Abstract
RDL receptors are GABA-activated inhibitory Cys-loop receptors found throughout the insect CNS. They are a key target for insecticides. Here, we characterize the GABA binding site in RDL receptors using computational and electrophysiological techniques. A homology model of the extracellular domain of RDL was generated and GABA docked into the binding site. Molecular dynamics simulations predicted critical GABA binding interactions with aromatic residues F206, Y254, and Y109 and hydrophilic residues E204, S176, R111, R166, S176, and T251. These residues were mutated, expressed in Xenopus oocytes, and their functions assessed using electrophysiology. The data support the binding mechanism provided by the simulations, which predict that GABA forms many interactions with binding site residues, the most significant of which are cation-π interactions with F206 and Y254, H-bonds with E204, S205, R111, S176, T251, and ionic interactions with R111 and E204. These findings clarify the roles of a range of residues in binding GABA in the RDL receptor, and also show that molecular dynamics simulations are a useful tool to identify specific interactions in Cys-loop receptors.
Abbreviations used: nACh, nicotinic acetylcholine; AChBP, acetylcholine binding protein; GABA, gamma-aminobutyric acid; MD, molecular dynamics; RDL, resistant to dieldrin; RMSD, root mean-square displacement; RMSF, root mean-square fluctuation
Introduction
Cys-loop receptors are pentameric ligand-gated ion channels that are involved in fast synaptic neurotransmission in the central and peripheral nervous systems. Members of the vertebrate Cys-loop receptor family include nACh, GABAA, glycine, and 5-HT3 receptors (1). Such receptors also exist in invertebrates; one of the best studied is the RDL (resistant to dieldrin) receptor, which is a GABA-gated chloride channel present in many insects, and is a major target site for insecticides. Understanding the specific amino acids important for agonist binding is thus of benefit for the development of RDL-targeting insecticides, facilitating the rational design of novel and more specific insecticides, in addition to clarifying similarities and differences between the mechanisms by which GABA binds in different GABA-gated Cys loop receptors.
The agonist binding site of Cys-loop receptors is located in the extracellular region of the receptor at the interface between adjacent subunits. It is composed of six discontinuous loops (A–F). The ligand binds between loops A, B, and C on the principal subunit; and loops D, E, and F on the complementary subunit (Fig. 1). Many residues involved in GABA binding in vertebrate GABAA and GABAρ (also known as GABAC) receptors have been identified within these extracellular binding-site loops, and roles for aromatic, hydroxyl-containing, and charged residues have been determined (2). Although there is a certain degree of similarity in GABA-binding site residues across different receptors, there are differences in ligand orientation and conformation. In GABAA receptors for example GABA prefers to bind in a partially folded conformation, whereas in GABAC and RDL receptors it appears to prefer an extended conformation (3–5). Specific binding-site interactions are also not always conserved: in GABAA receptors a loop A residue forms a cation-π interaction with GABA, whereas in GABAC receptors it is a loop B residue, and in RDL receptors two residues (in loops B and C) contribute (6–8).
Figure 1.
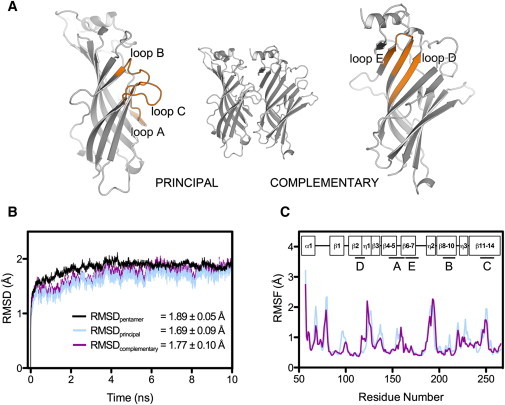
(A) Cartoon representation of the RDL-ECD dimer containing a single GABA binding site. The secondary structure elements (binding loops A–E) involved in agonist binding in RDL and other Cys-loop receptors are shown in orange. (B) Time evolution of the RMSD of protein backbone atoms for the pentameric RDL-ECD complex and constituent subunits of binding site BS2, during simulation 1. Values given (inset) represent the mean RMSD (± SD) calculated for the RDL-ECD pentamer (black), principal subunit (blue), and complementary subunit (magenta), over the 10 ns production run. (C) RMSF of protein CA atoms for the RDL-ECD principal subunit (blue) and complementary subunit (magenta), of binding-site BS2 following simulation 1. Boxes (inset) represent the secondary structure topology of a single RDL-ECD subunit relative to the residue positions. The locations of the binding regions (A–E) involved in GABA interactions are denoted by straight lines and labeled according to panel (A).
Computer-aided modeling of ligand-receptor interactions has proved helpful in understanding the nature of neurotransmitter binding to their receptor binding sites. In particular, MD simulations of structures derived from x-ray crystallography and homology modeling have allowed a quantitative description of ligand binding. Many of the earlier studies used models based on AChBP, a protein homologous to the extracellular domain (ECD) of nACh receptors, and which was the first such structure published (9). Since then many AChBP structures have been solved, and, although these proteins do not possess ligand-activated pore regions, a range of studies, including the structural determination of the nACh receptor α1 subunit and the functionality of a modified AChBP linked to a transmembrane domain (TMD), suggest that they provide a good representation of the ECD of Cys-loop receptors (10,11). The complete structures of two related bacterial ligand-gated channels, Erwinia chrysanthemi (ELIC) and Gloeobacter violaceus (GLIC), have also been published (12,13), and these have also proved useful for modeling Cys-loop receptors, although the pore of ELIC is unusual in having a bulky hydrophobic residue at the extracellular entrance. The only x-ray-derived structure of a Cys-loop receptor to date, however, is that of the glutamate-gated chloride channel from Caenorhabditis elegans (GluCl), which was cocrystallized with glutamate, ivermectin, and Fab fragments (14). Conclusions from homology models based on any of these structures, however, should be supported by experimental evidence, as none represent the perfect template: major concerns are that AChBP lacks a TMD, the bacterial channels encode neither a Cys-loop nor intracellular domain, and the presence of bulky ligands, particularly ivermectin and Fab, during GluCl crystallization, could have imposed unnatural structural restraints. Nevertheless, such models have provided a range of testable hypotheses, and have largely been shown to be reasonably accurate.
The availability of experimental structures and homology models has allowed the use of in silico methods to explore a range of Cys-loop receptor features, including ligand binding and conformational change at the extracellular domain (e.g., in AChBP (15–17), GABAC (18), nACh (19,20), and 5-HT3 receptors (21,22)), and characterization of antidepressant and anesthetic binding sites (23–30). Some of these data are supported by functional data: in the GABAC receptor, for example, MD simulations elucidated a key role for a loop D Arg stabilizing the carboxylate of GABA, and this role was confirmed using mutagenesis studies: when R104 is substituted with Ala or Glu an increase in GABA EC50 >10,000-fold was observed (18).
In previous studies on RDL receptors, we identified a range of potential residues involved in the constitution of the agonist binding site (4,7). The aim of this study was to clarify the specific molecular interactions involved in GABA binding, determining the chemical role of key amino acids involved in binding events, and to probe the spatio-temporal dynamics of ligand binding in this receptor.
Materials and Methods
Mutagenesis and preparation of mRNA and oocytes
RDL subunit cDNA was subcloned from pRmHa3-RDL into pGEMHE for oocyte expression as previously described (4,31). Site-directed mutagenesis was performed with the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). For insertion of unnatural amino acids the nonsense codon TAG was substituted at the desired location as previously described (32). The mMessage mMachine T7 kit (Ambion, Austin, TX) was used to generate capped mRNA for oocyte injection. Xenopus laevis (Nasco, WI) oocytes were prepared as previously described (8), and injected with 5 ng cRNA. After injection, oocytes were incubated for 24–48 h at 18°C.
Synthesis of tRNA and dCA amino acids
This was performed as previously described (8). In brief, unnatural amino acids were chemically synthesized as nitroveratryloxycarbonyl-protected cyanomethyl esters and coupled to the dinucleotide dC, which was enzymatically ligated to a 74-mer THG73 tRNACUA as previously described (32). Directly before coinjection with the mRNA, the aminoacyl tRNA was deprotected by photolysis (33). In a typical experiment, 10 ng of mRNA were injected with 25 ng of tRNA-aa in a total volume of 50 nl. In control experiments, mRNA was injected alone or together with THG74-dCA tRNA (with no unnatural amino acid attached).
Electrophysiology
Using a two-electrode voltage-clamp, Xenopus oocytes were clamped at −60 mV using an OC-725 amplifier (Warner Instruments, CT), Digidata 1322A, and the Strathclyde Electrophysiology Software Package (Department of Physiology and Pharmacology, University of Strathclyde, UK) or using the OpusXpress voltage-clamp system (Molecular Devices, Union City, CA). Currents were filtered at a frequency of 1 kHz. Microelectrodes were fabricated from borosilicate glass (GC120TF-10, Harvard Apparatus, Edenbridge, Kent, UK) using a one stage horizontal pull (P-87, Sutter Instrument, California) and filled with 3 M KCl. Pipette resistances ranged from 1.0 to 2.0 MΩ. Oocytes were perfused with ND96 (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 5 mM HEPES, pH 7.4). Drug application was via a simple gravity fed system or via the computer-controlled perfusion system of the OpusXpress.
Analysis and curve fitting was performed using Prism (GraphPad Software, San Diego, CA). Concentration-response data for each oocyte was normalized to the maximum current for that oocyte. The mean ± SE for a series of oocytes were plotted against agonist concentration and iteratively fitted to the following equation:
where C is the concentration of ligand present; IC is the current in the presence of ligand concentration C; Imin is the current when C = 0; Imax is the current when C = ∞, EC50 is the concentration of C, which evokes a current equal to (Imax + Imin)/2; and nH is the Hill coefficient. Significance was calculated using a one-way ANOVA.
Modeling
The crystal structure of C. elegans glutamate-gated chloride channel (GluCl; PDB ID: 3RIF (14)) was chosen as a template for building the GABA RDL extracellular domain model. A sequence alignment of GluCl (residues 1–212 [PDB numbering]) and GABA RDL (NCBI accession: NM_168321.1, residues 57–266) was produced with FUGUE (34), and used to model all five subunits simultaneously with MODELER 9v10 (35). The resulting 30 models were protonated with MolProbity (36) and ranked according to their energetic favorability (ANOLEA (37); QMEAN (38)) and stereo-chemical quality (PROCHECK (39); RAMPAGE (40)). Interatomic clashes were removed from the selected model (RDL-ECD) by 1500 steps of steepest descent minimization followed by 1000 steps of conjugate gradient minimization in a solvated, neutralized simulation box with positional restraints applied to the CA atoms.
Docking
The zwitterionic form of GABA was docked into the RDL-ECD binding site, which was defined as being within a 10 Å radius of F206, using GOLD v4.0 (CCDC, Cambridge, UK). In all 20 docking poses, the amine nitrogen of GABA was positioned consistent with the formation of a cation-π interaction with F206 and Y254, thus satisfying previous experimental criteria (7). Ten genetic algorithm runs were performed on each docking exercise using default parameters. The docking pose with the highest GoldScore fitness function was chosen for MD simulations. The structures were visualized using PyMOL v 1.3 (Schrödinger).
MD simulations
These were performed using the AMBER 2003 force field (41) and the GROMACS 4.5.4 suite of software (42). The charges of protein ionizable groups were found to be in the standard protonation state at neutral pH, as calculated using the Karlsberg webserver (43). The GABA ESP partial charges were calculated with the Density Functional Theory as implemented in the CPMD code, as previously described (18), and averaged over eight geometry configurations; the charges of equivalent atoms were also averaged. The protein-ligand complex was solvated with TIP3P water molecules in a periodically repeating truncated octahedral box. The net charge of the system was brought to neutrality and physiological ionic strength (0.15 M) by the addition of dissociated NaCl, resulting in a system comprising 38,611 water molecules, 121 Na+ ions, and 126 Cl− ions. An integration time step of 2 fs was used and all bonds were constrained using the LINCS algorithm (44). Long-range electrostatics was evaluated with the particle mesh Ewald method (45) and van der Waals forces were treated with a cutoff of 14 Å. Following 5000 steps of steepest descent energy minimization, the system was equilibrated with 100 ps of position-restrained MD under NVT conditions, followed by 100 ps under NPT conditions. Temperature and pressure were kept constant (T = 300 K, τt = 0.1 ps; P = 1 bar, τp = 2 ps) by coupling to a modified Berendsen thermostat based on stochastic velocity rescaling (46,47) and Parrinello-Rahman barostat (48), respectively. A 10 ns data production run was performed in the NPT ensemble with positional restraints imposed on the last three C-terminal CA atoms of each subunit (1000 kJ mol−1 nm−2), to mimic the presence of the TMD. The protein RMSD reached a plateau around 2 ns, and thus, ligand binding statistics were performed on the last 8 ns of the simulation. Hydrogen bonds were defined as having an acceptor-hydrogen distance ≤3.5 Å and an acceptor-donor-hydrogen angle ≤30°. Groups of opposite charge <6 Å apart were defined as ionic interactions. Cation-π interactions were identified as previously described (18): a distance cutoff (<6 Å) was applied between the GABA amine nitrogen and the center of mass of the phenyl ring, and an angle cutoff (<45°) was applied between the normal to the phenyl ring and the vector pointing from the ring center of mass to the GABA nitrogen. Cluster analysis was performed on each independent ligand trajectory (n = 20) using the gromos algorithm (49) (RMSD cutoff <1 Å), as implemented in the GROMACS program g_cluster: distance-RMSD (dRMSD) and atomic distance calculations were performed on each cluster member, data from equivalent clusters were pooled and mean (±SD) values were derived from each pool. The protein atom names used throughout correspond to the AMBER atom nomenclature and GABA atom naming is found in Fig. 2 A. Unless otherwise stated, statistics describing ligand motions and intermolecular interactions refer to the mean (±SEM) value, as averaged over all 20 independent binding sites.
Figure 2.
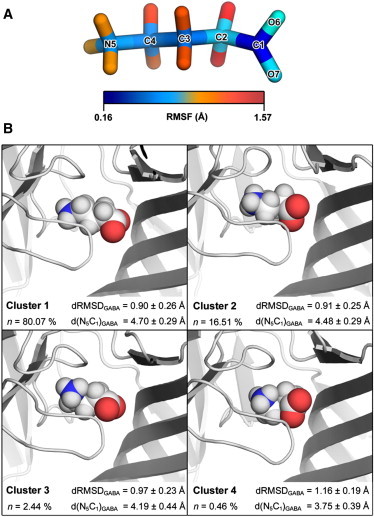
(A) Stick representation of GABA with heavy atoms color-coded to depict their RMSF, as calculated from the 10 ns simulation. (B) Cluster analysis of GABA conformations. Individual GABA structures were extracted from each 10 ns simulation trajectory (n = 20) and clustered according to their structural similarity (49). Data from equivalent clusters were pooled and the middle structure of each resultant cluster pool is presented. The degree to which GABA adopts an elongated conformation was determined from the distance between the amino nitrogen atom and carboxylate carbon atom of GABA (d(N5C1)GABA), averaged over equivalent clusters. The relative deviation of cluster members from the starting GABA conformation was determined without prior fitting and the root-square deviation of atom distances (dRMSD) were averaged over equivalent clusters. Statistics refer to the mean ± SD.
Results
Homology modeling and ligand docking
The crystal structure of the invertebrate Cys-loop receptor GluCl is the closest structural homolog of RDL currently available in the Protein Data Bank and these proteins have 38.1% extracellular domain sequence identity. This value is above the accepted 30% threshold for model reliability and is considerably higher than those for other related crystal structures (GLIC, 19.5%; ELIC, 21.0%; AChBPs, 17–20%). Because protein structure is more highly conserved at the tertiary level than at the primary level, we used an alignment generated by FUGUE (34), a program that performs profile analysis based on known sequence-structure compatibilities. Refinement of our model by energy minimization resulted in 99.7% of residues falling within the favored/allowed region of the Ramachandran plot. The number of rotamer deviations and bond angle distortions were within acceptable limits (1.5% and 0.4%, respectively), and residual bond length distortions and interatomic clashes were eliminated.
The GABA poses resulting from the docking procedure were highly clustered. In all cases, GABA was docked in a position consistent with the amine nitrogen atom of GABA making simultaneous cation-π interactions with F206 (mean distance = 5.87 ± 0.07 Å) and Y254 (mean distance 3.93 ± 0.05 Å). The mean pairwise RMSD for the GABA docking poses was 0.76 ± 0.35 Å without prior structure fitting. The positions of the amine nitrogen (N5) and carboxylate carbon (C1) atoms of GABA were largely invariant with an average displacement of 0.10 ± 0.06 Å and 0.15 ± 0.07 Å, respectively.
MD simulation
To ensure our simulations were representative, we performed four equivalent simulations and examined each of the five structurally identical binding sites. They were all broadly similar in terms of protein dynamics and bond occurrences between GABA and binding site residues, but we did observe some minor differences between the simulations, which were largest for residues R166, S176, F206, and T251, e.g., the ionic interaction between R166 and GABA was variable, as reflected in the high SEM (the mean ionic interaction occurrence was 19.3 ± 5.6%, Fig. 3). Relative to the initial energy minimized RDL-ECD structure, the RMSD of protein backbone atoms for the pentameric complex reached saturation within ∼2 ns of the simulation at ∼1.8 Å in all cases (Fig. 1 B). No significant secondary structure transitions were detected during the simulations using the program DSSP ((50); data not shown), thus further illustrating the relative stability of the model. In addition, the mean RMSD (calculated over 10 ns) for each of the five constituent subunits was within 0.22 Å of the pentameric complex in all of the simulations, suggesting that independent subunit motions had not significantly contributed to deviations from the initial structure (Fig. 1 B). We modeled RDL on a structure in which the classical agonist binding sites are occupied by agonist, and thus it is likely that our model resembles RDL in a ligand-bound state. This is supported by RMSF calculations for the protein CA atoms, which showed that the binding site loops (Fig. 1 A) underwent relatively modest atomic displacements (Fig. 1 C). Of these, the highest RMSF value of 1.47 ± 0.07 Å (averaged over 20 protein chains) corresponded to residue 250 within loop C (Fig. 1 C). This finding is not unexpected given that this loop shows considerable flexibility (see e.g. (51) and references therein).
Figure 3.
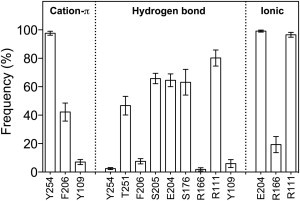
Percentage occurrence (± SEM) of interactions between GABA and binding site residues of RDL during the last 8 ns of simulation. Data were averaged over 20 independent binding sites.
Following equilibration, the position of each GABA molecule remained relatively close to its respective starting pose, with the GABA center of geometry deviating 1.9 ± 0.2 Å during the last 8 ns of the simulation, as averaged over 20 binding sites. The amine nitrogen (N5) deviated to a lesser degree (1.6 ± 0.1 Å), with the carboxylate carbon (C1) atom being positioned furthest from the initial pose (2.2 ± 0.2 Å). RMSF calculations for GABA showed that both termini of GABA were relatively stable (Fig. 2 A); mean RMSF values for N5 and C1 were 0.5 ± 0.02 Å and 0.2 ± 0.01 Å, respectively. The carboxylate moiety of GABA made hydrogen bonds with five different RDL residues (Y254, F206, S205, E204, Y109; Fig. 3), and ionic interactions with three others (E204, R166, R111; Fig. 3), thus explaining the high degree of stability at the C1 position. The carboxylate oxygen atoms underwent considerable atomic motion, and rotated on average by >90° about the C1–C2 bond for 44.4 ± 5.7% of the simulation.
Cluster analysis was performed to determine the most commonly adopted ligand conformation. As judged by the mean distance between the amino nitrogen atom and carboxylate carbon atom of GABA (d(N5C1)GABA), members of the most populous clusters (Cluster 1, n = 80.1%; Cluster 2, n = 16.5%) adopt a relatively elongated conformation (Fig. 2 B). Between these two clusters, members appear to differ with regard to the position of the carboxylate oxygen atoms about the C1–C2 bond; within each cluster, the highest degree of structural variability could be attributed to the C2 backbone carbon atom. Members of Cluster 3 and 4 deviated furthest from the starting GABA conformation, as determined from their mean RMSD values (calculated without prior fitting), and these cluster members appeared to adopt a more circularized conformation. Given that these type members are underrepresented in the simulation trajectory, it is unlikely that this type of conformation is stable.
During the simulations, the amino moiety of GABA made a number of hydrogen bonds with RDL, and an ionic interaction with E204 was evident throughout the simulations (Fig. 3). The amine nitrogen atom formed cation-π interactions with Y254 (97.6 ± 1.4%) and F206 (42.2 ± 6.3%) and, to a considerably lesser extent, with Y109 (7.0 ± 1.8%). The carboxylate group of GABA also made hydrogen bonds with a range of RDL residues, and ionic interactions with R111 (96.6 ± 1.7%) and R166 (19.3 ± 5.6%) were also detected (Fig. 3). Representative MD snapshots illustrating these interactions are given in Fig. 4.
Figure 4.
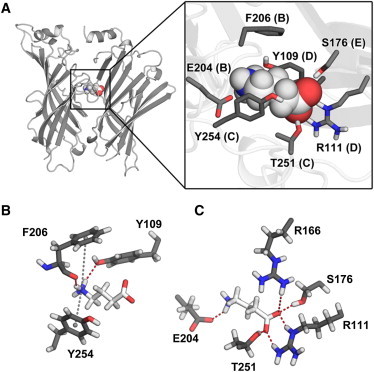
(A) Cartoon representation of the RDL-ECD binding site occupied by GABA (left) and the position of GABA interacting amino acid side chains within the binding site (inset). (B) Representative MD snapshot of the interaction between GABA and aromatic residues F206, Y109, and Y254. Gray dashed lines represent cation-π interactions and hydrogen bonds are represented by red dashed lines. Hydrogen bonds were also detected between the carboxylate oxygen atoms of GABA and the OH group of Y254 (not shown). (C) Hydrogen bonds involving nonaromatic amino acids. An additional ionic interaction is formed between the amino moiety of GABA and the carboxylate group of E204.
Specific interactions with loop A residues
The RDL homology model revealed that only one loop A residue, F146, contributes to the binding site. The simulations showed no direct F146 contacts with GABA in any of the simulations, but a persistent edge-to-face π-π interaction with F206, and a hydrophobic interaction with V148. Mutagenesis data showed that F146 is not critical for GABA binding and there is also no requirement for an aromatic residue at that position, as mutation to Ala only produced a moderate increase in EC50 (∼twofold; Table 1).
Table 1.
Parameters derived from concentration-response curves
| Loop | Mutant | pEC50 ± SE | EC50 (μM) | nH ± SE | n |
|---|---|---|---|---|---|
| WT | 4.72 ± 0.04 | 19 | 1.8 ± 0.2 | 9 | |
| D | Y109A | N/R | – | – | 6 |
| Y109F | 4.34 ± 0.04∗ | 46 | 1.7 ± 0.3 | 5 | |
| Y109S | 2.29 ± 0.10∗ | 5100 | 1.9 ± 0.3 | 5 | |
| R111A | N/R | – | – | 6 | |
| R111K | N/R | – | – | 6 | |
| A | F146A† | 4.14 ± 0.04∗ | 73 | 1.7 ± 0.2 | 3 |
| F147A | N/R | – | – | 6 | |
| E | R166A | N/R | – | – | 6 |
| R166G | N/R | – | – | 6 | |
| R166K | 4.22 ± 0.06∗ | 61 | 1.3 ± 0.2 | 4 | |
| S176A | 3.27 ± 0.06∗ | 540 | 1.6 ± 0.3 | 5 | |
| S176T | N/R | – | – | 6 | |
| R178A | N/R | – | – | 6 | |
| R178K | 4.82 ± 0.09 | 15 | 1.3 ± 0.3 | 5 | |
| B | E204A | N/R | – | – | 6 |
| E204D | N/R | – | – | 6 | |
| S205A | N/R | – | – | 6 | |
| S205T | N/R | – | – | 6 | |
| F206A | 2.88 ± 0.11∗ | 1300 | 0.7 ± 0.3 | 4 | |
| F206Y† | 6.94 ± 0.09∗ | 0.11 | 2.2 ± 0.3 | 6 | |
| Y208F | 4.15 ± 0.03∗ | 71 | 1.7 ± 0.3 | 6 | |
| Y208S | 3.84 ± 0.04∗ | 47 | 2.2 ± 0.3 | 5 | |
| C | T251A | N/R | – | - | 6 |
| T251S | 4.51 ± 0.06 | 31 | 1.2 ± 0.2 | 4 | |
| Y254F† | 4.05 ± 0.05∗ | 76 | 2.1 ± 0.4 | 5 | |
| Y254A† | N/R | – | – | 6 | |
| R256K | N/R | – | – | 6 | |
| R256A | N/R | – | – | 6 |
N/R indicates no response to 1 mM GABA.
Statistically different to wild-type (p < 0.05).
Data from (7).
We also investigated F147, as it has the potential to affect the adjacent residue. The RDL model showed that F147 points away from the binding site and makes several hydrophobic and π-π interactions within the protein interior (M89, F108 [π-π], F110 [π-π], L179, I181, I201, I203), suggesting that F147 is involved in maintaining the structural integrity of the loop A region of RDL. Construction and expression of F147A receptors yielded no GABA-induced responses, suggesting that either these receptors were not appropriately expressed, or they did not function; either of these scenarios supports our hypothesis.
Specific interactions with loop B residues
Loop B residues play important roles in a range of Cys-loop receptors, and our data indicate residues in this loop are critical for GABA binding in RDL receptors. The simulation data show H-bond (64.5 ± 4.5%) and ionic (99.1 ± 0.7%) interactions between the amino group of GABA and the carboxylate of E204 (Fig. 3). Mutagenesis data support an important role of this residue, as both conservative (Asp) and nonconservative (Ala) substitutions at E204 resulted in nonexpressed or nonfunctional receptors (Table 1).
The neighboring residue S205 had an H-bond between the carbonyl oxygen of S205 and the amino moiety of GABA (65.8 ± 3.7%); substitutions in this position to either Ala or Thr established a critical role for S205, as both mutant receptors were found to be not expressed or nonfunctional (Table 1).
Previous work has shown a cation-π interaction between GABA and F206. Here, the MD data revealed such an interaction for 42.2 ± 6.3% of the simulations. There was also an H-bond between the GABA amino group and carbonyl oxygen of F206 (7.6 ± 1.8%). As described previously, F206 formed an additional π-π interaction with F146 (100 ± 0.0%). Previous mutagenesis studies support a critical role for this residue in GABA binding and/or function: an F206A substitution results in a >200-fold increase in EC50, whereas substitution with Tyr decreases EC50 ∼15-fold (7).
The simulations showed that Y208 does not interact with GABA, but may form a hydrophobic interaction with I213 (87.0 ± 5.1%), and a π-π interaction with W141 (65.7 ± 8%). Mutagenesis data suggest that these interactions are not critical as substitution of Y208 with Phe or Ser resulted in only small changes (< fourfold) in EC50 values (Table 1).
Specific interactions with loop C residues
The simulations revealed H-bonds between T251 and the carboxylate moiety of GABA (46.7 ± 6.5%) and also between the Y254 and T251 hydroxyl groups (28.1 ± 4.3%). Mutagenesis data reveal the importance of the T251 hydroxyl as replacement of T251 with the chemically similar Ser had little effect on EC50, whereas a nonconservative replacement with Ala produced nonexpressed or nonfunctional receptors.
Previous work has shown a cation-π interaction between GABA and Y254 (7). The MD simulations supported this work in revealing such an interaction for 97.6 ± 1.4% of the time.
The simulations indicated that R256 does not interact with GABA, but does have ionic interactions with E202 (98.9 ± 0.7%) and E204 (99.5 ± 0.4%). Mutagenesis data suggest that these interactions are important for receptor function as both conservative (Asp) and nonconservative (Ala) substitutions of R256 resulted in nonexpressed or nonfunctional receptors.
Specific interactions with loop D residues
The simulations revealed multiple interactions of Y109, predominantly with other receptor residues: it formed a π-π interaction with Y90 (81.9 ± 3.7%) and a cation-π interaction with R111 (Loop D; NH1, 25.5 ± 5.4%; NH2, 47.8 ± 6.2%). For short periods Y109 also interacted with GABA via a cation-π interaction (7.0 ± 1.8%), and its hydroxyl oxygen formed a hydrogen bond (6.0 ± 2.8%) with the amino group of GABA. Mutagenesis data broadly support these observations as mutation to Ala (Y109A) abolished expression or function, mutation to Ser produced functional receptors with a >200-fold increase in EC50, and mutation to Phe revealed a small increase in EC50 (Table 1). Representative traces and concentration response curves are shown in Fig. 5.
Figure 5.
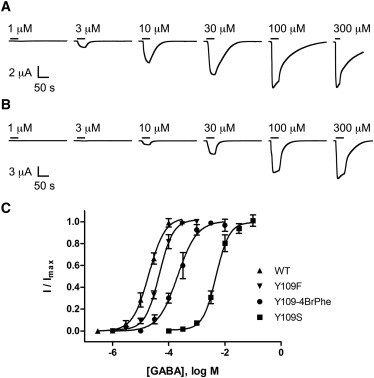
(A and B) Current traces indicating the response to application of GABA (as denoted by the horizontal black lines) in oocytes expressing WT RDL (A) and Y109F mutant receptors (B). Concentration-response curves from WT RDL and Y109F, Y109S, and Y109-4BrPhe mutants are shown in panel (C). Points represent the mean ± SEM, n = 3–5.
Data from unnatural amino acid mutagenesis experiments reveals more details (Table 2): There are increases in EC50 values with fluorinated Phes but no linear relationship with cation-π interaction energy (as observed with F206 and Y254) indicating a role of the π ring (also supported by the large increase in EC50 with cyclohexylalanine), but not a simple cation-π interaction. The difference in EC50 values for 4-Me Phe and 4-MeOPhe supports the role of a H-bond with this residue.
Table 2.
Concentration-response data for RDL with unnatural amino acids
| Mutant | pEC50 ± SE | EC50 (μM) | nH ± SE | n |
|---|---|---|---|---|
| WT | 4.77 ± 0.07 | 17 | 1.6 ± 0.3 | 7 |
| Y109F | 4.34 ± 0.04 | 46 | 1.7 ± 0.3 | 5 |
| Y109-FPhe | 3.24 ± 0.07∗ | 570 | 1.4 ±0.3 | 4 |
| Y109-F3Phe | 3.26 ± 0.04∗ | 550 | 1.6 ± 0.2 | 4 |
| Y109-4MePhe | 2.92 ± 0.09∗ | 1200 | 1.4 ±0.3 | 4 |
| Y109-4MeOPhe | 4.41 ± 0.11 | 34 | 1.9 ± 0.4 | 4 |
| Y109-4BrPhe | 3.44 ± 0.05∗ | 370 | 1.4 ± 0.2 | 4 |
| Y109-CHA | 3.05 ± 0.06∗ | 900 | 0.9 ± 0.4 | 4 |
| F206-FPhe† | 4.19 ± 0.05∗ | 65 | 1.8 ± 0.3 | 5 |
| F206-F2Phe† | 3.26 ± 0.05∗ | 550 | 1.8 ± 0.3 | 6 |
| F206-F3Phe† | 2.99 ± 0.04∗ | 1000 | 2.0 ± 0.4 | 6 |
| Y254F† | 4.05 ± 0.05∗ | 76 | 2.1 ± 0.4 | 4 |
| Y254-FPhe† | 3.99 ± 0.07∗ | 110 | 1.3 ± 0.4 | 6 |
| Y254-F2Phe† | 2.14 ± 0.08∗ | 730 | 1.1 ± 0.2 | 4 |
| Y254-F3Phe† | 1.60 ± 0.07∗ | 2500 | 1.2 ± 0.3 | 7 |
N/R indicates no response to 1 mM GABA; CHA = cyclohexylalanine.
Statistically different to wild-type (p < 0.05).
Data from (7).
R111 is another important loop D residue. The simulations revealed hydrogen bonds (80.2 ± 5.6%) and an ionic interaction (96.6 ± 1.7%) between R111 and the GABA carboxylate group. In addition, there were cation-π interactions with both Y90 (NH1, 34.6 ± 6.0%; NH2 35.0 ± 5.4%) and Y109, as mentioned previously. In support of these data, mutation of R111 to Ala or Lys resulted in not expressed or nonfunctional receptors, demonstrating the importance of this residue.
Specific interactions with loop E residues
The simulations revealed H-bonds between S176 and the GABA carboxylate moiety (63.2 ± 9.0%). Substitution of S176 with Ala showed an ∼25-fold increase in EC50, supporting these data. Interestingly, however, S176T receptors were not expressed or nonfunctional, indicating that the size and orientation of the side chain is important at this position.
R166 also interacts with GABA: the simulations revealed infrequent ionic interactions (19.3 ± 5.6%) and H-bonds (1.9 ± 1.2%) with the GABA carboxylate moiety. Although unlikely to be essential for GABA binding, the requirement for a positively charged residue at this position is exemplified by the mutagenesis data, in which substitutions for either Ala or Gly resulted in nonexpressed or nonfunctional receptors, whereas the EC50 for the R166K mutant was only moderately increased (∼threefold).
R178 is located in the model at the bottom of loop E, where it forms an ionic interaction with D107 (92.1 ± 2.3%). R187 is positioned too far from GABA to interact with it, but may be important in holding loop E in the correct position. In support of this hypothesis, substitution with Ala resulted in not expressed or nonfunctional receptors, whereas a Lys substitution produced receptors with a near wild-type EC50.
Discussion
There are a range of GABA-activated Cys-loop receptors in vertebrates and invertebrates, and similar receptors are also present in prokaryotes. Data suggest that the GABA binding sites in these receptors are broadly similar, but GABA adopts unique orientations in each. These conclusions are largely based on mutagenesis and docking studies, but previous work on GABAC and other receptors have shown that MD can be a useful tool to provide further and more detailed information about specific interactions of agonists at Cys-loop receptor binding sites. Although efforts to obtain the crystal structure of a GABA-bound Cys-loop receptor remain ongoing, homology modeling and MD simulations provide an effective and computationally affordable alternative. Such analyses also provide insight into the dynamic nature of protein-ligand interactions, a feature often not readily apparent from crystal structures. Many studies have used these techniques to probe the interactions of small ligands with their binding sites in the extracellular domain (15–30), and simulations have also revealed information on the mechanism of peptide neurotoxin action and the role of cholesterol in nACh receptor function (52–55). MD studies have also been used to probe the structural characteristics of the TMD, and as a means to elucidate the structural determinants governing channel opening, mostly in nACh receptors, and in the bacterial homologs ELIC and GLIC (56–76). Here, we have used this technique to explore the interactions of GABA in the insect GABA-activated RDL receptor, and show that many residues in the binding site contribute to GABA binding. Some of these residues are conserved with those in other GABA receptor binding sites, whereas others are not; our data therefore provide an explanation for the subtly different orientation of GABA in the RDL binding pocket compared to GABAA and GABAC receptors, and emphasizes the range of interactions that can occur, even with a relatively simple ligand, in a Cys-loop receptor binding site.
Our template for examining the RDL binding site was the ECD of the GluCl receptor, which is the only Cys-loop receptor whose x-ray crystal structure is currently available. GluCl was crystallized in the presence of ivermectin, a partial allosteric agonist, which stabilizes the agonist binding site in an activated conformation (14); thus, although our model is likely to reflect ligand interactions of an activated receptor, there may be some inaccuracies as RDL is not similarly modulated by ivermectin. Because we were concerned primarily with ligand binding, we did not consider the drawbacks of using a structure that has no TMD to be significant, and indeed studies with AChBP, which also has no TMD, suggest this is a reasonable assumption. We did consider using ELIC as a template, as this ligand-gated ion channel is gated by GABA, but the % sequence identity of these two proteins is poor (21%) compared to GluCl (38%), and the pharmacology of ELIC is quite distinct to that of GABAA receptors (77). Interestingly, however, the locations of ACh and GABA in the binding site shows some similarities with that of GABA in the RDL binding site, for example two residues contribute to cation-π interactions to stabilize the tertiary amine (78,79). The agonist binding site of RDL receptors, like all Cys-loop receptors, contains aromatic residues from different parts of the extracellular domain (loops A, B, C, and D), which form a so-called aromatic box (2,6,8,18,80–82). In RDL receptors, two of these aromatic residues interact with GABA via a cation-π interaction: a loop B Phe (F206) and a loop C Tyr (Y254) (7). Data from the MD simulations support these findings, as they reveal that both F206 and Y254 form a cation-π interaction with GABA. The data indicate Y254 is closer to GABA than F206, suggesting it has a stronger interaction. This slightly conflicts with the functional data, as fluorination plots have revealed slopes of 0.13 and 0.10 for Phe-206 and Tyr-254, respectively, indicating a marginally weaker cation-π interaction with the latter. Thus, it may be that other interactions that were not considered in the simulations also play a role; for example, in the nACh receptor the cation-π interaction (which is with the loop B Trp) is influenced by a hydrogen bond between ACh and the backbone carbonyl of this residue, and residues located immediately behind the aromatic also influence such an interaction (e.g. (82,83)).
The simulations indicated that the loop D aromatic, Tyr-109, could contribute to a cation-π interaction with GABA, but only for ∼7% of the simulations. Up to four residues have been shown to contribute to cation-π interactions in some proteins, e.g., glucoamylase (84), but in the majority of Cys-loop receptors studied to date there appears to be only one such interaction; thus RDL is unusual in having two residues contribute, and would be even more unusual if a third (Y109) was also involved. This, combined with the short interaction profile, leads us to suggest that if a cation-π occurs with Y109, it is not a significant influence on GABA binding. The simulations also indicate a range of other interactions in which Tyr-109 is involved: a π-π interaction with Y90 (81.9 ± 3.7%), a cation-π interaction with R111 (NH1, 25.5 ± 5.4%; NH2, 47.8 ± 6.2%), and an H-bond with GABA (6.0 ± 2.8%). Data from unnatural amino acid substitutions at this location show the importance of both the π ring and the hydroxyl group, and, combined with the simulation data, provide an explanation for the large decreases in potency in Y109A (nonfunctional) and Y109S (250-fold EC50 increase) mutant receptors. Mutation of the homologous residue in GABAA receptors (α1Phe-65) to Leu increased GABA EC50 from 6 to 1260 μM, with the IC50 values of bicuculline and SR95531 (competitive antagonists) increasing by similar amounts, again suggesting an important role for the residue in this location in GABA binding (85) and similarly studies using unnatural amino acids have confirmed the importance of an aromatic residue at this location, possibly by increasing the general hydrophobicity of the region (8).
Other residues in the RDL receptor that play important roles in the binding site are E204, S176, T251, and R111. The loop B Glu (E204) is especially sensitive to mutation as, even when replaced by the similarly charged Asp, the receptor is not expressed or is nonfunctional. This can be explained by the multiple roles of this residue as revealed by the simulation: an ionic interaction and a H-bond with the amino group of GABA, and an ionic interaction with R256. Mutagenesis reveals that R111 is similarly a critical residue, and the simulations revealed hydrogen bonds with both of the two carboxylate oxygens of GABA, and cation-π interactions with Y90 and Y109. S176 and T251 form H-bonds with GABA in the simulations, and their importance was also demonstrated in the mutagenesis data, even though they have essentially opposite effects when substituted conservatively or with Ala: Thr cannot be substituted for Ser at position 176 but Ser can replace Thr at position 251, whereas Ala substitutions result in nonfunctional receptors at 251, but not at 176. These data indicate the H-bond at 251 is more important, whereas the size/orientation of the residue at 176 is more critical. The simulations also revealed an interaction between loop E residue R166 and GABA, albeit for limited periods (H-bond, 1.9 ± 1.2%; ionic interaction: 19.3 ± 5.6%). The requirement for a positively charged residue in this position was confirmed by mutagenesis, however, we speculate that due to the relatively high variability in bond occurrences between the simulations, R166 may be involved in maintaining a more general charge distribution within the agonist site, rather than being directly required for GABA binding.
Hydroxylated and charged residues are also involved in GABA binding in GABAA and GABAC receptors through direct salt-bridge or hydrogen bonding interactions. In GABAA receptors the hydroxylated residues α1S68, β2T160, β2T202, β2S204, and β2S209, and the charged residues α1R120, α1D183, α1R66, and β2R207 are involved in ligand binding (86–90). A smaller repertoire of residues is involved in GABA binding in the related GABAC receptor: R104, F138, R158, and T244 (6,91–94). There are also differences in ligand orientation and conformation: In GABAA receptors GABA likely binds in a partially folded conformation, in GABAC receptors and in the RDL receptor GABA has been described as binding in an extended or elongated conformation (3–5). The current data support this and reveal specific interactions that may occur during the binding process.
Conclusions
GABA binds to GABA-activated Cys-loop receptors in a variety of different orientations. Here, we use MD simulations, combined with mutagenesis and functional data, to explore the different interactions with binding-site residues in the GABA-activated RDL receptor. The data reveal a range of interactions, predominantly with aromatic, charged, and polar residues and suggest that interactions with R111, E204, F206, Y254, and R256 are the most critical.
Acknowledgments
We acknowledge financial support from The Wellcome Trust (WT 81925 to SCRL; SCRL is a Wellcome Trust Senior Research Fellow in Basic Biomedical Science), the European Union 7th Framework Program No. HEALTH-F2-2007-202088 ("NeuroCypres" project) to SCRL; the Medical Research Council (a studentship to I.McG.), the U. S. National Institutes of Health (NS34407 to D.A.D.), and the EPSRC (EP/F037457/1 “Support for the UK Car-Parrinello Consortium”) to CM.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.Thompson A.J., Lester H.A., Lummis S.C.R. The structural basis of function in Cys-loop receptors. Q. Rev. Biophys. 2010;43:449–499. doi: 10.1017/S0033583510000168. [DOI] [PubMed] [Google Scholar]
- 2.Lummis S.C.R. Locating GABA in GABA receptor binding sites. Biochem. Soc. Trans. 2009;37:1343–1346. doi: 10.1042/BST0371343. [DOI] [PubMed] [Google Scholar]
- 3.Jones M.V., Sahara Y., Westbrook G.L. Defining affinity with the GABAA receptor. J. Neurosci. 1998;18:8590–8604. doi: 10.1523/JNEUROSCI.18-21-08590.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGonigle I., Lummis S.C.R. Molecular characterization of agonists that bind to an insect GABA receptor. Biochemistry. 2010;49:2897–2902. doi: 10.1021/bi901698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodward R.M., Polenzani L., Miledi R. Characterization of bicuculline/baclofen-insensitive (ρ-like) γ-aminobutyric acid receptors expressed in Xenopus oocytes. II. Pharmacology of γ-aminobutyric acidA and γ-aminobutyric acidB receptor agonists and antagonists. Mol. Pharmacol. 1993;43:609–625. [PubMed] [Google Scholar]
- 6.Lummis S.C.R., Beene D.L., Dougherty D.A. A cation-π binding interaction with a tyrosine in the binding site of the GABAC receptor. Chem. Biol. 2005;12:993–997. doi: 10.1016/j.chembiol.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Lummis S.C.R., McGonigle I., Dougherty D.A. Two amino acid residues contribute to a cation-π binding interaction in the binding site of an insect GABA receptor. J. Neurosci. 2011;31:12371–12376. doi: 10.1523/JNEUROSCI.1610-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Padgett C.L., Hanek A.P., Lummis S.C. Unnatural amino acid mutagenesis of the GABA(A) receptor binding site residues reveals a novel cation-π interaction between GABA and β 2Tyr97. J. Neurosci. 2007;27:886–892. doi: 10.1523/JNEUROSCI.4791-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brejc K., van Dijk W.J., Sixma T.K. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 10.Grutter T., Prado de Carvalho L., Changeux J.P. A chimera encoding the fusion of an acetylcholine-binding protein to an ion channel is stabilized in a state close to the desensitized form of ligand-gated ion channels. C. R. Biol. 2005;328:223–234. doi: 10.1016/j.crvi.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Dellisanti C.D., Yao Y., Chen L. Crystal structure of the extracellular domain of nAChR alpha1 bound to alpha-bungarotoxin at 1.94 A resolution. Nat. Neurosci. 2007;10:953–962. doi: 10.1038/nn1942. [DOI] [PubMed] [Google Scholar]
- 12.Hilf R.J.C., Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 13.Bocquet N., Nury H., Corringer P.J. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 14.Hibbs R.E., Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geitmann M., Retra K., Danielson U.H. Interaction kinetic and structural dynamic analysis of ligand binding to acetylcholine-binding protein. Biochemistry. 2010;49:8143–8154. doi: 10.1021/bi1006354. [DOI] [PubMed] [Google Scholar]
- 16.Gao F., Bren N., Sine S.M. Agonist-mediated conformational changes in acetylcholine-binding protein revealed by simulation and intrinsic tryptophan fluorescence. J. Biol. Chem. 2005;280:8443–8451. doi: 10.1074/jbc.M412389200. [DOI] [PubMed] [Google Scholar]
- 17.Stober S.T., Abrams C.F. Enhanced meta-analysis of acetylcholine binding protein structures reveals conformational signatures of agonism in nicotinic receptors. Protein Sci. 2012;21:307–317. doi: 10.1002/pro.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melis C., Lummis S.C.R., Molteni C. Molecular dynamics simulations of GABA binding to the GABAC receptor: the role of Arg-104. Biophys. J. 2008;95:4115–4123. doi: 10.1529/biophysj.107.127589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pérez E.G., Cassels B.K., Zapata-Torres G. Molecular modeling of the alpha9alpha10 nicotinic acetylcholine receptor subtype. Bioorg. Med. Chem. Lett. 2009;19:251–254. doi: 10.1016/j.bmcl.2008.10.094. [DOI] [PubMed] [Google Scholar]
- 20.Henchman R.H., Wang H.L., McCammon J.A. Ligand-induced conformational change in the alpha7 nicotinic receptor ligand binding domain. Biophys. J. 2005;88:2564–2576. doi: 10.1529/biophysj.104.053934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melis C., Chau P.L., Molteni C. Exploring the binding of serotonin to the 5-HT3 receptor by density functional theory. J. Phys. Chem. B. 2006;110:26313–26319. doi: 10.1021/jp063762a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson A.J., Lummis S.C.R. 5-HT3 receptors. Curr. Pharm. Des. 2006;12:3615–3630. doi: 10.2174/138161206778522029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanghvi M., Hamouda A.K., Arias H.R. Identifying the binding site(s) for antidepressants on the Torpedo nicotinic acetylcholine receptor: [3H]2-azidoimipramine photolabeling and molecular dynamics studies. Biochim. Biophys. Acta. 2008;1778:2690–2699. doi: 10.1016/j.bbamem.2008.08.019. [DOI] [PubMed] [Google Scholar]
- 24.Nury H., Van Renterghem C., Corringer P.J. X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature. 2011;469:428–431. doi: 10.1038/nature09647. [DOI] [PubMed] [Google Scholar]
- 25.Brannigan G., LeBard D.N., Klein M.L. Multiple binding sites for the general anesthetic isoflurane identified in the nicotinic acetylcholine receptor transmembrane domain. Proc. Natl. Acad. Sci. USA. 2010;107:14122–14127. doi: 10.1073/pnas.1008534107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Q.A., Cheng M.H., Tang P. Anesthetic binding in a pentameric ligand-gated ion channel: GLIC. Biophys. J. 2010;99:1801–1809. doi: 10.1016/j.bpj.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willenbring D., Xu Y., Tang P. The role of structured water in mediating general anesthetic action on alpha4beta2 nAChR. Phys. Chem. Chem. Phys. 2010;12:10263–10269. doi: 10.1039/c003573d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mowrey D., Haddadian E.J., Tang P. Unresponsive correlated motion in alpha7 nAChR to halothane binding explains its functional insensitivity to volatile anesthetics. J. Phys. Chem. B. 2010;114:7649–7655. doi: 10.1021/jp1009675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L.T., Haddadian E.J., Tang P. Higher susceptibility to halothane modulation in open- than in closed-channel alpha4beta2 nAChR revealed by molecular dynamics simulations. J. Phys. Chem. B. 2010;114:626–632. doi: 10.1021/jp908944e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L.T., Willenbring D., Tang P. General anesthetic binding to neuronal alpha4beta2 nicotinic acetylcholine receptor and its effects on global dynamics. J. Phys. Chem. B. 2009;113:12581–12589. doi: 10.1021/jp9039513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Millar N.S., Buckingham S.D., Sattelle D.B. Stable expression of a functional homo-oligomeric Drosophila GABA receptor in a Drosophila cell line. Proc. Biol. Sci. 1994;258:307–314. doi: 10.1098/rspb.1994.0178. [DOI] [PubMed] [Google Scholar]
- 32.Nowak M.W., Gallivan J.P., Lester H.A. In vivo incorporation of unnatural amino acids into ion channels in Xenopus oocyte expression system. Methods Enzymol. 1998;293:504–529. doi: 10.1016/s0076-6879(98)93031-2. [DOI] [PubMed] [Google Scholar]
- 33.Kearney P.C., Zhang H., Lester H.A. Determinants of nicotinic receptor gating in natural and unnatural side chain structures at the M2 9′ position. Neuron. 1996;17:1221–1229. doi: 10.1016/s0896-6273(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 34.Shi J., Blundell T.L., Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 35.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 36.Chen V.B., Arendall W.B., 3rd, Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melo F., Feytmans E. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 1998;277:1141–1152. doi: 10.1006/jmbi.1998.1665. [DOI] [PubMed] [Google Scholar]
- 38.Benkert P., Schwede T., Tosatto S.C. QMEANclust: estimation of protein model quality by combining a composite scoring function with structural density information. BMC Struct. Biol. 2009;9:35. doi: 10.1186/1472-6807-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laskowski R.A., Macarthur M.W., Thornton J.M. Procheck - a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 40.Lovell S.C., Davis I.W., Richardson D.C. Structure validation by Calpha geometry: φ,ψ and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 41.Ponder J.W., Case D.A. Force fields for protein simulations. Adv. Protein Chem. 2003;66:27–85. doi: 10.1016/s0065-3233(03)66002-x. [DOI] [PubMed] [Google Scholar]
- 42.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 43.Kieseritzky G., Knapp E.W. Optimizing pKa computation in proteins with pH adapted conformations. Proteins. 2008;71:1335–1348. doi: 10.1002/prot.21820. [DOI] [PubMed] [Google Scholar]
- 44.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 45.Cerutti D.S., Duke R.E., Lybrand T.P. Staggered Mesh Ewald: an extension of the Smooth Particle-Mesh Ewald method adding great versatility. J. Chem. Theory Comput. 2009;5:2322. doi: 10.1021/ct9001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 47.Bussi G., Zykova-Timan T., Parrinello M. Isothermal-isobaric molecular dynamics using stochastic velocity rescaling. J. Chem. Phys. 2009;130:074101. doi: 10.1063/1.3073889. [DOI] [PubMed] [Google Scholar]
- 48.Parrinello M., Rahman A. Polymorphic transitions in single-crystals: a new molecular-dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 49.Daura X., Gademann K., Mark A.E. Peptide folding: when simulation meets experiment. Angew. Chem. Int. Ed. 1999;38:236–240. [Google Scholar]
- 50.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 51.Sander T., Bruun A.T., Balle T. Docking to flexible nicotinic acetylcholine receptors: a validation study using the acetylcholine binding protein. J. Mol. Graph. Model. 2010;29:415–424. doi: 10.1016/j.jmgm.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 52.Yu R., Kaas Q., Craik D.J. Delineation of the unbinding pathway of α-conotoxin ImI from the α7 nicotinic acetylcholine receptor. J. Phys. Chem. B. 2012;116:6097–6105. doi: 10.1021/jp301352d. [DOI] [PubMed] [Google Scholar]
- 53.Yi M., Tjong H., Zhou H.X. Spontaneous conformational change and toxin binding in alpha7 acetylcholine receptor: insight into channel activation and inhibition. Proc. Natl. Acad. Sci. USA. 2008;105:8280–8285. doi: 10.1073/pnas.0710530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu R., Craik D.J., Kaas Q. Blockade of neuronal α7-nAChR by α-conotoxin ImI explained by computational scanning and energy calculations. PLOS Comput. Biol. 2011;7:e1002011. doi: 10.1371/journal.pcbi.1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brannigan G., Hénin J., Klein M.L. Embedded cholesterol in the nicotinic acetylcholine receptor. Proc. Natl. Acad. Sci. USA. 2008;105:14418–14423. doi: 10.1073/pnas.0803029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hung A., Tai K., Sansom M.S. Molecular dynamics simulation of the M2 helices within the nicotinic acetylcholine receptor transmembrane domain: structure and collective motions. Biophys. J. 2005;88:3321–3333. doi: 10.1529/biophysj.104.052878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saladino A.C., Xu Y., Tang P. Homology modeling and molecular dynamics simulations of transmembrane domain structure of human neuronal nicotinic acetylcholine receptor. Biophys. J. 2005;88:1009–1017. doi: 10.1529/biophysj.104.053421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu Y., Barrantes F.J., Jiang H. Conformational dynamics of the nicotinic acetylcholine receptor channel: a 35-ns molecular dynamics simulation study. J. Am. Chem. Soc. 2005;127:1291–1299. doi: 10.1021/ja044577i. [DOI] [PubMed] [Google Scholar]
- 59.Corry B. Theoretical conformation of the closed and open states of the acetylcholine receptor channel. Biochim. Biophys. Acta. 2004;1663:2–5. doi: 10.1016/j.bbamem.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 60.Corry B. An energy-efficient gating mechanism in the acetylcholine receptor channel suggested by molecular and Brownian dynamics. Biophys. J. 2006;90:799–810. doi: 10.1529/biophysj.105.067868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saiz L., Klein M.L. The transmembrane domain of the acetylcholine receptor: insights from simulations on synthetic peptide models. Biophys. J. 2005;88:959–970. doi: 10.1529/biophysj.104.049726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu F., Hummer G. Gating transition of pentameric ligand-gated ion channels. Biophys. J. 2009;97:2456–2463. doi: 10.1016/j.bpj.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taly A., Delarue M., Changeux J.P. Normal mode analysis suggests a quaternary twist model for the nicotinic receptor gating mechanism. Biophys. J. 2005;88:3954–3965. doi: 10.1529/biophysj.104.050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Law R.J., Henchman R.H., McCammon J.A. A gating mechanism proposed from a simulation of a human alpha7 nicotinic acetylcholine receptor. Proc. Natl. Acad. Sci. USA. 2005;102:6813–6818. doi: 10.1073/pnas.0407739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng X., Ivanov I., McCammon J.A. Nanosecond-timescale conformational dynamics of the human alpha7 nicotinic acetylcholine receptor. Biophys. J. 2007;93:2622–2634. doi: 10.1529/biophysj.107.109843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng X., Lu B., McCammon J.A. Channel opening motion of alpha7 nicotinic acetylcholine receptor as suggested by normal mode analysis. J. Mol. Biol. 2006;355:310–324. doi: 10.1016/j.jmb.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 67.Cheng X., Wang H., McCammon J.A. Targeted molecular dynamics study of C-loop closure and channel gating in nicotinic receptors. PLOS Comput. Biol. 2006;2:e134. doi: 10.1371/journal.pcbi.0020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu X., Xu Y., Barrantes F.J. Mechanics of channel gating of the nicotinic acetylcholine receptor. PLOS Comput. Biol. 2008;4:e19. doi: 10.1371/journal.pcbi.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haddadian E.J., Cheng M.H., Tang P. In silico models for the human alpha4beta2 nicotinic acetylcholine receptor. J. Phys. Chem. B. 2008;112:13981–13990. doi: 10.1021/jp804868s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Law R.J., Lightstone F.C. Modeling neuronal nicotinic and GABA receptors: important interface salt-links and protein dynamics. Biophys. J. 2009;97:1586–1594. doi: 10.1016/j.bpj.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Melis C., Bussi G., Molteni C. Trans-cis switching mechanisms in proline analogues and their relevance for the gating of the 5-HT3 receptor. J. Phys. Chem. B. 2009;113:12148–12153. doi: 10.1021/jp9046962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng X., Ivanov I., McCammon J.A. Molecular-dynamics simulations of ELIC-a prokaryotic homologue of the nicotinic acetylcholine receptor. Biophys. J. 2009;96:4502–4513. doi: 10.1016/j.bpj.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu F., Hummer G. Pore opening and closing of a pentameric ligand-gated ion channel. Proc. Natl. Acad. Sci. USA. 2010;107:19814–19819. doi: 10.1073/pnas.1009313107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nury H., Poitevin F., Baaden M. One-microsecond molecular dynamics simulation of channel gating in a nicotinic receptor homologue. Proc. Natl. Acad. Sci. USA. 2010;107:6275–6280. doi: 10.1073/pnas.1001832107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang H.L., Cheng X., Sine S.M. Intramembrane proton binding site linked to activation of bacterial pentameric ion channel. J. Biol. Chem. 2012;287:6482–6489. doi: 10.1074/jbc.M111.305839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Prevost M.S., Sauguet L., Corringer P.J. A locally closed conformation of a bacterial pentameric proton-gated ion channel. Nat. Struct. Mol. Biol. 2012;19:642–649. doi: 10.1038/nsmb.2307. [DOI] [PubMed] [Google Scholar]
- 77.Thompson A.J., Alqazzaz M., Lummis S.C. The pharmacological profile of ELIC, a prokaryotic GABA-gated receptor. Neuropharmacology. 2012;63:761–767. doi: 10.1016/j.neuropharm.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pan J., Chen Q., Tang P. Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine. Nat Commun. 2012;3:714. doi: 10.1038/ncomms1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spurny R., Ramerstorfer J., Ulens C. Pentameric ligand-gated ion channel ELIC is activated by GABA and modulated by benzodiazepines. Proc. Natl. Acad. Sci. USA. 2012 doi: 10.1073/pnas.1208208109. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bartos M., Corradi J., Bouzat C. Structural basis of activation of cys-loop receptors: the extracellular-transmembrane interface as a coupling region. Mol. Neurobiol. 2009;40:236–252. doi: 10.1007/s12035-009-8084-x. [DOI] [PubMed] [Google Scholar]
- 81.Pless S.A., Millen K.S., Dougherty D.A. A cation-π interaction in the binding site of the glycine receptor is mediated by a phenylalanine residue. J. Neurosci. 2008;28:10937–10942. doi: 10.1523/JNEUROSCI.2540-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiu X., Puskar N.L., Dougherty D.A. Nicotine binding to brain receptors requires a strong cation-π interaction. Nature. 2009;458:534–537. doi: 10.1038/nature07768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cashin A.L., Torrice M.M., Dougherty D.A. Chemical-scale studies on the role of a conserved aspartate in preorganizing the agonist binding site of the nicotinic acetylcholine receptor. Biochemistry. 2007;46:630–639. doi: 10.1021/bi061638b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aleshin A.E., Stoffer B., Honzatko R.B. Crystallographic complexes of glucoamylase with maltooligosaccharide analogs: relationship of stereochemical distortions at the nonreducing end to the catalytic mechanism. Biochemistry. 1996;35:8319–8328. doi: 10.1021/bi960321g. [DOI] [PubMed] [Google Scholar]
- 85.Sigel E., Baur R., Malherbe P. Point mutations affecting antagonist affinity and agonist dependent gating of GABAA receptor channels. EMBO J. 1992;11:2017–2023. doi: 10.1002/j.1460-2075.1992.tb05258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boileau A.J., Evers A.R., Czajkowski C. Mapping the agonist binding site of the GABAA receptor: evidence for a beta-strand. J. Neurosci. 1999;19:4847–4854. doi: 10.1523/JNEUROSCI.19-12-04847.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Amin J., Weiss D.S. GABAA receptor needs two homologous domains of the β-subunit for activation by GABA but not by pentobarbital. Nature. 1993;366:565–569. doi: 10.1038/366565a0. [DOI] [PubMed] [Google Scholar]
- 88.Newell J.G., Czajkowski C. The GABAA receptor α 1 subunit Pro174-Asp191 segment is involved in GABA binding and channel gating. J. Biol. Chem. 2003;278:13166–13172. doi: 10.1074/jbc.M211905200. [DOI] [PubMed] [Google Scholar]
- 89.Westh-Hansen S.E., Witt M.R., Nielsen M. Arginine residue 120 of the human GABAA receptor α 1, subunit is essential for GABA binding and chloride ion current gating. Neuroreport. 1999;10:2417–2421. doi: 10.1097/00001756-199908020-00036. [DOI] [PubMed] [Google Scholar]
- 90.Wagner D.A., Czajkowski C., Jones M.V. An arginine involved in GABA binding and unbinding but not gating of the GABA(A) receptor. J. Neurosci. 2004;24:2733–2741. doi: 10.1523/JNEUROSCI.4316-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Harrison N.J., Lummis S.C.R. Locating the carboxylate group of GABA in the homomeric rho GABA(A) receptor ligand-binding pocket. J. Biol. Chem. 2006;281:24455–24461. doi: 10.1074/jbc.M601775200. [DOI] [PubMed] [Google Scholar]
- 92.Harrison N.J., Lummis S.C.R. Molecular modeling of the GABA(C) receptor ligand-binding domain. J. Mol. Model. 2006;12:317–324. doi: 10.1007/s00894-005-0034-6. [DOI] [PubMed] [Google Scholar]
- 93.Zhang J., Xue F., Chang Y. Agonist- and antagonist-induced conformational changes of loop F and their contributions to the ρ1 GABA receptor function. J. Physiol. 2009;587:139–153. doi: 10.1113/jphysiol.2008.160093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Amin J., Weiss D.S. Homomeric rho 1 GABA channels: activation properties and domains. Receptors Channels. 1994;2:227–236. [PubMed] [Google Scholar]


