic and extrinsic fluorescence, circular dichroism (CD), and dynamic light scattering (DLS), was measured at 3°C in the presence of different GdnHCl concentrations. Then, the sample temperature was increased to 20°C, until reaching a stable signal for the measured properties (at least 6 h) to get equilibrium conditions.
Model analysis of the transitions obtained by varying the GdnHCl concentration or temperature
The stability curves obtained in the presence of GdnHCl were analyzed according to a two-state I↔U or three-state N2↔2I↔2U model (4).
The dependence of ΔG on the temperature is given by the Gibbs-Helmholtz equation,
where ΔH(Tg) is the enthalpy of the reaction at Tg (the temperature for which ΔG(T) is zero) and T is the measured temperature. ΔCp is the change of specific heat of the reaction assumed to be constant in the temperature interval studied (9). For each temperature, ΔG(T) was calculated using a bimolecular two-state model of unfolding N2↔2I. Additional relationships between the observed dichroic signal at 220 nm and ΔG(T) were derived as indicated in several publications (18,26).
Results
Thermal unfolding of Pfk-2
Fig. 2 shows the temperature dependence of the CD signal of Pfk-2 at 222 nm. Clearly, the enzyme undergoes two thermal transitions at low (cold-unfolding) and elevated (heat-unfolding) temperatures within a short temperature interval (3–53°C). These transitions were reversible and were obtained under equilibrium conditions (see section S1 in the Supporting Material). As should be noted, cold and heat unfolding of Pfk-2 were captured completely because both transitions end with a substantial decrease of the CD signal at 222 nm defined by a clear nonnative plateau below 4°C. The observation of this behavior is uncommon in the absence of additional perturbations such as chaotropic agents (11,20), high pressure (21), extreme pH (22), or point mutations (16).
Figure 2.
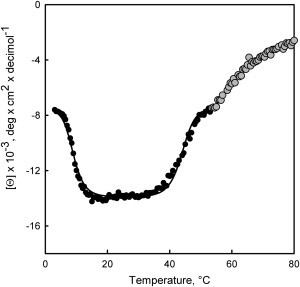
Cold and heat transitions of Pfk-2. The mean-residue molar ellipticity obtained at 220 nm is plotted as a function of the temperature. Above 53°C, heat unfolding becomes irreversible (shaded symbols). To reach equilibrium conditions, spectra were taken by increasing the temperature from 3°C at a velocity of 2°C/h. Representative results of three independent experiments are presented.
As shown in Fig. 3 A, the CD spectra taken between 3 and 23°C show a clear isosbestic point at ∼207 nm, suggesting that cold unfolding of Pfk-2 is a two-state process. However, the CD spectra taken during the heat-induced transition did not show a clear isosbestic point (Fig. 3 B), which could be due to irreversible reactions. The refolding yield after heating the protein to 53°C was 80% (see section S2 in the Supporting Material), after which temperature, the thermal unfolding becomes completely irreversible (Fig. 2, shaded symbols; Fig. 3 B, shaded lines).
Figure 3.
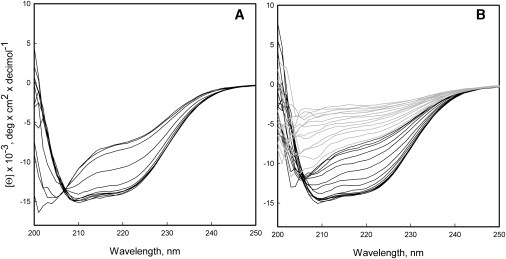
Cold and heat denaturation of Pfk-2 followed by circular dichroism (CD). CD spectra obtained from 3 to 23°C (A) and from 23 to 80°C (B). In panel B, the CD spectra obtained above 53°C are shown (shaded representation) where heat denaturation of Pfk-2 becomes irreversible.
GdnHCl-induced dissociation of Pfk-2 is coupled with an extensive unfolding of the subunits (4,28) so the effect of the protein concentration on both cold and heat transitions was determined. As indicated in Fig. 4, increasing the protein concentration shifts the cold-induced transition toward lower temperatures and the heat-induced transition toward elevated temperatures (inset, Fig. 4). This behavior is expected for a dissociative equilibrium and has been reported for other dimeric proteins (18,29) supporting the fact that both transitions involve subunits dissociation.
Figure 4.
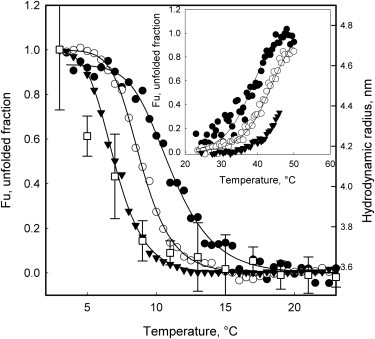
Effect of the protein concentration on heat- and cold-induced transitions and variation of Rh as measured by DLS. (Main graph) Fraction of unfolded protein within the cold temperature regime as estimated from the CD signal at 220 nm for several protein concentrations: 0.36 μM (●), 3.6 μM (○), and 45 μM (▾). (Inset) Effect of protein concentration on the heat denaturation transition of Pfk-2 obtained at 0.36 μM (●), 3.6 μM (○), and 30 μM (▾). The presence of visible aggregates prevented the evaluation of the entire heat transition using 30 μM of protein (▾, inset). (Main graph, □) Hydrodynamic radius estimated by DLS as a function of the temperature. Rh values were taken by increasing the temperature in 2°C steps after equilibration for 1 h of a 30 μM sample previously unfolded at 3°C.
Analytical ultracentrifugation measurements performed at 4°C confirm that Pfk-2 is dissociated at lower temperatures. As shown in Fig. 5, the sedimentation coefficient of Pfk-2 at this temperature is nearly half that observed at 25°C, although there is no complete dissociation. This can be seen from the presence of a small peak that matches the sedimentation coefficient for the dimer and accounts for 18% of the total signal (Table 1).
Figure 5.
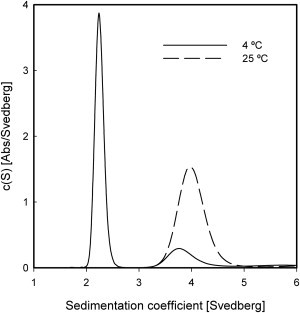
Dissociation of Pfk-2 induced by cold. Sedimentation velocity experiments were performed at 4°C (continuous line) and 25°C (dotted line) and protein samples were spun at 41,000 rpm. Pfk-2 was incubated on ice for 24 h before centrifugation. Sedimentation coefficient distributions were generated using a continuous size-distribution model.
Table 1.
Fractional population of the species observed by sedimentation velocity
| Monomer (%) | Dimer (%) | Other signals (%) | |
|---|---|---|---|
| 25°C | - | 85 ± 2 | 15 ± 2 |
| 4°C | 74 ± 2 | 18 ± 3 | 8 ± 2 |
Relative amounts of all species were calculated by integration of the area under each peak. The errors represent the standard deviation calculated from three independent experiments.
To gain further insight into the hydrodynamic changes that occur on cold treatment, the hydrodynamic radius of Pfk-2 was measured by DLS at several temperatures using a protein concentration of 30 μM (Fig. 4). As can be seen, the cold-induced transition revealed by CD was accompanied by a substantial increase to the hydrodynamic radius (Rh) reaching values of 4.6 ± 0.3 nm at 3°C.
Thermodynamic analysis
Taking into account the isosbestic point of the cold-induced unfolding together with the protein concentration dependence of the observed thermal transitions, the Gibbs-Helmholtz equation for a dissociative two-state system (see Materials and Methods) was used to fit both the heat- and cold-induced transitions. The continuous line in Fig. 2 represents the best fit for a dissociative two-state mechanism of unfolding operating at low and high temperatures. Nevertheless, there is a deviation at high temperatures, suggesting that a two-state model could be too simple.
The thermodynamic parameters are shown in Table 2 and were used to build the stability curve of Pfk-2 (Fig. 6). It is worth noting that the stability of Pfk-2 at 20°C predicted by the adjustment (∼11 kcal/mol) is similar to previous estimates calculated from the dissociation and unfolding step N2↔2I obtained in the presence of GdnHCl (12 ± 0.6 kcal/mol (4)). As expected from the large curvature of the stability curve (Fig. 6), thermal unfolding of Pfk-2 is dominated by a large ΔCp value (∼7.8 kcal/mol/°K), one of the largest reported in literature to date for a protein (30).
Table 2.
Thermodynamic parameters for the thermal unfolding of Pfk-2
| ΔH at Tg = 57°C∗ | 247 ± 10 kcal/mol |
| ΔH at Tg = 0°C† | −206 ± 10 kcal/mol |
| ΔCp‡ | 7.8 ± 0.5 kcal/mol |
| ΔASA | 56,021 Å2 |
| ΔCp§ | 7.8–10.0 kcal/mol |
Values where the ΔG is zero at high temperatures.
Values where the ΔG is zero at low temperatures.
Measured from the experimental data.
Expected values calculated from the literature parameterization of ΔASA. Standard errors were obtained from the fit procedure.
Figure 6.
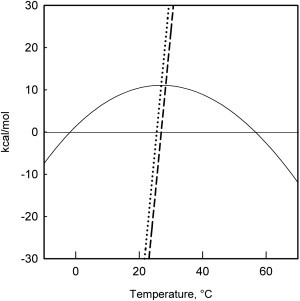
Stability curve of Pfk-2. (Curved continuous line) Free-energy dependence for bimolecular equilibrium calculated from variation of the enthalpies (dotted line) and entropy (dashed line) with the temperature using the thermodynamic parameters of Table 2.
GdnHCl-induced unfolding of the cold denatured state of Pfk-2
Further insights about the structural properties of the cold unfolded state of Pfk-2 were obtained by comparing the GdnHCl-induced unfolding transitions starting from the native dimer at 20°C (Fig. 7, solid symbols) or from the cold unfolded state at 3°C (Fig. 7, open symbols). GdnHCl-induced transitions obtained from the heat-unfolded state were not pursued, due to the presence of aggregates that formed during the large incubation times required to perform these experiments.
Figure 7.
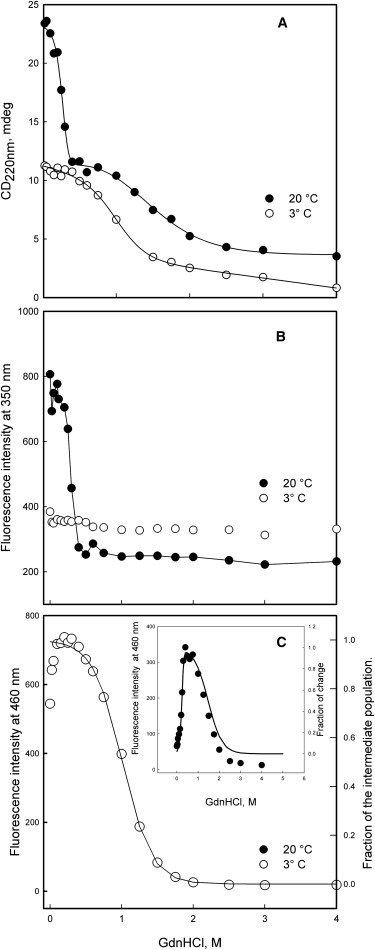
Unfolding of Pfk-2 induced by GdnHCl. Protein samples were incubated at 3°C (○) or at 20°C (●) at several GdnHCl concentrations: CD measurements (A), intrinsic fluorescence intensity (B), and fluorescence intensity of the hydrophobic probe ANS (C). (Continuous lines in panel A) Fit to a N2↔2I↔2U mechanism at 20°C and to a I↔U mechanism at 3°C. (Continuous lines in panel C) Variation of the intermediate species population predicted at 3°C (main graph) and 20°C (inset) using the thermodynamic values obtained by CD (see Table 3). The protein concentration was 3.6 μM. For comparison purposes, the sign of the CD signal was inverted.
Under equilibrium conditions, GdnHCl-induced unfolding of the native dimer has been described by a three-state equilibrium process (N2 ↔ 2I ↔ 2U) characterized by a poorly structured and expanded monomeric intermediate, “I.” The presence of a monomeric intermediate can be deduced from the two transitions separated by a wide plateau when the secondary structure change is monitored by CD at 222 nm (Fig. 7 A, solid symbols). The sharper CD transition observed at low GdnHCl concentrations represents the N2↔2I step whereas the second one represents the I↔U step of unfolding (4). This intermediate was not observed during the GdnHCl-induced unfolding at 3°C, which shows a unique transition whose amplitude and shape resemble that of the I↔U step measured at 20°C. Indeed, the native baseline measured by CD at 3°C was coincident with the plateau that separates both transitions at 20°C (Fig. 7 A), suggesting that the cold-induced species may be similar or identical to the intermediate “I.”
The similarity of the intermediate I to the cold-induced state is also supported by the GdnHCl-induced changes of the Trp-88 fluorescence and extrinsic fluorescence of ANS measured at 3°C. The monomeric intermediate of Pfk-2 must have a nonnative environment of its single Trp similar to that of the fully unfolded state (4) because the equilibrium transition observed by intrinsic fluorescence at 20°C fails to reproduce the I↔U step but describes quite well the sharp N2↔2I transition (Fig. 7 B, solid symbols). In agreement with this behavior, the intrinsic fluorescence of Pfk-2 measured at 3°C did not show major changes as a function of the GdnHCl concentration (Fig. 7 B, open symbols) suggesting that the intermediate induced by cold also lacks the native environment of Trp-88.
Another feature of the monomeric intermediate is the presence of solvent-exposed hydrophobic regions (4). The extrinsic fluorescence of the hydrophobic probe ANS (31) shows an asymmetrical bell-shape curve that follows the fractional population variation of the monomeric intermediate deduced from the three-state equilibrium (N2↔2I↔2U) at 20°C (Fig. 7 C, inset, solid symbols). Conversely, the ANS intensity measured at 3°C shows a decrease as the GdnHCl concentration increases, suggesting that the cold-induced intermediate has solvent-exposed hydrophobic regions in the absence of GdnHCl (Fig. 7 C, open symbols). The fractional population change of Pfk-2, extracted upon fitting the single CD transition observed at 3°C to a two-state mechanism (Table 3), is in good agreement with this observation and matches quite well with the decrease of the ANS intensity induced by GdnHCl at 3°C (Fig. 7 C, open symbols).
Table 3.
Thermodynamic parameters for the chemical unfolding of Pfk-2 at 20 and 3°C
| 20°C |
3°C |
||
|---|---|---|---|
| N2↔2I | I↔U | I↔U | |
| ΔG | 12.1 ± 2.1 | 1.7 ± 1.5 | 2.7 ± 0.8 |
| m | 18 ± 7 | 1.3 ± 0.4 | 2.6 ± 0.7 |
Values of ΔG in kcal/mol and m-values in kcal/mol/°K. Standard errors were obtained from the fit procedure.
Discussion
Cold- and heat-induced unfolding of Pfk-2 resembles the GdnHCl-induced unfolding/dissociation step
As indicated in Fig. 2, the amplitudes of both transitions (cold and heat-induced) are similar in terms of the molar ellipticity, suggesting that the cold- and heat-denatured states of Pfk-2 share some properties in terms of their secondary structure content. Moreover, both transitions are sensitive to the protein concentration, indicating that unfolding occurs concomitantly with dimer dissociation. Sedimentation measurements confirm this fact by demonstrating the presence of a monomer at low temperature with an Rh value of 4.6 ± 0.3 nm as measured by DLS. Taking into account that a fully unfolded chain of 33 kDa is expected to have an Rh value of ∼5.3 nm (32), we conclude that the cold-induced transition produces largely unfolded subunits. Rh values of several proteins obtained by NMR measurements have shown that cold-induced unfolding can be accompanied by an expansion of the polypeptide chain reaching values close to the fully unfolded polypeptide in GdnHCl (33).
However, cold unfolding of Pfk-2 does not lead to full unfolding. Treatment of the cold-unfolded state of Pfk-2 with GdnHCl generates an additional sigmoidal transition when monitored by CD (Fig. 7 A) whose amplitude was similar to that which describes the I↔U equilibrium observed at 20°C. The CD transition induced by GdnHCl at 3°C was also accompanied by a large decrease of the ANS fluorescence without changes to the intrinsic fluorescence of Pfk-2. These characteristics match those described for the expanded intermediate “I” characterized under equilibrium conditions in the presence of GdnHCl at 20°C (4), but there are some differences with respect to the fluorescence obtained upon ANS binding. The amplitude of the ANS fluorescence at 3°C was larger than the amplitude of the I↔U step measured at 20°C. These changes cannot be attributed to the temperature dependence of the ANS fluorescence because, in absence of protein, the quantum yield and maximum of emission of the probe remained unaltered (data not shown). Hence, because the overall properties of the monomeric state observed at 3°C seem similar to those of the intermediate “I” measured at 20°C, it is proposed that the affinity of the probe for hydrophobic regions of the protein is increased at lower temperatures.
From a thermodynamic point of view, the two-state fit to the thermal unfolding curves of Pfk-2 predicts free energy values of ∼11 kcal/mol at 20°C, which is similar to that calculated from the dissociation and unfolding step N2↔2I obtained in the presence of GdnHCl (12 kcal/mol ± 0.6 (4)). Nevertheless, this analysis should be taken with caution because the unfolding transition induced by heat shows deviations from a two-state mechanism. Although we cannot discard the presence of intermediates upon heat denaturation, it is plausible that side reactions leading to irreversible aggregates could account for the absence of an isosbestic point during the heat transition. Therefore, from a thermodynamic and structural point of view, cold-induced unfolding of Pfk-2 resembles the cooperative dissociation and unfolding represented by the N2↔2I equilibrium characteristic of the effect of GdnHCl.
Why is Pfk-2 unfolded upon cold treatment?
From the classic thermodynamic point of view, cold denaturation of proteins results as a natural consequence of a large ΔCp value arising from the hydrophobic surfaces exposed to the solvent upon protein unfolding (7,8). This value has been parameterized in terms of the polar and hydrophobic contributions for a particular solvent-exposed protein surface (see Spolar et al. (30), Myers et al. (34), and Whitten et al. (35), and references therein). The Pfk-2 dimer hides a surface area of 56,021 Å2 upon folding (see Materials and Methods), which corresponds to a ΔCp value between 7.8 and 10 kcal depending on the parameterization used (see Spolar et al. (30), Myers et al. (34), and Whitten et al. (35), and references therein). Parameterized values are in agreement with the observed ΔCp of 7.8 kcal/mol/°K obtained with the assumption of a two-state mechanism for the thermal unfolding of Pfk-2 (Table 2). Also, there are no unusual thermodynamic values obtained from the cold- and heat-denaturation processes, as the enthalpies calculated at some temperatures (e.g., 110°C, 650 kcal/mol) can be predicted adequately using the parameterization of Freire (110°C, 760 kcal/mol; see Whitten et al. (35) and references therein). Therefore, the simplest explanation for the observed cold unfolding of Pfk-2 is the large size of the cooperative unit, i.e., almost the entire dimeric structure.
The large size of the cooperative unit of Pfk-2 can be rationalized from within the framework of the hierarchic model (see Freire et al. (19) and Whitten et al. (35), and references therein). As pointed out by Freire et al. (19), cold denaturation can be seen as a special kind of perturbation simply because the intrinsic stabilities of different domains or parts of the protein structure and their coupling energy of interaction vary in such a way as a function of temperature. Interaction among their cooperative units is favorable at high temperatures yet unfavorable at low temperatures. Thus, to show unbiased cold unfolding, the unfolding coupling energy among several parts of the Pfk-2 structure should remain larger than their intrinsic stabilities either at low or high temperatures. In the absence of quantitative knowledge about the energies of coupling and intrinsic stability, an unbiased cold-unfolding mechanism can be best explained by a mutual constraint imposed between the β-clasp and the α/β/α domain. Such domain is created by an interrupted chain connectivity of both domains (discontinuous domains) together with the intertwined nature of the β-clasp that stabilizes both subunits (see Fig. 1).
Acknowledgments
We thank Richard C. Garratt for critical reading of the manuscript and Avram Slovic for technical assistance. CD measurements and light-scattering experiments were performed at the Grupo de Cristalografía, Instituto de Física de São Carlos, Universidade de São Paulo, Brazil. Sedimentation velocity experiments were performed at the Biophysics Facility, Department of Chemistry and Biochemistry, University of California, San Diego, CA.
This work was supported by a grant from the Comisión Nacional de Investigación Científica y Tecnológica (CONICYT), Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT) No. 1090336, Chile. Christian A. M. Wilson was a recipient of a CONICYT graduate fellowship, a Beca de Apoyo a Tesis Doctoral award (No. 24090160), and a travel prize from Fermelo Chile S.A. César A. Ramírez-Sarmiento was the recipient of a CONICYT graduate fellowship, a joint Mejoramiento de la Calidad y Equidad de la Educación and Sciences, Technology and Innovation Program for the Americas fellowship No. UCH0717, and was partially supported by a FONDECYT grant.
Footnotes
Christian A. M. Wilson’s present address is QB3 Institute, University of California at Berkeley, Berkeley, CA.
Supporting Material
References
- 1.Warringer J., Blomberg A. Evolutionary constraints on yeast protein size. BMC Evol. Biol. 2006;6:61. doi: 10.1186/1471-2148-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabrera R., Babul J., Guixé V. Ribokinase family evolution and the role of conserved residues at the active site of the PfkB subfamily representative, Pfk-2 from Escherichia coli. Arch. Biochem. Biophys. 2010;502:23–30. doi: 10.1016/j.abb.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 3.Cabrera R., Ambrosio A.L., Babul J. Crystallographic structure of phosphofructokinase-2 from Escherichia coli in complex with two ATP molecules. Implications for substrate inhibition. J. Mol. Biol. 2008;383:588–602. doi: 10.1016/j.jmb.2008.08.029. [DOI] [PubMed] [Google Scholar]
- 4.Baez M., Babul J. Reversible unfolding of dimeric phosphofructokinase-2 from Escherichia coli reveals a dominant role of inter-subunit contacts for stability. FEBS Lett. 2009;583:2054–2060. doi: 10.1016/j.febslet.2009.05.034. [DOI] [PubMed] [Google Scholar]
- 5.Baez M., Wilson C.A.M., Babul J. Folding kinetic pathway of phosphofructokinase-2 from Escherichia coli: a homodimeric enzyme with a complex domain organization. FEBS Lett. 2011;585:2158–2164. doi: 10.1016/j.febslet.2011.05.041. [DOI] [PubMed] [Google Scholar]
- 6.Luque I., Leavitt S.A., Freire E. The linkage between protein folding and functional cooperativity: two sides of the same coin? Annu. Rev. Biophys. Biomol. Struct. 2002;31:235–256. doi: 10.1146/annurev.biophys.31.082901.134215. [DOI] [PubMed] [Google Scholar]
- 7.Privalov P.L., Gill S.J. Stability of protein structure and hydrophobic interaction. Adv. Protein Chem. 1988;39:191–234. doi: 10.1016/s0065-3233(08)60377-0. [DOI] [PubMed] [Google Scholar]
- 8.Dill K.A. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 9.Privalov P.L. Cold denaturation of proteins. Crit. Rev. Biochem. Mol. Biol. 1990;25:281–305. doi: 10.3109/10409239009090612. [DOI] [PubMed] [Google Scholar]
- 10.Gursky O., Atkinson D. High- and low-temperature unfolding of human high-density apolipoprotein A-2. Protein Sci. 1996;5:1874–1882. doi: 10.1002/pro.5560050913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griko Y.V., Kutyshenko V.P. Differences in the processes of β-lactoglobulin cold and heat denaturations. Biophys. J. 1994;67:356–363. doi: 10.1016/S0006-3495(94)80488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griko Y.V., Privalov P.L., Venyaminov S.Y. Cold denaturation of staphylococcal nuclease. Proc. Natl. Acad. Sci. USA. 1988;85:3343–3347. doi: 10.1073/pnas.85.10.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griko Y.V., Privalov S.Y., Venyaminov P.L. Heat and cold denaturation of phosphoglycerate kinase (interaction of domains) FEBS Lett. 1989;244:276–278. doi: 10.1016/0014-5793(89)80544-7. [DOI] [PubMed] [Google Scholar]
- 14.Richardson J.M., 3rd, Lemaire S.D., Makhatadze G.I. Difference in the mechanisms of the cold and heat induced unfolding of thioredoxin H from Chlamydomonas reinhardtii: spectroscopic and calorimetric studies. Biochemistry. 2000;39:11154–11162. doi: 10.1021/bi000610b. [DOI] [PubMed] [Google Scholar]
- 15.Lopez C.F., Darst R.K., Rossky P.J. Mechanistic elements of protein cold denaturation. J. Phys. Chem. B. 2008;112:5961–5967. doi: 10.1021/jp075928t. [DOI] [PubMed] [Google Scholar]
- 16.Chen B.L., Schellman J.A. Low-temperature unfolding of a mutant of phage T4 lysozyme. 1. Equilibrium studies. Biochemistry. 1989;28:685–691. doi: 10.1021/bi00428a041. [DOI] [PubMed] [Google Scholar]
- 17.Pastore A., Martin S.R., Temussi P.A. Unbiased cold denaturation: low- and high-temperature unfolding of yeast frataxin under physiological conditions. J. Am. Chem. Soc. 2007;129:5374–5375. doi: 10.1021/ja0714538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitakuni E., Kuroda Y., Nakamura H. Thermodynamic characterization of an artificially designed amphiphilic α-helical peptide containing periodic prolines: observations of high thermal stability and cold denaturation. Protein Sci. 1994;3:831–837. doi: 10.1002/pro.5560030512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freire E., Murphy K.P., Privalov P.L. The molecular basis of cooperativity in protein folding. Thermodynamic dissection of interdomain interactions in phosphoglycerate kinase. Biochemistry. 1992;31:250–256. doi: 10.1021/bi00116a034. [DOI] [PubMed] [Google Scholar]
- 20.Mizuguchi M., Hashimoto D., Nitta K. Cold denaturation of α-lactalbumin. Proteins. 2000;38:407–413. [PubMed] [Google Scholar]
- 21.Kunugi S., Tanaka N. Cold denaturation of proteins under high pressure. Biochim. Biophys. Acta. 2002;1595:329–344. doi: 10.1016/s0167-4838(01)00354-5. [DOI] [PubMed] [Google Scholar]
- 22.Azuaga A.I., Galisteo M.L., Mateo P.L. Heat and cold denaturation of β-lactoglobulin B. FEBS Lett. 1992;309:258–260. doi: 10.1016/0014-5793(92)80784-e. [DOI] [PubMed] [Google Scholar]
- 23.Babul J. Phosphofructokinases from Escherichia coli. Purification and characterization of the nonallosteric isozyme. J. Biol. Chem. 1978;253:4350–4355. [PubMed] [Google Scholar]
- 24.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowie J.U., Sauer R.T. Equilibrium dissociation and unfolding of the Arc repressor dimer. Biochemistry. 1989;28:7139–7143. doi: 10.1021/bi00444a001. [DOI] [PubMed] [Google Scholar]
- 27.Reference deleted in proof.
- 28.Baez M., Cabrera R., Babul J. Unfolding pathway of the dimeric and tetrameric forms of phosphofructokinase-2 from Escherichia coli. Biochemistry. 2007;46:6141–6148. doi: 10.1021/bi7002247. [DOI] [PubMed] [Google Scholar]
- 29.Thompson K.S., Vinson C.R., Freire E. Thermodynamic characterization of the structural stability of the coiled-coil region of the bZIP transcription factor GCN4. Biochemistry. 1993;32:5491–5496. doi: 10.1021/bi00072a001. [DOI] [PubMed] [Google Scholar]
- 30.Spolar R.S., Livingstone J.R., Record M.T., Jr. Use of liquid hydrocarbon and amide transfer data to estimate contributions to thermodynamic functions of protein folding from the removal of nonpolar and polar surface from water. Biochemistry. 1992;31:3947–3955. doi: 10.1021/bi00131a009. [DOI] [PubMed] [Google Scholar]
- 31.Daniel E., Weber G. Cooperative effects in binding by bovine serum albumin. I. The binding of 1-anilino-8-naphthalenesulfonate. Fluorimetric titrations. Biochemistry. 1966;5:1893–1900. doi: 10.1021/bi00870a016. [DOI] [PubMed] [Google Scholar]
- 32.Uversky V.N. Use of fast protein size-exclusion liquid chromatography to study the unfolding of proteins which denature through the molten globule. Biochemistry. 1993;32:13288–13298. doi: 10.1021/bi00211a042. [DOI] [PubMed] [Google Scholar]
- 33.Li Y., Shan B., Raleigh D.P. The cold denatured state is compact but expands at low temperatures: hydrodynamic properties of the cold denatured state of the C-terminal domain of L9. J. Mol. Biol. 2007;368:256–262. doi: 10.1016/j.jmb.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 34.Myers J.K., Pace C.N., Scholtz J.M. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitten S.T., Kurtz A.J., Hilser V.J. Revealing the nature of the native state ensemble through cold denaturation. Biochemistry. 2006;45:10163–10174. doi: 10.1021/bi060855+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


