Abstract
Endothelial-to-mesenchymal transition (EndMT) is now widely considered a pivotal contributor to cancer progression. In this study, we show that the Kaposi’s sarcoma (KS)–associated herpesvirus (KSHV) is a sufficient cause of EndMT, potentially helping to explain the aggressiveness of KS that occurs commonly in AIDS patients. Upon KSHV infection, primary dermal microvascular endothelial cells lost expression of endothelial markers and acquired expression of mesenchymal markers, displaying new invasive and migratory properties along with increased survival. KSHV activated Notch-induced transcription factors Slug and ZEB1, and canonical Notch signaling was required for KSHV-induced EndMT. In contrast, KSHV did not utilize the TGFβ signaling pathway, which has also been linked to EndMT. Within KS lesions, KSHV-infected spindle cells displayed features compatible with KSHV-induced EndMT including a complex phenotype of endothelial and mesenchymal properties, Notch activity, and nuclear ZEB1 expression. Our results show that KSHV engages the EndMT program to increase the invasiveness and survival of infected endothelial cells, traits that likely contribute to viral persistence and malignant progression. One important implication of our findings is that therapeutic approaches to disrupt the Notch pathway may offer novel approaches for KS treatment.
Introduction
Kaposi’s sarcoma (KS)-associated herpesvirus (KSHV), also known as human herpesvirus 8, is a gamma herpesvirus discovered in tissue of AIDS-associated KS, and found to be the etiologic agent of all types of KS (1). KSHV is also linked to primary effusion lymphoma (2), a subset of multicentric Castleman’s disease (3) and an interleukin-6 (IL-6)-related inflammatory syndrome in patients with AIDS (4).
KS is a multicentric angioproliferative disease often localized in the skin (5). Histologically, KS lesions are complex (6). The characteristic KS spindle cells are elongated KSHV-infected fibroblast-like cells, which represent the main proliferating cell in the lesions. All KS lesions also contain aberrant small vessels and form slit-like leaky spaces replete with red cells. Infiltration with inflammatory lymphocytes, plasma cells, and monocytes is common (5, 6). KS lesions often transition from a stage in which inflammatory and vascular elements are more prominent to a stage in which spindle cells are more prominent (5).
The origin of the KS spindle cells has been controversial, in part, because KS cells express heterogeneous markers. Several studies have concluded that the spindle cells are of endothelial lineage, as they express the vascular endothelial cell markers CD31, CD34, CD36, and others, but the staining for endothelial markers is inconsistent (5, 6). Other studies have proposed that KS cells derive from mesenchymal precursors or mature pericytes, smooth-muscle cells or fibroblasts, as they express smooth-muscle actin (SMA; ref. 7). More recently, KS spindle cells have been thought to be of lymphatic endothelial cell lineage because they express LYVE-1, VEGFC, and VEGF-R3, markers for the lymphatic endothelium (8). The difficulty at defining the origins of KS cells is explained, in part, by studies in vitro showing that KSHV infection causes phenotypic changes in endothelial cells (9, 10). In vascular endothelial cells, KSHV promotes the expression of lymphatic markers, and, in lymphatic endothelial cells, KSHV promotes the expression of vascular endothelial markers (11, 12), suggesting that viral infection can reprogram endothelial cells.
Endothelial cells display considerable plasticity manifested by the ability to morph (transform) into mesenchymal cells. Endothelial-to-mesenchymal transformation (EndMT), similar to epithelial-to-mesenchymal (EMT) transformation, is critical to certain steps in development and contributes to disease. EndMT is required for endothelial/endocardial cells to form the mesenchymal heart cushion cells that generate portions of the heart septa and valves (13, 14). In the adult, EndMT contributes to pathologic fibrosis in the heart and kidneys (15–17), to cancer progression by generating carcinoma-associated fibroblasts (18), and to heterotopic bone formation in patients with fibrodysplasia ossificans progressiva (19).
The heterogeneity and evolution of KS tissue biopsies, the uncertainties surrounding the origin and phenotype of KS cells, as well as the observation that KSHV infection promotes change in gene expression and marker profiles raised the possibility that KSHV reprograms endothelial cells to acquire a mesenchymal phenotype. In this study, we examined the possibility that KSHV is an inducer of EndMT. We report that KSHV infection induces transcriptional, phenotypic, and functional changes in endothelial cells, consistent with their transformation into mesenchymal-like cells, and that such changes are recapitulated in KS tissues.
Materials and Methods
Virus production
rKSHV.219, a recombinant KSHV that expresses the green fluorescent protein (GFP) during latent replication and the red fluorescent protein during lytic replication were used for all experiments. The construction and production of rKSHV.219 have been previously published (20).
Cell lines
EAHY926 cells (American Type Culture Collection) were maintained in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO; Invitrogen) with 10% heat-inactivated FBS and 1 mmol/L glutamine; neonatal human Dermal Microvascular Endothelial Cells (DMVEC; Lonza) were maintained in EBM-2 basal endothelial growth medium supplemented with EGM-2MV Bullet Kit (Clonetics). Human bone marrow–derived mesenchymal stem cells (BM-MSC; Cambrex BioScience) were propagated in DMEM, low glucose (Invitrogen) with 10% FBS.
Tissues
Biopsies of cutaneous KS lesions were obtained from 8 patients (7/8 patients HIV-positive; 3 stage T0, 5 stage T1; 3 with <50 and 5 with >50 cutaneous lesions; all lesions were nodular) volunteering on protocols in the HIV and AIDS Malignancy Branch of the National Cancer Institute (NCI). Skin was obtained from autopsies of cancer patients without HIV infection or KS. The biopsies were utilized fresh or snap-frozen and kept at −80°C until utilized. The Institutional Review Board of the NCI approved the protocols; all patients gave written informed consent.
KSHV infection
rKSHV.219 (2.5 mL supernatant of virus producing VERO cells) preincubated (30 minutes, room temperature) with 2 μg/mL polybrene was added onto cells in 60-mm plates and centrifuged (1 hour; 2,500 rpm; 30°C). Viral supernatant was replaced with culture medium after 18 hours; KSHV-infected cells were selected with 0.5 μg/mL or 1 μg/mL puromycin (Invitrogen).
Cell culture
KSHV-infected endothelial cells (48 hours after infection) were cultured in DMEM low glucose, glutamine 2 mmol/L, heparin 2 mg/mL, and 10% FBS, 48 hours after infection. The γ-secretase inhibitor DAPT (N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester; Sigma-Aldrich) dissolved in dimethylsulfoxide (DMSO) (Sigma-Aldrich) was added to KSHV-infected (48 hours after infection) and noninfected endothelial cell cultures at the final concentrations of 1, 5, 8, and 10 μmol/L. DMSO (0.1%) was used as vehicle control. Fresh DAPT was added at every culture passage for the length of the 12-day culture.
Adipocyte differentiation
Confluent MSCs were subjected to 3 cycles of induction/maintenance. Each cycle consists of 3-day culture in Adipogenesis Induction medium (Lonza) followed by 1 to 3 days culture in maintenance medium. Cultures were fixed (10% buffered formalin) and stained with Oil Red O. Bright-field and fluorescence microscopy images were acquired through a Nikon Eclipse E600 microscope equipped with Plan Apo 4× and photographed with a digital camera (Retiga 1300; QImaging). Images obtained with IPLab for Windows software (Scanalytics) were imported into Adobe Photoshop (Adobe Systems).
Immunoblotting
Protein extracts for phosphorylated proteins prepared in SDS lysis buffer with protease inhibitor cocktail set III (Calbiochem), 50 mmol/L NaF, and 1 mmol/L sodium orthovanadate were resolved in NuPAGE 4% to 12% Bis-Tris Gel or 6% tris-glycine(Invitrogen) and transferred to protran membranes (Whatman GmbH). Antibodies for immunoblotting: SMA (1A4; Dako); CD31 (R&D Systems); TGFBRII, Tie-2 and actin (c11; all from Santa Cruz Biotechnology); and VE cadherin, Vimentin, PDGFRβ, SMAD3, and P-SMAD2/3 (all from Cell Signaling Technologies). Horseradish peroxidase (HRP)–conjugated rabbit anti–goat IgG-Fc was from Cal-biochem; and HRP-conjugated donkey anti–mouse IgG and donkey anti–rabbit IgG were from Amersham Pharmacia Biotech. Bound secondary antibodies were visualized by enhanced chemiluminescence (Amersham).
Immunofluorescence
DMVECs were grown on Labtek chamber slides (Nunc; Thermo Scientific), fixed (cold 4% paraformaldehyde in PBS) and stained with antibodies to Vimentin (CST); F-actin was detected with Alexa Fluor 568–conjugated antibody to phalloidin (Molecular Probes). KS biopsies (no. 8) were fixed (cold 4% paraformaldehyde in PBS), cryoprotected in 15% and 30% sucrose, embedded in OCT, and processed for histology. Sections were immunostained with combinations of the following antibodies: CD31 (R&D Systems), CD146 (BD Pharmingen), PDGFRβ (Cell Signaling Technologies) SMA (clone 1A4; Dako), ZEB-1 (clone H102; Santa Cruz Biotechnology), Hey2 (Santa Cruz Biotechnology), ZEB-2 (Bethyl Laboratories), and LANA (ABI). Secondary antibodies were combinations of spectrally compatible Alexa Fluor–conjugated antibodies (all from Molecular Probes). Nuclei were visualized with DAPI (1:2,000; Invitrogen). Confocal microscopy images were acquired using either a Zeiss LSM 510 confocal microscope (Carl Zeiss) or a Zeiss Axiovert 100M inverted microscope fitted with a 20×/0.3 NA Plan-Neofluar dry objective or a 40×/0.8 NA Plan-Neofluar oil immersion objective. Wide field fluorescence microscopy images were taken using an Axiovert 200M fluorescence microscope (Carl Zeiss) through a ×32 objective and Velocity Acquisition software (Improvision). Images were imported into Adobe Photoshop (Adobe Systems). Fluorescent images were quantified with Image Pro-Plus software (MediaCybernetics, Inc).
Cell growth assay
EAHY926 cells were seeded (24,000 cells per well; 24-well plates) in culture medium supplemented with 0.1% FBS. Growth curves were obtained from time lapse images of live cells obtained every 30 minutes over 50 hours of culture using the IncuCyte system (Essen Instruments).
Scratch assay
Confluent cultures of EAHY926 cells in 24-well plates were scratched by using standard protocols and rinsed 3 times to remove cell debris. Plates were placed in the IncuCyte system and imaged overnight every 30 minutes. Wound closure curves are constructed automatically from kinetic imaging data points. Each data point represents the average (±SD) wound closure from 3 fields per well in 3 independent wells.
Chemotaxis assay
Chemotaxis ibiTreat μ–Slides (ibidi, GmbH) consisting of 2 reservoirs (40 μL each) separated by a thin channel (for maintenance of a stable chemotactic gradient up to 48 hours) were used. Noninfected or KSHV-infected EAHY926 or DMVECs (4 × 106 and 2 × 106 cells/mL, respectively; in a volume of 6 μL) were seeded into the μ-Slides channel in medium without serum. Slides were incubated (37°C, 8 hours) to allow cell attachment. A gradient of chemoattractant was created in the channel by applying a solution of 50 ng/mL PDGF-BB (R&D Systems) or 10% FBS medium onto one reservoir. Cells were imaged continuously for 16 hours using a Zeiss microscope. Images were analyzed using ImageJ 1.38× software (Rasband, W.S., ImageJ; US NIH, Bethesda, MD). Chemotaxis plots of individual cells were generated with the Chemotaxis and Migration Tool (ibidi).
RNA, microarray analysis, and Real-time RT-PCR
RNA was extracted using TRI Reagent (Molecular Research Center). cRNA synthesis, hybridization with Affymetrix HG133A, and analyses were done by the Affymetrix Core Service in Frederick, Maryland, Center for Cancer Research, NCI (GEO accession number GSE33984). cDNA was synthesized from 1 μg total RNA (High Capacity cDNA Reverse Transcription Kit; Applied Biosystems). mRNA was measured by real-time PCR with 1 μL cDNA and TaqMan PCR Universal Master Mix (Applied Biosystems). TaqMan Gene expression probes for the human mRNAs CD31, VE cadherin, NG-2, PDGFRβ, acta2, Hey1, Hey2, and ALK2 were from Applied Biosystems. PCR reaction conditions were those recommended by the manufacturer. The mRNAs of CD34, CD146, SMA, Snail, Slug, Twist, Zeb1, and Zeb2 were measured by real-time PCR with 1 μL cDNA and power SYBER green PCR Master Mix (Applied Biosystems). Primer sequences are as follows: CD34 F: CAT CAC TGG CTA TTT CCT GAT G and CD34 R: AGC CGA ATG TGT AAA GGA CAG; CD146 F: AAG GCA ACC TCA GCC ATG TCG and CD146 R: CTC GAC TCC ACA GTC TGG GAC;ZEB-1 F: GCA CAA CCA AGT GCA GAA GA and ZEB-1 R: GCC TGG TTC AGG AGA AGA TG; ZEB-2 F: CAA GAG GCG CAA ACA AGC and ZEB-2 R: GGT TGG CAA TAC CGT CAT CC; Twist-1 F: AAG AAA CAG GGC GTG GGG CG and Twist-1 R: CCG CTG CCC GTC TGG GAA TC; SNAIL F: GCT GCA GGA CTC TAA TCC AGA and SNAIL R: ATC TCC GGA GGT GGG ATG; SLUG F: TGG TTG CTT CAA GGA CAC AT and SLUG R: GTT GCA GTG AGG GCA AGA A; SMA F: CCA CTA TTG GCA ATG AGC GC and SMA R: CCA ATG AAG GAG GGC TGG AA. Thermal cycling conditions for amplification were set at 50°C for 30 minutes and 95°C for 10 minutes, respectively. PCR denaturing was set at 95°C for 15 seconds and annealing/extending at 60°C for 60 seconds for 40 cycles. The results were quantified with the comparative Ct method (known as the 2−[delta][delta]Ct method).
Cytokines measurements
Human (h)TGFβ1 and hTGFβ2 levels were measured by ELISA (Quantikine; R&D Systems).
Statistical analysis
Group differences were evaluated by 2-tailed Student t test. P values less than 0.05 were considered significant.
Results
KSHV represses expression of endothelial markers and increases expression of mesenchymal markers in endothelial cells
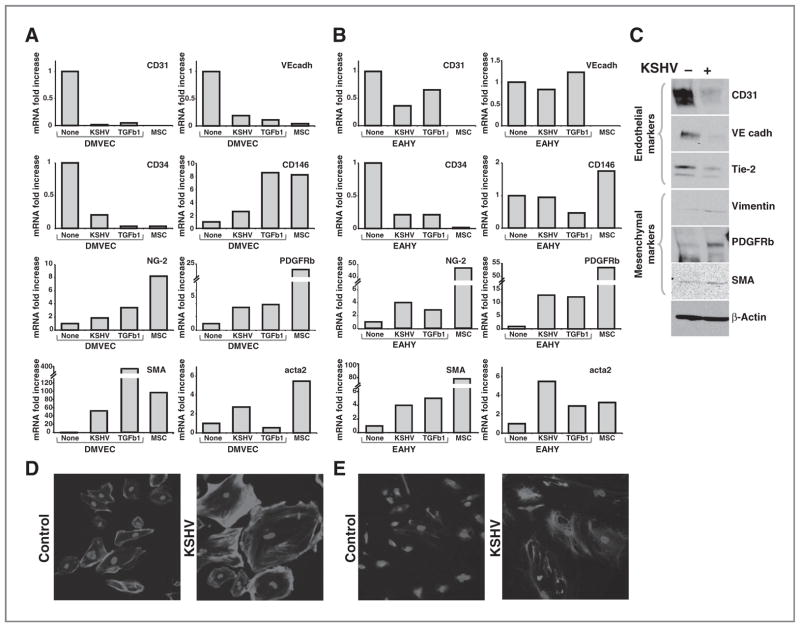
We infected primary DMVECs and the endothelial-like cell line EAHY926 (EAHY) with a recombinant KSHV (rKSHV.219) (20). After 12-day culture with puromycin selection achieving more than 90% infection (as judged by % GFP+ cells), we examined expression levels of selected endothelial and mesenchymal cell–related markers by quantitative PCR. We used as controls noninfected primary endothelial cells (DMVECs and EAHY), primary bone marrow–derived MSCs, and endothelial cells (DMVECs and EAHY) cultured in vitro with TGFβ1 (5 ng/mL; 12 days) under conditions reported to promote EndMT (18). In DMVECs (Fig. 1A, representative experiment), KSHV infection reduced mRNA levels of the endothelial markers (21) CD31, VE-cadherin, and CD34 similar to the reduction induced by TGFβ1. As a result, levels of expression of CD31, VE-cadherin, and CD34 in KSHV-infected DMVECs were comparable with those detected in MSCs. KSHV increased mRNA levels of the mesenchymal markers (21) NG-2, PDGFRβ, SMA, and acta2 (smooth muscle a-actin) in DMVECs (Fig. 1A), which was similar to that achieved by TGFβ1 stimulation or found in MSCs. We tested for expression of CD146, a progenitor-type marker expressed in MSCs isolated from multiple organs (21). We found that KSHV augments somewhat CD146 expression in DMVECs, but the mRNA levels are lower than those detected in MSCs or DMVECs cultured with TGFβ1 (Fig. 1A). Similar results were derived from EAHY cells, albeit the degree of change induced by KSHV or TGFβ was somewhat lower than in DMVECs (Fig. 1B, representative experiment). For comprehensive analysis of changes in gene expression induced by KSHV in EAHY cells, Affymetrix GeneChip arrays were used. These results (Supplementary Tables S1 and S2 and GEO accession number GSE33984) showed that KSHV alters expression of a large number of genes: downregulates expression of numerous endothelial related genes and adhesion molecules, and upregulates expression of genes linked to cell motility and cytoskeleton rearrangement.
Figure 1.
Repression of endothelial markers and upregulation of mesenchymal markers in endothelial cells infected with KSHV. Relative mRNA levels of CD31, VE-cadherin, CD34, CD146, NG-2, PDGFRB, SMA, and acta2 in (A) DMVECs and (B) EAHY926 cell line (EAHY) control (none), infected with KSHV (KSHV), or cultured with TGFβ1 (TGFβ1); mRNA levels were measured by quantitative PCR. RNAs were extracted 12 days after KSHV infection or treatment with 5 ng/mL of TGFβ1. RNA from MSCs is tested as a control. mRNA levels normalized for GAPDH are expressed as fold change relative to control cells. Experiments are representative of 8 (DMVECs) and 6 (EAHY) conducted. C, cell lysates from control and KSHV-infected DMVECs (12 days after infection) were immunoblotted for CD31, VE cadherin, Tie-2, Vimentin, PDGFRB, SMA proteins, and β-actin (loading control). The experiment is representative of 3 conducted. D, fluorescence microscopy images of uninfected DMVECs (left) and KSHV-infected (12 days postinfection) DMVECs (right) stained with phalloidin and DAPI or (E) immunostained for vimentin and DAPI; original magnification 25×. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
By immunoblotting using DMVECs, we found that KSHV infection reduces protein levels of the endothelial markers CD31, VE-cadherin, and Tie-2 and increases protein levels of the mesenchymal markers PDGFRβ, SMA, and vimentin (Fig. 1C). By confocal microscopy, we found the KSHV infection causes an increase and redistribution of polymerized actin, as revealed by phalloidin staining (Fig. 1D) and vimentin (Fig. 1E). These results indicated that the changes in gene expression induced by KSHV in endothelial cells are consistent with the occurrence of EndMT.
Functional changes in endothelial cells infected with KSHV
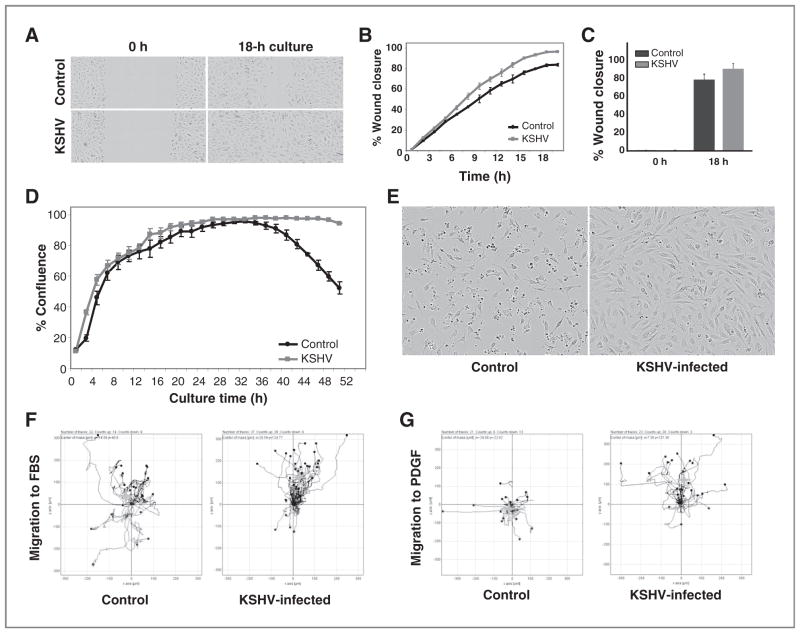
Mesenchymal cells are locally invasive and can migrate to distant sites (22); epithelial cells acquire invasive and migratory properties as they transition into mesenchymal cells (EMT; ref. 23). We examined whether endothelial cell reprogramming by KSHV is associated with increased cell motility. Using in vitro wound healing assays, we found that KSHV-infected EAHY cells are significantly more efficient than uninfected EAHY cells at closing the wound (P = 0.002 slope curve difference; P = 0.02 difference at the 18-hour time point; Fig. 2A–C). To test whether these differences are attributable to differences in cell proliferation, we compared cell growth of control and KSHV-infected EAHY cells. KSHV-infected EAHY cells grew at a similar rate as the control noninfected cells up to 30 to 32 hours of incubation (Fig. 2D), suggesting that the difference in wound closure (occurring during this time frame) is likely attributable to increased cell motility induced by KSHV. Continued observation beyond 32 hours showed that whereas control EAHY cells became significantly less (P < 0.05 at time points 43–51 hours) confluent approximately 24 hours after reaching full cell confluency, due to increased cell death, KSHV-infected EAHY maintained full cell confluency over this period and showed little morphologic evidence of death (Fig. 2D and E). This result is consistent with the observation that KSHV-infected endothelial cells lose growth contact inhibition (9) and with evidence that resistance to cell death is a characteristic trait acquired during EMT (22, 23).
Figure 2.
KSHV promotes functional change in endothelial cells. A, wound closure by KSHV-infected EAHY cells compared with uninfected EAHY cells. Images from scratch assay; original magnification 10×. B, wound closure (% wound area) as a function of time by uninfected (control) and KSHV-infected (KSHV) EAHY cells. Each data point is the average (±SD) wound closure from 3 fields per well in 3 independent experiments (±SD); C, mean relative wound closure (± SD) by control and KSHV-infected EAHY at time 0- and 18-hour incubation (3 experiments). D, growth curves of uninfected (control) and KSHV-infected (KSHV) EAHY cells over 48-hour culture (0.1% FBS) from IncuCyte imaging [expressed as % mean (±SD) confluency (3 fields per well; 3 wells; representative experiment]. E, bright-field images of uninfected and KSHV-infected EAHY (50 hours after seeding at equal density); original magnification 10×. Chemotaxis of (F) EAHY cells toward FBS (10%) and (G) DMVECs toward PDGF (50 ng/mL). Cells were seeded (IBIDI chambers) and cell movement was imaged overnight every 10 minutes. Lines reflect tracks of cells that moved away or toward the chemoattractant; grey symbol (+) represent the average movement of the cell population. Representative results from 3 experiments. Cells were used on day 10 postinfection.
We used a modified Boyden chamber assay to evaluate cell migration in response to FBS, selected as a nonspecific stimulator of cell movement, and to platelet-derived growth factor (PDGF), selected as a mesenchymal selective stimulator of cell movement (24). In response to FBS, KSHV-infected EAHY cells migrated less randomly and further toward the stimulant in comparison with noninfected control EAHY cells (Fig. 2F). In response to PDGF, KSHV-infected DMVECs migrated mostly selectively toward the stimulant, whereas control noninfected DMVECs showed no specific migration (Fig. 2G). Thus, KSHV infection promotes increased motility and resistance to cell death in endothelial cells.
MSCs have self-renewal potential and can differentiate into cells of bone, fat, and cartilage lineages (25). If KSHV can reprogram cells into mesenchymal cells, such reprogrammed cells would be expected to differentiate into bone, fat, and cartilage cells with appropriate stimulation. To test for this, we first induced endothelial cell reprogramming by infecting DMVECs with KSHV, and 12 days after infection, we induced osteogenic and adipogenic differentiation. As expected, control MSCs from bone marrow differentiated into bone and fat cells over 21 days culture; by contrast, KSHV-infected cells did not (not shown).
Previous studies showed that the KSHV nuclear protein LANA blocks differentiation of 3T3L1 cells into fat cells (26). Because KSHV-LANA is expressed in cells infected with KSHV, we examined whether cells reprogrammed by KSHV may be differentiation inhibited. To this end, we infected with KSHV bone marrow–derived MSCs and examined their differentiation into fat cells in comparison with uninfected MSCs. As expected, uninfected MSCs progressively accumulated lipid droplets starting on or about day 10 after culture with adipocyte differentiation medium (Supplementary Figs. SA and SB). By contrast, KSHV-infected MSCs displayed a markedly reduced lipid accumulation throughout the 21-day culture (Supplementary Figs. SC and SD; demonstration of KSHV infection, Supplementary Figs. SE and SF). These results showed that KSHV blocks adipogenic differentiation in MSCs, suggesting that KSHV-infected endothelial cells may be similarly impaired.
Analysis of endothelial and mesenchymal markers in Kaposi sarcoma tissues
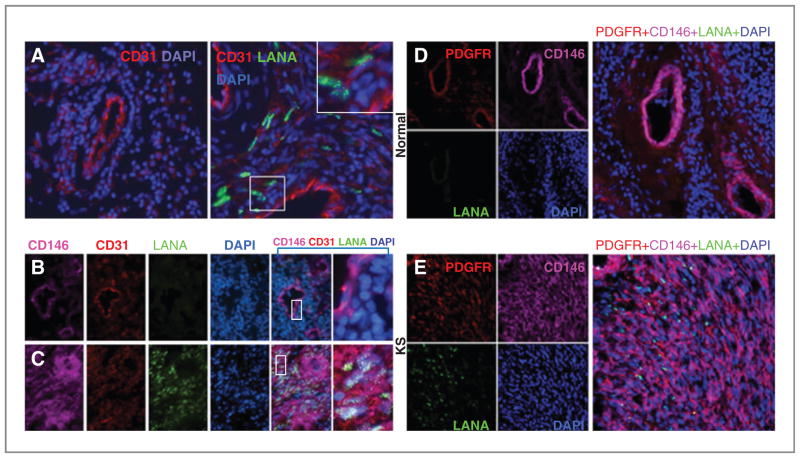
Because KSHV infection causes reprogramming of endothelial cell in vitro resulting in the loss of endothelial cell markers and acquisition of mesenchymal markers, we investigated whether a similar process occurs in KS tissues. Immunofluorescence staining (Fig. 3A) showed that the endothelium lining of the capillaries from the normal skin (left panel) and from KS tissue (right panel) stains similarly intensely for CD31 (mean staining intensity 20 and 21 units, respectively). However, KS spindle cells that are LANA+ (KSHV infected) generally display a dimmer CD31 staining in comparison with the capillary endothelium (mean staining intensity 6.6 units). A higher magnification (larger boxed area) allows visualization of the typical, punctate LANA immunostaining.
Figure 3.
Analysis of CD31, CD146, and PDGFR expression in KS tissues. A, fluorescence microscopy images of normal skin tissue (left) and KS (stage T1) tissue (right) immunostained for CD31 (red) and LANA (green); nuclei are visualized with DAPI (blue). CD31bright endothelial cells lining vessels are visualized in the normal skin and in KS tissue. The spindle-like cells visualized in the KS tissue (LANA-positive) are typically CD31low or CD31−. Original magnification 32×; the larger boxed panel represents a magnification of the smaller boxed panel. Images from normal skin tissue (B) and KS (stage T1) tissue (C) immunostained for CD146 (pink), CD31 (red), and LANA (green); nuclei are visualized with DAPI (blue). In normal skin, CD31+CD146+ cells are LANA− and mark endothelial cells lining blood vessels. In KS tissue, the LANA+CD146+CD31low/neg cells are widely distributed in the tumor tissue; original magnification 40×; the merged images are magnified in the panel furthest to the right. Normal skin (D) and KS (stage T1) tissue (E) immunostained for PDGFRβ (red), CD146 (pink), and LANA (green); nuclei are stained with DAPI (blue). In normal skin, CD146+ PDGFRβ+ cells mark vessels and do not express LANA. In KS tissue, CD146+PDGFRβ+LANA+ cells are widely distributed within the tumor tissue. The right panel reflects merged images; the inset reflects a magnification within the panel; original magnification 40×.
As markers for mesenchymal cells, we used CD146, PDGFRβ, and SMA (21). Because CD146 is also expressed in the vascular endothelium (21), we used CD31 costaining to distinguish between these cell types. In the normal skin, the endothelial cells lining blood vessels coexpress (coexpression 98%) CD146 and CD31 (Fig. 3B). Instead, in KS tissue, CD146 is intensely and broadly expressed by cells that do not stain or stain faintly for CD31, are often LANA+ (CD146 and LANA coexpression 96%; CD31, CD146, and LANA coexpression 6.1%) and are not associated with a capillary (Fig. 3C). Immunostaining of the normal skin (Fig. 3D) revealed that PDGFRβ expression coincides with CD146 expression and marks capillaries (CD31, PDGFRβ, and CD146 coexpression 95%). Instead, in KS tissue (Fig. 3E), PDGFRβ is widely expressed and mostly coincides with LANA and CD146 expression (PDGFRβ, CD146, and LANA coexpression 87%).
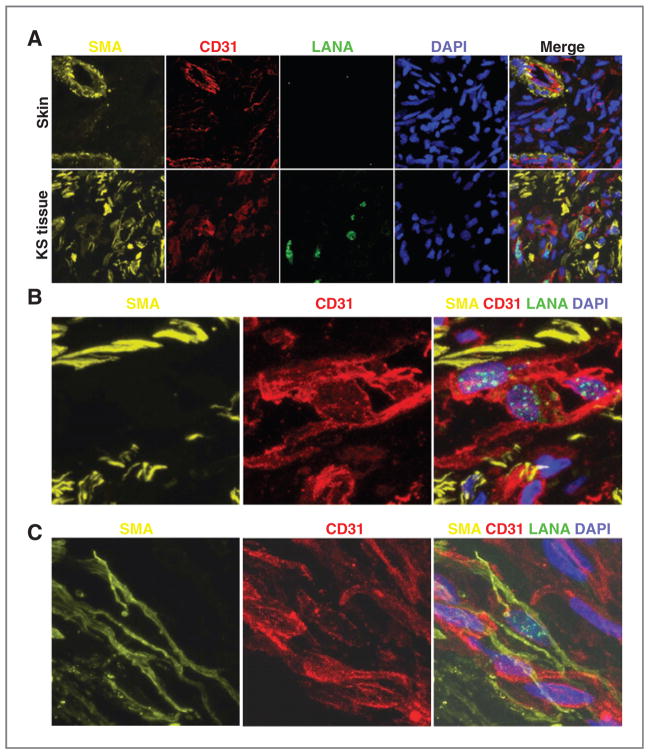
SMA is expressed in cells of mesenchymal origin, including vascular smooth muscle cells. In the normal skin (Fig. 4A, top panels) SMA and CD31 expression is mostly limited to vessels, with SMA generally surrounding the inner CD31+ endothelium. By contrast, in KS tissue (Fig. 4A, bottom panels), SMA expression is broadly detected with or without CD31 expression. Selected areas within various KS biopsies show the presence of occasional KSHV-infected cells that expressed CD31 at high levels, but lack expression of SMA (representative fields, Fig. 4B), and occasional LANA+ cells that coexpress CD31 and SMA (Fig. 4C). Thus, a proportion of KSHV-infected cells within KS tissues display a mesenchymal-like phenotype; other KSHV-infected cells display an endothelial phenotype and occasionally a transitional endothelial–mesenchymal phenotype.
Figure 4.
Expression of SMA and CD31 in KS tissue and normal skin. A, fluorescence microscopy images of normal skin (top panels) and KS (stage T0) tissue (bottom) immunostained for SMA (yellow), CD31 (red), and LANA (green); nuclei are stained with DAPI (blue). In normal skin, SMA+CD31+ cells mark the vessel wall. In KS tissue, SMA+ cells are widely distributed. B, microscopy images of KS (stage T0) tissue displaying a cluster of CD31bright cells expressing nuclear LANA+. C, microscopy images showing coexpression of SMA (yellow) and low-level CD31 (red) in a LANA+ cell. Original magnification of all images, 60×.
Analysis of TGFβ and Notch signaling in KSHV-induced endothelial–mesenchymal transformation
During cardiac development, the TGFβ/Smad and Notch signaling pathways are implicated in the control of EndMT that accompanies the generation of the mesenchymal cushion cells from endothelial/endocardial cells (27, 28). Studies in vitro have supported the importance of TGFβ and Notch signaling in EndMT (19, 29, 30). We examined the potential role of these signaling pathways in EndMT induced by KSHV.
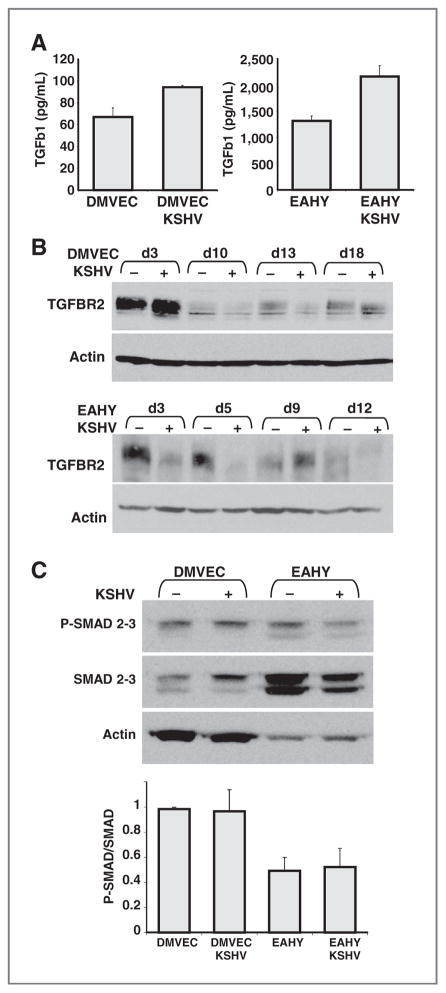
First, we tested whether KSHV infection alters TGFβ secretion by endothelial cells. TGFβ2 was not detected in the conditioned media (36-hour conditioning) of DMVECs and EAHY cells infected with KSHV (not shown), whereas TGFβ1 was detected at higher levels in supernatants from KSHV-infected cells compared with uninfected cells (Fig. 5A). Because TGFβ signaling is initiated by the engagement of TGFβ type II receptor (TGFβR2), we tested TGFβR2 protein expression during the time interval in which KSHV induces EndMT. By immunoblotting (Fig. 5B), we observed that TGFβR2 levels oscillate in DMVECs (top panel) and EAHY cells (bottom panel) during 12- to 18-day culture with or without KSHV infection. In general, KSHV-infected endothelial cells displayed lower levels of TGFβR2 than uninfected cells.
Figure 5.
Effects of KSHV infection on TGFβ1 secretion and signaling. A, levels of TGFβ1 were measured by ELISA in supernatants from 36-hour cultures of uninfected or KSHV-infected cells (DMVECs and EAHY). Results are expressed as means (± SD) of 2 experiments tested in duplicate. B, TGFβ receptor 2 (TGFBR2) in cell lysates of DMVECs (top) and EAHY (bottom) cells at the indicated time points (d3 = day3) after KSHV infection or culture without infection detected by immunoblotting. The membranes were reprobed for actin. The experiment is representative of 5 (DMVECs) or 3 (EAHY) conducted. C, phosphorylated (P) SMAD2/3, total SMAD2/3, and actin detected by immunoblotting in cell lysates of DMVECs and EAHY cultured for 9 days with or without infection with KSHV. Experiment representative of 3 conducted. The bar graph reflects the mean relative ratios (± SD) of P-SMAD2/3/total SMAD2/3 band intensities detected by immunoblotting in 3 experiments.
To evaluate the potential contribution of TGFβ1/TGFβR2 signaling, we examined the phosphorylation status of the downstream effectors Smad2 and Smad3. We found that the levels of phosphorylated Smad2/3 (P-Smad2/3) were similar in KSHV-infected (9 days postinfection) and uninfected DMVECs and EAHY cells, as were the ratios between P-Smad2/3 and total Smad2/3 (Fig. 5C). Furthermore, the Smad3 inhibitor SIS3 (31), which inhibited SMAD3 phosphorylation in KSHV-infected endothelial cells, did not prevent KSHV-induced EndMT, as judged by expression levels of endothelial and mesenchymal markers (not shown). These results provided evidence that TGFβ signaling is not critical to KSHV-induced EndMT.
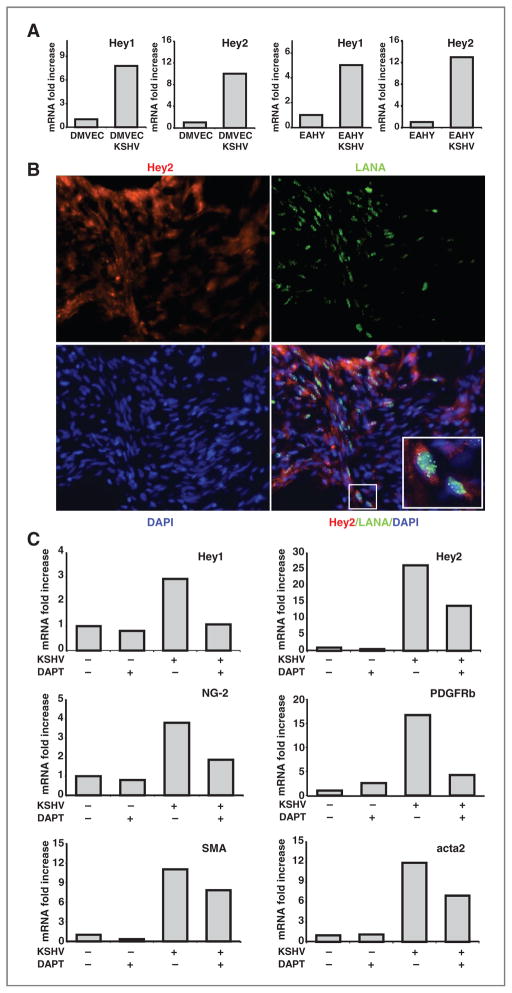
KSHV activates the Notch pathway directly as KSHV-RTA binds to the Notch signaling component RPBj, and indirectly by inducing the Notch ligands Jag1 and Dll4 through KSHV-vFLIP and KSHV-GCPR (32, 33). We investigated the potential contribution of Notch signaling to KSHV-induced EndMT. First, we tested whether Notch signaling is active in KSHV-infected DMVECs by measuring levels of expression of the Notch target genes Hey1 and Hey2 (34). We found that expression levels of Hey1 and Hey2 mRNAs were increased in KSHV-infected DMVECs and EAHY cells in comparison with the uninfected controls (Fig. 6A). Second, we examined whether KSHV-infected cells within KS tissues have evidence of Notch activation by immunostaining for LANA and Hey2. Fluorescence microscopy images showed that Hey2 is expressed in KS tissue, particularly where LANA is coexpressed (Fig. 6B). Third, we tested whether blocking Notch signaling affects KSHV-induced EndMT in endothelial cells, by using the compound DAPT, which is an inhibitor of γ-secretase, the enzyme that cleaves the intracellular domain of Notch initiating signaling (34). EAHY cells (KSHV infected or uninfected) were treated with DAPT (5 μmol/L throughout the culture period) or left untreated. RNA was extracted at the end of a 12-day culture. DAPT reduced expression of the Notch target genes Hey1 and Hey2 in KSHV-infected EAHY cells, indicative of successful inhibition of Notch signaling. In addition, DAPT reduced expression of the mesenchymal markers NG-2, PDGFRβ, SMA, and acta2 in comparison with control cells in which Notch signaling was not blocked (Fig. 6C). These observations confirmed that KSHV induces Notch signaling in endothelial cells, showed that KSHV-infected KS lesions have evidence of Notch activation, and that Notch signaling is a critical contributor to KSHV-induced EndMT.
Figure 6.
KSHV infection promotes activation of the Notch pathway. A, expression levels of Hey1 and Hey2 mRNAs in (DMVECs) and EAHY cells uninfected or infected with KSHV. RNAs were extracted 12 days after KSHV infection or culture without infection. The results from quantitative PCR are expressed as fold change relative to the uninfected cells. The experiment is representative of 3 conducted. B, fluorescence microscopy images of KS (stage T0) tissue immunostained for Hey2 (red) and LANA (green); nuclei are stained with DAPI (blue); original magnification 32×. The inset reflects a magnification showing the coexpression of LANA and Hey2 immunostaining. C, effects of the γ-secretase inhibitor DAPT (5 μmol/L throughout the culture period) on the levels of Hey1, Hey2, NG-2, PDGFRB, SMA, and acta2 mRNAs in the EAHY926 cells uninfected or KSHV-infected (12-day culture after achieving more than 90% cell infection as detected by GFP). The experiment is representative of 4 conducted.
Transcription factors expression in KSHV-infected cells and KS tissue
The transcription factors Slug and Snail play a role in the regulation of EndMT during the development of the cardiac cushion (28, 35). Snail was implicated in TGFβ-induced EndMT of human cutaneous microvascular endothelial cells (19). Besides Snail (SNAI1; ref. 36) and Slug (SNAI2; ref. 37), Twist (38), ZEB1 (δEF1; ref. 39), and ZEB2 (SIP1; ref. 40) have been linked to EMT. We investigated whether KSHV-induced EndMT is associated with changes in the expression levels of these transcriptions factors.
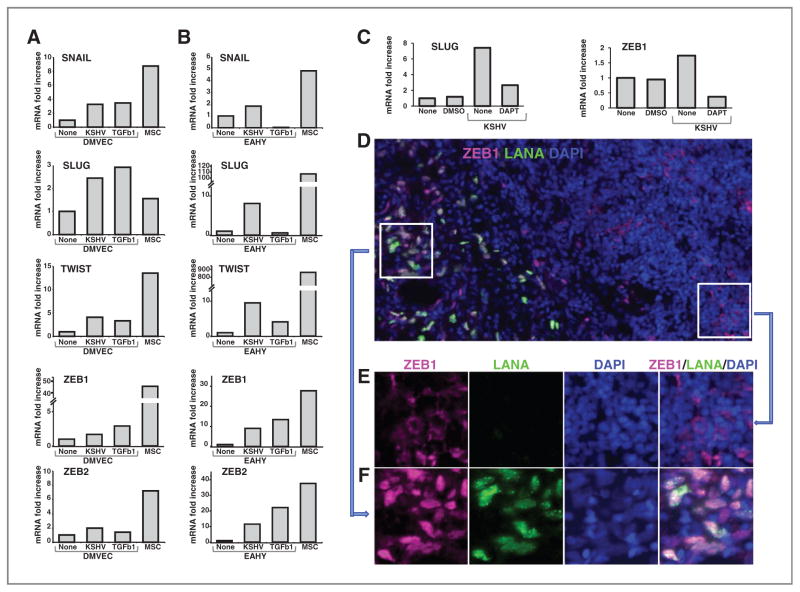
Expression of Snail, Slug, Twist, ZEB1, and ZEB2 was consistently increased in DMVECs (Fig. 7A) and EAHY cells (Fig. 7B) infected with KSHV (12 days postinfection) compared with control uninfected cells. In kinetic experiments, ZEB1 and ZEB2 expression was markedly increased (as much ~500-fold) in DMVECs and EAHY cells 7 days post-KSHV infection to the uninfected control cells (not shown). Thus, KSHV regulates expression of transcription factors that have been linked to EndMT or EMT. In additional experiments, we tested whether KSHV promotes expression of these transcription factors acting through Notch. The γ-secretase inhibitor DAPT (10 μg/mL) reduced Slug, ZEB1, and ZEB2 expression induced by KSHV (Fig. 7C, ZEB2 expression not shown), but not the expression of Jag1, Jag2, Dll4, or Snail (not shown), providing evidence for a role of Notch signaling in the selective regulation of expression of these transcription factors.
Figure 7.
KSHV infection promotes the expression of SNAIL, SLUG, TWIST, ZEB1, and ZEB2 transcription factors. RNA was extracted from DMVECs (A) and EAHY cells (B) cultured (12 days) in medium only or with TGFβ (5 ng/mL) or after KSHV infection (~90% cells infected). RNA from MSCs was a control. Results reflect fold increase in mRNA (quantitative PCR) relative to control cells after normalization for GAPDH. The results in A and B are representative of 3 to 5 experiments conducted. C, effects of the γ-secretase inhibitor DAPT (10 μg/mL) on KSHV-induced expression of SLUG and ZEB1 in EAHY cells (experimental conditions described in the legend to Fig. 6C). D, images of KS (stage T1) tissue immunostained for ZEB1 (pink) and LANA (green); nuclei are stained with DAPI (blue); original magnification (32×). E, magnification of the inset linked by the arrow showing an area of the tissue in which LANA− cells express mostly cytoplasmic ZEB1 staining. F, magnification of the inset linked by the arrow showing an area of KS tissue with LANA+ cells, in which both LANA and ZEB1 are mostly coexpressed in the nuclei; individual stains in E and F are merged in the right panel. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
We examined whether ZEB1 is expressed in KSHV-infected cells within KS tissues. Costaining for LANA and ZEB1 (Fig. 7D) showed strong ZEB1 expression in areas of the biopsy where the cells are infected with KSHV, and much lower or no ZEB1 expression in areas of the biopsy where LANA is not detected. A magnification of the KS tissue specimen showed that LANA and ZEB1 are expressed in the same cells within the nuclei (Fig. 7F), whereas ZEB1 staining is mostly cytoplasmic in LANA− cells (Fig. 7E). The nuclear localization of ZEB1 is consistent with ZEB1 activity in these cells. These results provided evidence for a previously unrecognized link between the transcription factor ZEB1 and EndMT.
Discussion
KSHV is the causative agent of KS, a malignancy characterized by the presence of virus-infected spindle cells, mixed in with inflammatory cells and aberrant slit-like capillaries. The nature of KS spindle cells and exactly how KSHV promotes KS development is still controversial. Here we show that endothelial cells infected with KSHV lose expression of mature endothelial phenotypic markers and acquire phenotypic and functional markers of mesenchymal cells. We find that this transdifferentiation program is associated with activation of a group of transcription factors linked to EMT and is critically driven by KSHV-induced Notch signaling. Consistent with these observations, KSHV-infected spindle cells within KS biopsies display a complex and variable phenotype that includes endothelial and mesenchymal cell markers, express the EMT-inducing transcription factor ZEB1, and have evidence of Notch activation. These novel findings clarify the cell source of KS spindle cells and explain the reasons for previous controversy, provide the first example of virus-induced EndMT, and contribute new insights into KS pathogenesis and the mechanisms and potential roles of EndMT.
Activation of Notch signaling was previously reported in KSHV-infected cells (41) and was attributed to a direct stimulatory effect by KSHV-RTA (32), or to a Jag1/Dll4-mediated effect by vGPCR and vFLIP gene products (33), but its role as a mediator of KSHV infection and disease has remained unclear. Notch signaling can regulate the expression of KSHV genes (42) and promote quiescence in uninfected cells neighboring the KSHV-infected cells, thus providing a growth advantage to the KS cells (33). In this view, KS cells provide the Notch ligands Jag1 and Dll4, which induce Notch signaling in uninfected cells. Other studies using the KS cell lines SLK or KS IMM, which are not infected with KSHV, or using a KSHV-infected, but E6-E7 immortalized lymphatic endothelial cell line suggested that Notch signaling provides a survival advantage to the cells (41, 43). The current results show that Notch signaling plays a critical role in promoting KSHV-induced EndMT and that this transformation confers on endothelial cells increased motility and resistance to death, a set of traits that likely underline the characteristic ability of KS cells to infiltrate locally. Increased VEGF (5) likely contributes to KSHV-induced EndMT but does not explain the increased cell motility to the mesenchymal-selective promigratory factor PDGF (24). Interestingly, VEGF contributes to EndMT during heart development (44).
EndMT is deployed during morphogenesis of the heart enabling endocardial cells (with an endothelial phenotype) to generate the heart cushion cells from which the mesenchymal portion of cardiac septa and valves is formed (13). Here, the EndMT process results in the generation of terminally differentiated cells. Similarly, the EndMT program has been linked to organ fibrosis and to ectopic tissue ossification from endothelial cells (15–17, 19). In cancer, EndMT was implicated as a source of cancer-associated fibroblasts, which may promote tumor progression by modifying the tumor micro-environment (18). By contrast, EndMT induced by KSHV reprograms endothelial cells to acquire either a mixed endothelial–mesenchymal phenotype or predominantly mesenchymal phenotype, without progression to mature fibroblasts, fat, cartilage, or bone cells. This maturation block is determined by KSHV itself, and it is likely attributable to KSHV-LANA inactivation of glycogen synthase kinase 3, which regulates key participants in cell differentiation (26). Consistent with this phenotype, KS lesions do not normally include fibrous, cartilage, bone, or fat tissue, and the virus-infected KS cells display a complex endothelial–mesenchymal phenotype that is not encountered in normal tissues. Thus, KSHV-induced EndMT is more similar to EMT occurring in epithelial malignancies in displaying different stages of mesenchymal cell differentiation within a tumor (23). Occasionally, aggressive KS has been associated with “woody” cutaneous edema and fibroma-like nodules (45). The pathogenesis of these lesions has remained unclear. Our results suggest the possibility that EndMT may lead KS cells to differentiate into fibrous tissue, perhaps in conjunction with loss of viral infection.
TGFβ2 is an inducer of EntMT in development and in mature endothelial cells (18, 19, 46), but we find no evidence that TGFβ signaling is deployed in KSHV-induced EndMT. This is consistent with the observation that the TGFβ signaling is blocked in KSHV-infected cells by epigenetic mechanisms (47). Other factors from the KS microenvironment (perhaps the inflammatory component) may contribute to KSHV-induced EndMT, as KS spindle cells are not fully autonomous and require growth factors from the microenvironment for growth (5). Activation of an EMT program in tumor cells often relies on the interplay between cancer cells and the neighboring stroma (23), and many factors contribute to EMT, including fibroblast growth factor, hepatocyte growth factor, IL-6, and insulin-like growth factors (22, 23).
Signaling downstream of TGFβ converge in the stimulation of the EndMT-inducing transcription factors Slug and Snail, which coordinate change in lineage marker expression and function (19, 35, 46). These and other transcription factors, including ZEB1, ZEB2, and Twist are effectors of EMT (22). Notch signaling can induce expression of Snail, Slug, and ZEB1 (28, 48, 49). ZEB1 can reciprocally activate Notch signaling, and Snail and Twist cooperate in the regulation of expression and stability of ZEB1 during EMT (50). We find increased expression of Snail, Slug, Twist, ZEB1, and ZEB2 in KSHV-induced EndMT, and link expression of Snail, ZEB1, and ZEB2 to Notch signaling regulation. ZEB1 is abundant in KS lesions, including the KSHV-infected spindle cells where the nuclear localization is consistent with activity.
In spite of often displaying an indolent clinical course, KS can be aggressive and difficult to manage in patients with AIDS. We find that KS cells have phenotypic traits consistent with EndMT, and that KSHV can induce Notch-dependent EndMT in vitro. Mesenchymal transformation likely plays an important role in KS pathogenesis as it does in many cancer types, suggesting that inhibition of Notch signaling should be considered for the treatment of KS.
Supplementary Material
Supplementary Figure. KSHV inhibits adipocytes maturation in mesenchymal stem cells. Uninfected (MSC control) and KSHV-infected mesenchymal stem cells (MSC KSHV) were treated with adipocyte differentiation medium for 14 days. Control MSC unstained (A) and stained with red oil after fixation (B); insets show magnifications of differentiated adipocytes. KSHV-infected MSC unstained (C) and stained with red oil after fixation (D). Images in A–D are captured in bright field. KSHV-infected MSC are GFP positive (E) demonstrating viral infection. (F): Merged image from bright field in (C) and fluorescent field in (E); Original magnification 4x.
Supplementary Table 1. mRNAs strongly decreased in KSHV-infected EAHY cells compared to control EAHY cells
Acknowledgments
The authors thank the patients who volunteered for this study; Drs. D. Lowy, M. Boehm, S. Sakakibara, M. Segarra, and K. Jiang for contributions to this work; R. Braveman, S. Garfield, and L. Li for technical help; K. Aleman and K. M. Wyvill for patient care and organizational aspects of this work.
Grant Support
The work was supported by Intramural research program of CCR/NCI/NIH and NINDS/NIH.
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Moore PS, Chang Y. Detection of herpesvirus-like DNA sequences in Kaposi’s sarcoma in patients with and without HIV infection. N Engl J Med. 1995;332:1181–5. doi: 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–91. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 3.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–80. [PubMed] [Google Scholar]
- 4.Uldrick TS, Wang V, O’Mahony D, Aleman K, Wyvill KM, Marshall V, et al. An interleukin-6-related systemic inflammatory syndrome in patients co-infected with Kaposi sarcoma-associated herpesvirus and HIV but without Multicentric Castleman disease. Clin Infect Dis. 2010;51:350–8. doi: 10.1086/654798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganem D. KSHV infection and the pathogenesis of Kaposi’s sarcoma. Annu Rev Pathol. 2006;1:273–96. doi: 10.1146/annurev.pathol.1.110304.100133. [DOI] [PubMed] [Google Scholar]
- 6.Regezi JA, MacPhail LA, Daniels TE, DeSouza YG, Greenspan JS, Greenspan D. Human immunodeficiency virus-associated oral Kaposi’s sarcoma. A heterogeneous cell population dominated by spindle-shaped endothelial cells. Am J Pathol. 1993;143:240–9. [PMC free article] [PubMed] [Google Scholar]
- 7.Albini A, Mitchell CD, Thompson EW, Seeman R, Martin GR, Wittek AE, et al. Invasive activity and chemotactic response to growth factors by Kaposi’s sarcoma cells. J Cell Biochem. 1988;36:369–76. doi: 10.1002/jcb.240360406. [DOI] [PubMed] [Google Scholar]
- 8.Skobe M, Brown LF, Tognazzi K, Ganju RK, Dezube BJ, Alitalo K, et al. Vascular endothelial growth factor-C (VEGF-C) and its receptors KDR and fit-4 are expressed in AIDS-associated Kaposi’s sarcoma. J Invest Dermatol. 1999;113:1047–53. doi: 10.1046/j.1523-1747.1999.00798.x. [DOI] [PubMed] [Google Scholar]
- 9.Moses AV, Fish KN, Ruhl R, Smith PP, Strussenberg JG, Zhu L, et al. Long-term infection and transformation of dermal microvascular endothelial cells by human herpesvirus 8. J Virol. 1999;73:6892–902. doi: 10.1128/jvi.73.8.6892-6902.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poole LJ, Yu Y, Kim PS, Zheng QZ, Pevsner J, Hayward GS. Altered patterns of cellular gene expression in dermal microvascular endothelial cells infected with Kaposi’s sarcoma-associated herpesvirus. J Virol. 2002;76:3395–420. doi: 10.1128/JVI.76.7.3395-3420.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang HW, Trotter MW, Lagos D, Bourboulia D, Henderson S, Makinen T, et al. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat Genet. 2004;36:687–93. doi: 10.1038/ng1384. [DOI] [PubMed] [Google Scholar]
- 12.Hong YK, Foreman K, Shin JW, Hirakawa S, Curry CL, Sage DR, et al. Lymphatic reprogramming of blood vascular endothelium by Kaposi sarcoma-associated herpesvirus. Nat Genet. 2004;36:683–5. doi: 10.1038/ng1383. [DOI] [PubMed] [Google Scholar]
- 13.Icardo JM, Manasek FJ. An indirect immunofluorescence study of the distribution of fibronectin during the formation of the cushion tissue mesenchyme in the embryonic heart. Dev Biol. 1984;101:336–45. doi: 10.1016/0012-1606(84)90147-7. [DOI] [PubMed] [Google Scholar]
- 14.Nakajima Y, Yamagishi T, Hokari S, Nakamura H. Mechanisms involved in valvuloseptal endocardial cushion formation in early cardiogenesis: roles of transforming growth factor (TGF)-beta and bone morphogenetic protein (BMP) Anat Rec. 2000;258:119–27. doi: 10.1002/(SICI)1097-0185(20000201)258:2<119::AID-AR1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 15.Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med. 2007;13:952–61. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- 16.Zeisberg EM, Potenta SE, Sugimoto H, Zeisberg M, Kalluri R. Fibroblasts in kidney fibrosis emerge via endothelial-to-mesenchymal transition. J Am Soc Nephrol. 2008;19:2282–7. doi: 10.1681/ASN.2008050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kizu A, Medici D, Kalluri R. Endothelial-mesenchymal transition as a novel mechanism for generating myofibroblasts during diabetic nephropathy. Am J Pathol. 2009;175:1371–3. doi: 10.2353/ajpath.2009.090698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeisberg EM, Potenta S, Xie L, Zeisberg M, Kalluri R. Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res. 2007;67:10123–8. doi: 10.1158/0008-5472.CAN-07-3127. [DOI] [PubMed] [Google Scholar]
- 19.Medici D, Shore EM, Lounev VY, Kaplan FS, Kalluri R, Olsen BR. Conversion of vascular endothelial cells into multipotent stem-like cells. Nat Med. 2010;16:1400–6. doi: 10.1038/nm.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vieira J, O’Hearn PM. Use of the red fluorescent protein as a marker of Kaposi’s sarcoma-associated herpesvirus lytic gene expression. Virology. 2004;325:225–40. doi: 10.1016/j.virol.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 21.Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–13. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 24.Hellstrom M, Kalen M, Lindahl P, Abramsson A, Betsholtz C. Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development. 1999;126:3047–55. doi: 10.1242/dev.126.14.3047. [DOI] [PubMed] [Google Scholar]
- 25.Ferrari G, Cusella-De Angelis G, Coletta M, Paolucci E, Stornaiuolo A, Cossu G, et al. Muscle regeneration by bone marrow-derived myogenic progenitors. Science. 1998;279:1528–30. doi: 10.1126/science.279.5356.1528. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Martin H, Shamay M, Woodard C, Tang QQ, Hayward SD. Kaposi’s sarcoma-associated herpesvirus LANA protein downregulates nuclear glycogen synthase kinase 3 activity and consequently blocks differentiation. J Virol. 2007;81:4722–31. doi: 10.1128/JVI.02548-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanford LP, Ormsby I, Gittenbergerde Groot AC, Sariola H, Friedman R, Boivin GP, et al. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124:2659–70. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timmerman LA, Grego-Bessa J, Raya A, Bertrán E, Pérez-Pomares JM, Díez J, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18:99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noseda M, McLean G, Niessen K, Chang L, Pollet I, Montpetit R, et al. Notch activation results in phenotypic and functional changes consistent with endothelial-to-mesenchymal transformation. Circ Res. 2004;94:910–7. doi: 10.1161/01.RES.0000124300.76171.C9. [DOI] [PubMed] [Google Scholar]
- 30.Zeisberg M, Yang C, Martino M, Duncan MB, Rieder F, Tanjore H, et al. Fibroblasts derive from hepatocytes in liver fibrosis via epithelial to mesenchymal transition. J Biol Chem. 2007;282:23337–47. doi: 10.1074/jbc.M700194200. [DOI] [PubMed] [Google Scholar]
- 31.Jinnin M, Ihn H, Tamaki K. Characterization of SIS3, a novel specific inhibitor of Smad3, and its effect on transforming growth factor-beta1-induced extracellular matrix expression. Mol Pharmacol. 2006;69:597–607. doi: 10.1124/mol.105.017483. [DOI] [PubMed] [Google Scholar]
- 32.Liang Y, Chang J, Lynch SJ, Lukac DM, Ganem D. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 2002;16:1977–89. doi: 10.1101/gad.996502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emuss V, Lagos D, Pizzey A, Gratrix F, Henderson SR, Boshoff C. KSHV manipulates Notch signaling by DLL4 and JAG1 to alter cell cycle genes in lymphatic endothelia. PLoS Pathog. 2009;5:e1000616. doi: 10.1371/journal.ppat.1000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shawber CJ, Das I, Francisco E, Kitajewski J. Notch signaling in primary endothelial cells. Ann N Y Acad Sci. 2003;995:162–70. doi: 10.1111/j.1749-6632.2003.tb03219.x. [DOI] [PubMed] [Google Scholar]
- 35.Carmona R, Gonzalez-Iriarte M, Macias D, Perez-Pomares JM, Garcia-Garrido L, Munoz-Chapuli R. Immunolocalization of the transcription factor Slug in the developing avian heart. Anat Embryol (Berl) 2000;201:103–9. doi: 10.1007/pl00008230. [DOI] [PubMed] [Google Scholar]
- 36.Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 37.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–54. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, et al. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–85. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 40.Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–78. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 41.Curry CL, Reed LL, Golde TE, Miele L, Nickoloff BJ, Foreman KE. Gamma secretase inhibitor blocks Notch activation and induces apoptosis in Kaposi’s sarcoma tumor cells. Oncogene. 2005;24:6333–44. doi: 10.1038/sj.onc.1208783. [DOI] [PubMed] [Google Scholar]
- 42.Lu J, Verma SC, Cai Q, Robertson ES. The single RBP-J{kappa} site within the LANA promoter is crucial for establishing Kaposi’s sarcoma-associated herpesvirus latency during primary infection. J Virol. 2011;85:6148–61. doi: 10.1128/JVI.02608-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu R, Li X, Tulpule A, Zhou Y, Scehnet JS, Zhang S, et al. KSHV-induced notch components render endothelial and mural cell characteristics and cell survival. Blood. 2010;115:887–95. doi: 10.1182/blood-2009-08-236745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Armstrong EJ, Bischoff J. Heart valve development: endothelial cell signaling and differentiation. Circ Res. 2004;95:459–70. doi: 10.1161/01.RES.0000141146.95728.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bayley AC. Aggressive Kaposi’s sarcoma in Zambia, 1983. Lancet. 1984;1:1318–20. doi: 10.1016/s0140-6736(84)91818-x. [DOI] [PubMed] [Google Scholar]
- 46.Kokudo T, Suzuki Y, Yoshimatsu Y, Yamazaki T, Watabe T, Miyazono K. Snail is required for TGFbeta-induced endothelial-mesenchymal transition of embryonic stem cell-derived endothelial cells. J Cell Sci. 2008;121(Pt 20):3317–24. doi: 10.1242/jcs.028282. [DOI] [PubMed] [Google Scholar]
- 47.Di Bartolo DL, Cannon M, Liu YF, Renne R, Chadburn A, Boshoff C, et al. KSHV LANA inhibits TGF-beta signaling through epigenetic silencing of the TGF-beta type II receptor. Blood. 2008;111:4731–40. doi: 10.1182/blood-2007-09-110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leong KG, Niessen K, Kulic I, Raouf A, Eaves C, Pollet I, et al. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J Exp Med. 2007;204:2935–48. doi: 10.1084/jem.20071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z, Li Y, Kong D, Banerjee S, Ahmad A, Azmi AS, et al. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 2009;69:2400–7. doi: 10.1158/0008-5472.CAN-08-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brabletz S, Bajdak K, Meidhof S, Burk U, Niedermann G, Firat E, et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011;30:770–82. doi: 10.1038/emboj.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure. KSHV inhibits adipocytes maturation in mesenchymal stem cells. Uninfected (MSC control) and KSHV-infected mesenchymal stem cells (MSC KSHV) were treated with adipocyte differentiation medium for 14 days. Control MSC unstained (A) and stained with red oil after fixation (B); insets show magnifications of differentiated adipocytes. KSHV-infected MSC unstained (C) and stained with red oil after fixation (D). Images in A–D are captured in bright field. KSHV-infected MSC are GFP positive (E) demonstrating viral infection. (F): Merged image from bright field in (C) and fluorescent field in (E); Original magnification 4x.
Supplementary Table 1. mRNAs strongly decreased in KSHV-infected EAHY cells compared to control EAHY cells