Abstract
Genetic correlations between quantitative traits measured in many breeding programs are pervasive. These correlations indicate that measurements of one trait carry information on other traits. Current single-trait (univariate) genomic selection does not take advantage of this information. Multivariate genomic selection on multiple traits could accomplish this but has been little explored and tested in practical breeding programs. In this study, three multivariate linear models (i.e., GBLUP, BayesA, and BayesCπ) were presented and compared to univariate models using simulated and real quantitative traits controlled by different genetic architectures. We also extended BayesA with fixed hyperparameters to a full hierarchical model that estimated hyperparameters and BayesCπ to impute missing phenotypes. We found that optimal marker-effect variance priors depended on the genetic architecture of the trait so that estimating them was beneficial. We showed that the prediction accuracy for a low-heritability trait could be significantly increased by multivariate genomic selection when a correlated high-heritability trait was available. Further, multiple-trait genomic selection had higher prediction accuracy than single-trait genomic selection when phenotypes are not available on all individuals and traits. Additional factors affecting the performance of multiple-trait genomic selection were explored.
Keywords: GenPred, shared data resources, genomic selection, plant breeding, low-heritability traits, genetic correlation, full hierarchical modeling
THE principle of genomic selection is to estimate simultaneously the effect of all markers in a training population consisting of phenotyped and genotyped individuals (Meuwissen et al. 2001). Genomic estimated breeding values (GEBVs) are then calculated as the sum of estimated marker effects for genotyped individuals in a prediction population. Fitting all markers simultaneously ensures that marker-effect estimates are unbiased, small effects are captured, and there is no multiple testing.
Current genomic prediction models usually use only a single phenotypic trait. However, new varieties of crops and animals are evaluated for their performance on multiple traits. Crop breeders record phenotypic data for multiple traits in categories such as yield components (e.g., grain weight or biomass), grain quality (e.g., taste, shape, color, nutrient content), and resistance to biotic or abiotic stress. To take advantage of genetic correlation in mapping causal loci, multi-trait QTL mapping methods have been developed using maximum-likelihood (Jiang and Zeng 1995) and Bayesian (Banerjee et al. 2008; Xu et al. 2009) methods. Calus and Veerkamp (2011) recently presented three multiple-trait genomic selection (MT-GS) models: ridge regression (GBLUP), BayesSSVS, and BayesCπ. The authors ranked the performances of these MT-GS methods (BayesSSVS > BayesCπ > GBLUP) based on simulated traits under a single genetic architecture. Genetic correlation was shown to be a key factor determining the MT-GS advantage over single-trait genomic selection (ST-GS). A few issues for these MT-GS methods still need attention. First, genetic architecture has been shown to affect the performance of different ST-GS methods differently (Daetwyler et al. 2010). Only a single genetic architecture was tested to rank these MT-GS methods. Second, the performance of these MT-GS methods on real breeding data were not shown since only simulated data were tested. Third, heritability is a key factor affecting GS performance. How heritability of multiple traits affects the performance of MT-GS has not been evaluated. Finally, no MT-GS packages are publicly available yet.
In addressing these issues, we also note and deal with a statistical issue identified by Gianola et al. (2009) in the BayesA and BayesB models of Meuwissen et al. (2001). In particular, the posterior inverse-χ2 distribution of marker effects has only one more degree of freedom than its prior distribution, which restricts Bayesian learning from the data by allowing the prior to dominate the posterior (Gianola et al. 2009). One solution, called BayesCπ (Habier et al. 2011), combines all markers with nonzero effects and estimates for them a common variance. This approach pools evidence from the markers and enables Bayesian learning. The solution we propose here considers the parameters of the marker effect variance prior as random variables and estimates them in a full hierarchical BayesA.
Our objectives in this study are to: (1) solve the statistical issue in conventional BayesA directly by the development of full hierarchical Bayesian modeling; (2) develop and extend two multiple-trait models (i.e., BayesA and BayesCπ); (3) test different MT-GS methods using simulated and real data and compare them to ST-GS methods; and (4) investigate factors affecting the performance of MT-GS methods.
Materials and Methods
Data simulation
Genomic selection models were compared using simulated data. Under the default simulation scenario, a pedigree consisting of six generations (generation 0–5) was simulated with an effective population size (Ne) of 50 haploids and starting from a base population with 5000 SNPs obtained using the coalescence simulation program GENOME (Liang et al. 2007). Value 0 or 1 was assigned to the two possible homozygote genotypes. This coalescent simulator assumes a standard neutral model and provides whole-genome haplotypes from a population in mutation–recombination–drift equilibrium. The census population size from base to generation 4 was equal to Ne but increased to 500 in generation 5. The simulated genome was similar to that of barley (Hordeum vulgare L.) with seven chromosomes, each of 150 cM. In total, 2020 SNPs were randomly selected from all polymorphic SNPs and 20 of those SNPs were randomly selected as QTL. QTL effects on two phenotypic traits were sampled from a standard bivariate normal distribution with correlation 0.5. This choice assumes some level of pleiotropy at all loci. The true breeding value for each individual was the sum of the QTL effects for each trait. Normal error deviates were added to achieve heritabilities of 0.1 for trait 1 and 0.5 for trait 2. All individuals have phenotypes on both traits. The covariance of errors between traits was zero. A single simulation parameter at a time was perturbed from the default scenario. Perturbed parameters included trait heritability (using values 0.1, 0.5, and 0.8), genetic correlation between traits (0.1, 0.3, 0.5, 0.7, and 0.9), error correlation (−0.2, 0, and 0.2), and number of QTL (20 and 200). Each simulation scenario was repeated 24 times for each prediction model to estimate the standard deviation of the prediction performance. All simulated data are available in supporting information, File S1.
Pine breeding data
Previously published pine breeding data (Resende et al. 2012) were used for model comparison. Deregressed estimated breeding values (EBVs) given in this study for disease resistance Rust_bin (presence or absence of rust) and Rust_gall_vol (Rust gall volume) were fit in different models. A total of 769 individuals had phenotypes for both traits and genotypes. We filtered genotype data to retain polymorphic SNPs with <50% missing data resulting in 4755 SNPs for analysis. Missing SNP scores were imputed with the corresponding mean for that SNP. As for the simulated data, value 0 or 1 was assigned to the two possible homozygote genotypes and 0.5 to the heterozygote genotypes.
Linear regression model
Marker effects on phenotypic traits were estimated from the mixed linear model:
In univariate models, y is a vector (n × 1) of phenotypes on n individuals, u is the overall population mean, X is a design matrix (n × p) allocating the p marker genotypes to n individuals, αj is the allele substitution effect for marker j assumed normally distributed αj ∼ N(0, ), is an indicator variable with value 1 if marker j is in the model and value 0 otherwise, e is a vector (n × 1) of identically and independently distributed residuals with e ∼ N(0, ).
In multivariate models with m traits, marker effects on phenotypic traits were estimated from the mixed linear model below.
where y is a matrix (n × m) of m phenotypes on n individuals, is a vector (1 × m) for the effects of molecular marker j on all m traits and assumed normally distributed ∼N(0, ), is the variance–covariance matrix (m × m) for marker j, e is a matrix (n × m) of residuals with each row having variance (m × m).
Single-trait and multi-trait pedigree-BLUP and GBLUP models
The numerator relationship matrix calculated from pedigree and the realized relationship matrix derived from SNPs were fit in ASReml (Gilmour et al. 2009) to predict the breeding values of individuals for validation. For multivariate pedigree-BLUP and GBLUP estimation, the breeding values of multiple traits for individuals for validation were predicted from a multi-trait model in ASReml in which an unstructured covariance matrix among traits was assumed.
Single-trait BayesA (ST-BayesA) model
In the BayesA method, all = 1 so that all markers are fit in the model. The prior distribution of marker substitution effect αj is normal N(0, ) and the prior distribution for marker variance is a scaled inverse-χ2 distribution with χ-2(ν, s). The prior distribution of the error variance, , is χ−2(−2, 0). The univariate BayesA developed in this study is different from the BayesA in Meuwissen et al. (2001) in that the parameters of the χ−2(ν, s) prior for were treated as unknown instead of being fixed. Below, we call the BayesA model in Meuwissen et al. (2001) “conventional BayesA” and the one developed in this study “full hierarchical BayesA.” Both ν and s were given improper flat priors and estimated from the data using the Metropolis algorithm to sample from the joint posterior distribution (see Appendix). Estimation for other parameters were the same as for conventional BayesA (Meuwissen et al. 2001). In total, 50,000 MCMC iterations were conducted and the first 5000 iterations were discarded as burn-in for all ST-GS Bayesian models. All Bayesian models were coded in C using the GNU Scientific Library. The source code is available upon request.
Multi-trait BayesA (MT-BayesA) model
The prior of the marker substitution effect vector, aj, was normal, N(0, ), and the prior of was a scaled inverse-Wishart distribution inv-Wis(ν, Sm×m). The prior distribution of the error variance, Σe, was inv-Wis(−2, ), where is a symmetric zero matrix. Like univariate BayesA, the (ν, Sm×m) were given a flat prior and estimated from the data using the Metropolis algorithm to sample from the joint posterior distribution (see Appendix). Full conditional distributions used for Gibbs sampling of parameters were as follows.
For the variance of marker j’s effect, , a scaled inverse-Wishart distribution,
For the residual variance, Σe, a scaled inverse-Wishart distribution,
Given the error variance and the marker effects, the overall mean u was sampled from the multivariate normal distribution,
In total, 110,000 MCMC iterations were conducted for all MT-GS Bayesian models and the first 10,000 iterations were discarded as burn-in.
Single-trait BayesCπ (ST-BayesCπ) model
The second Bayesian approach estimates the marker effects by variable selection and has been named BayesCπ (Habier et al. 2011). We present the algorithm briefly. In BayesCπ, marker effects on phenotypic traits were sampled from a mixture of null and normal distributions,
where δj = 0 with probability π and δj = 1 with probability 1 – π. The markers in the model shared a common variance . The prior for the genetic effect of each molecular marker, αj, depends on the variance and the probability π that markers do not have a genetic effect. The procedures for variable selection and parameter estimation are shown in the Appendix.
Multi-trait BayesianCπ (MT-BayesCπ) model
In MT-BayesCπ, marker effects on the phenotypic traits were estimated by the same mixed linear model as univariate BayesCπ,
where now y is a n × m matrix for m trait values on n individuals, u is a n × m matrix representing the overall mean for m traits in the population, aj is a 1 × m vector for the genetic effects of marker j on the m traits, e is the n × m matrix of residuals, and δj is the indicator variable as in ST-BayesCπ. The procedures for variable selection and parameter estimation are shown in the Appendix.
Imputation of missing phenotypic data were implemented in each MCMC iteration in MT-BayesCπ. As in Calus and Veerkamp (2011) for individual i, denote the set of missing traits by m and the set of observed traits by o. The expectation of yim can be split into two components, one that depends only on the genotype of i and one that depends on the residuals of the observed traits eio. The first component is
while the mean and variance of the second component comes from multivariate regression of the missing on the observed and is given by Calus and Veerkamp (2011):
Estimation of trait genetic parameter from MT-GS modeling
Three genetic parameters were calculated and compared for multiple traits: (1) genetic correlation between traits; (2) error correlation between traits; (3) heritability for each trait. Genetic correlation between trait t1 and t2 was calculated as , where is the genetic variance–covariance matrix for multiple traits. The σg was calculated as , where var(SNPi) is the genotype variance for SNPi and is the estimated marker effect vector for SNPi in iteration k for an analysis run over k2 iterations and with k1 burn-in iterations. The error correlation was calculated as , where is the estimated error variance–covariance matrix of multiple traits in MCMC iteration k. The heritability of trait t was calculated as .
Model validation for simulated and real data
For each simulated data set of 500 individuals, a randomly selected 400 formed the training set and the remaining 100 were for validation.
For the pine data set, 10-fold cross validation with a two-step analysis scheme was applied. First, after removal of the validation fold, the 4755 SNPs were ranked based on their association with the traits of interest, quantified as the P-value from a multivariate analysis of variance procedure. Second, the 500 SNPs with the smallest P-values from this analysis were used for ST- and MT-GS model fitting. The two-step analysis was repeated for each of the 10 validation folds.
For simulated (real breeding) data, the prediction accuracy was defined as the correlation between the simulated true breeding values (observed phenotype data) and the predicted GEBV values in the validation population. The standard deviation of the prediction accuracy was reported.
Results
Estimating variance hyperparameters in Bayesian genomic selection models
To implement the Bayesian learning in the prior selection for marker variance, the parameters in the inverse-χ2 (ST-BayesA) or inverse-Wishart (MT-BayesA) distribution were treated as unknowns. The conventional ST-BayesA model assumed the same prior for marker variance with v = 4.012 and s = 0.002 used in Meuwissen et al. (2001). For comparison, in conventional MT-BayesA v was set to 4.012 and S to a diagonal matrix with 0.002 on the diagonal. For the two sets of simulated phenotypic traits controlled by 20 or 200 QTL, both conventional and full hierarchical ST-BayesA and MT-BayesA were applied. Prediction accuracies were similar between conventional and full hierarchical models for the traits controlled by the 20 QTL genetic architecture (Table 1). In contrast, for the traits controlled by 200 QTL, the full hierarchical models exhibited higher prediction accuracy for either one or both traits than the conventional BayesA methods for both ST- and MT-BayesA. For the low-heritability trait 1, the prediction accuracy (0.33) of MT-BayesA with fixed prior was significantly lower than the conventional ST-BayesA model. In contrast, the full hierarchical MT-BayesA increased the prediction accuracy by 51% (from 0.33 to 0.50). A similar significant increase was observed for the high-heritability trait 2 (from 0.53 to 0.73). The different estimated priors for the marker variance in full hierarchical models (Table 1) compared to the conventional BayesA methods reflected the Bayesian learning process from the data. To take advantage of the full hierarchical ST- and MT-BayesA method, all BayesA analyses in all later sections of this study adopted the corresponding full hierarchical models.
Table 1 . Prediction accuracies of conventional (fixed hyperparameter) and full-hierarchical BayesA methods for ST- and MT-GS models.
| Degree of freedom |
Scaled |
Prediction accuracye |
||||||
|---|---|---|---|---|---|---|---|---|
| BayesA typea | Datab | Model typec | Trait 1 | Trait 2 | Trait 1 | Trait 2 | Trait 1 | Trait 2 |
| ST | GA20 | Fixed | 4.012 | 4.012 | 0.002 | 0.002 | 0.49 ± 0.15 | 0.80 ± 0.07 |
| ST | GA20 | Full | 4.041 | 2.509 | 0.002 | 0.002 | 0.49 ± 0.15 | 0.81 ± 0.06 |
| ST | GA200 | Fixed | 4.012 | 4.012 | 0.002 | 0.002 | 0.53 ± 0.10 | 0.61 ± 0.10 |
| ST | GA200 | Full | 4.380 | 2.060 | 0.002 | 0.002 | 0.51 ± 0.11 | 0.70 ± 0.07 |
| MT | GA20 | Fixed | 4.012 | 4.012 | 0.002 | 0.002 | 0.54 ± 0.15 | 0.80 ± 0.08 |
| MT | GA20 | Full | 3.235 | 3.235 | 0.002 | 0.003 | 0.60 ± 0.14 | 0.83 ± 0.06 |
| MT | GA200 | Fixed | 4.012 | 4.012 | 0.002 | 0.002 | 0.33 ± 0.13 | 0.53 ± 0.10 |
| MT | GA200 | Full | 3.088 | 3.088 | 0.004 | 0.012 | 0.50 ± 0.10 | 0.73 ± 0.06 |
ST, single-trait BayesA; MT, multiple-trait BayesA.
Two data sets simulated for traits controlled by either 20 QTL (GA20) or 200 QTL (GA200).
Fixed, fixed hyperparameter BayesA; Full, full hierarchical BayesA model.
Scale parameter in ST-BayesA or scale matrix for MT-model in which only the values on diagonal were shown here for comparison.
Mean ± standard deviation of the prediction accuracy was reported.
Prediction of breeding values using different ST- and MT-GS methods
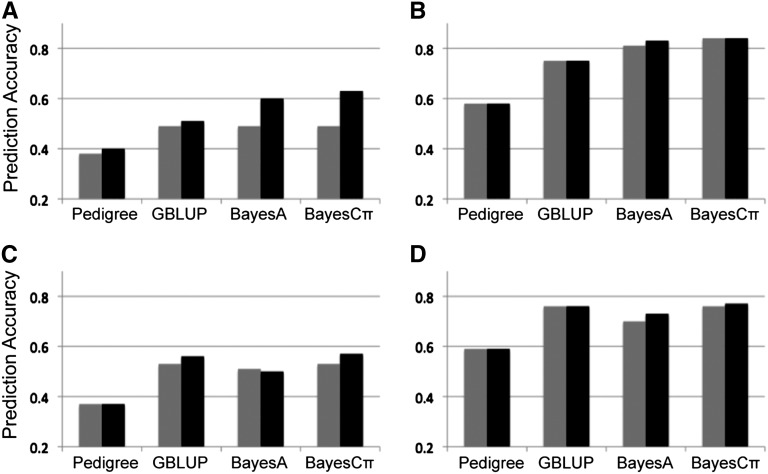
For comparison between the ST- and MT-GS methods, the simulated data sets with 20 QTL and 200 QTL were analyzed with four sets of ST- and MT-GS models: (1) pedigree-BLUP; (2) GBLUP based on SNP; (3) BayesA, and (4) BayesCπ. In all cases, SNP-based genomic selection model performed better than pedigree-based BLUP method for both ST-GS and MT-GS methods for all simulated data (Figure 1). With 20 QTL (Figure 1, A and B), the prediction accuracies of low-heritability trait 1 increased 5, 4, 22, and 36% using the MT-GS compared to ST-GS for pedigree-BLUP, GBLUP, BayesA, and BayesCπ, respectively. In both ST- and MT-GS analysis, Bayesian methods outperformed both pedigree-BLUP and GBLUP with the 20 QTL scenario and BayesA was slightly better than BayesCπ. For the high-heritability trait 2, the prediction accuracies of ST-GS and MT-GS were almost the same. In contrast, under the 200 QTL scenario (Figure 1, C and D), neither the ST or MT Bayesian methods outperformed GBLUP and within each type of method, the prediction accuracies between ST- and MT-GS were very similar.
Figure 1 .
Comparison of ST-GS (shaded) and MT-GS (solid) for correlated low-heritability (h2 = 0.1) trait 1 (A and C) with high heritability (h2 = 0.5) trait 2 (B and D) under the genetic architecture of 20 QTL (A and B) and 200 QTL (C and D). Genetic correlation between the two traits under each of genetic architectures is 0.5.
Effect of heritability on predictions using multi-trait GS
Four combinations of trait heritability were simulated to test the effect of heritability on MT-GS accuracy. MT-BayesCπ was used for this comparison. Under the ST-BayesCπ analysis, the prediction accuracy for the low-heritability trait (h2 = 0.1) was 0.49. Given the genetic correlation of 0.5, the MT-BayesCπ prediction accuracy of the low-heritability trait 1 was 0.67 and 0.70 when the heritability of correlated trait 2 was 0.5 and 0.8, respectively (Table 2). In contrast, the prediction accuracy for the medium- (h2 = 0.5) or high- (h2 = 0.8) heritability traits did not change as the heritability of the correlated trait changed.
Table 2 . Prediction accuracy for traits with different heritabilities.
| Heritability |
Prediction accuracya |
||
|---|---|---|---|
| Trait 1 | Trait 2 | Trait 1 | Trait 2 |
| 0.1 | 0.5 | 0.63 ± 0.10 | 0.86 ± 0.05 |
| 0.1 | 0.8 | 0.70 ± 0.08 | 0.94 ± 0.02 |
| 0.5 | 0.8 | 0.89 ± 0.04 | 0.93 ± 0.03 |
| 0.8 | 0.8 | 0.93 ± 0.03 | 0.94 ± 0.03 |
Accuracy from the MT-BayesCπ for traits simulated with the parameters under the default simulation except different heritabilities.
Effect of genetic correlation between traits on the prediction of multi-trait GS
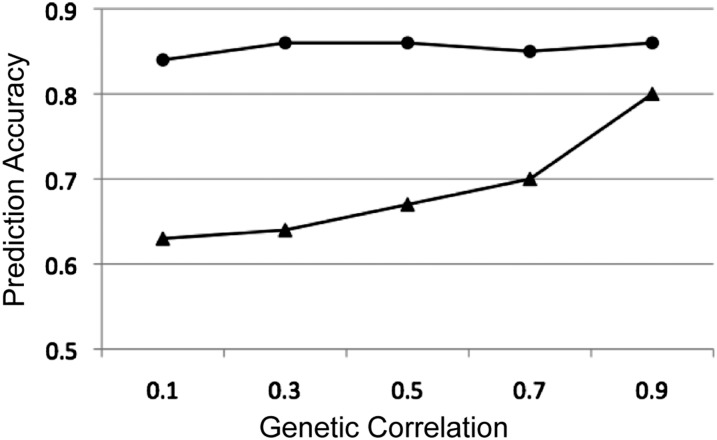
As genetic correlation increased between traits, the prediction accuracies increased for the low-heritability trait 1 (Figure 2). When the genetic correlation was 0.1 between the two traits, the prediction accuracy for the low-heritability trait was 0.63, which was already higher than the prediction accuracy based on the univariate analysis (0.49). As the genetic correlation increased, the prediction accuracies for the low-heritability trait also increased. In contrast, for the high-heritability trait 2, no obvious change in prediction accuracy was observed as the genetic correlation increased from 0.1 to 0.9.
Figure 2 .
Effect of genetic correlation (x-axis) on the prediction accuracy (y-axis) of low-heritability trait 1 (▴) and high-heritability trait 2 (●) using MT-BayesCπ.
Effect of error correlation between traits on the prediction of multi-trait GS
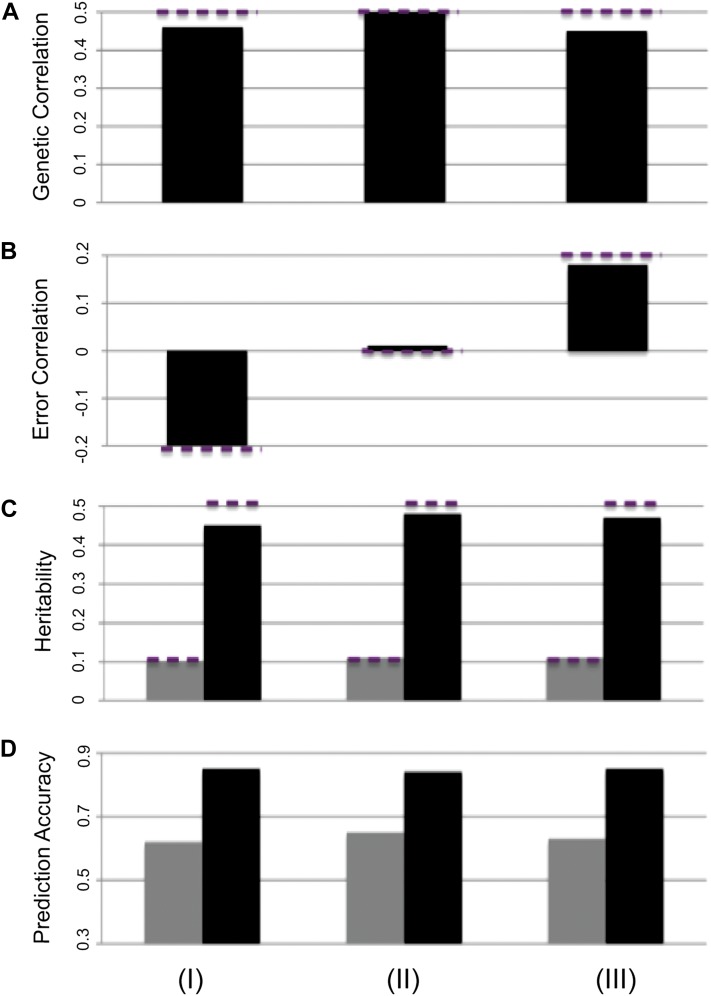
Phenotypic correlation between traits contains both genetic and error correlations. The error correlation under the default simulation scenario was zero (Materials and Methods). Three data sets were simulated with different error correlations (−0.2, 0, and 0.2), while keeping other parameters at their default settings (Figure 3). The MT-GS model was able to separate error correlation from genetic correlation and estimate the heritability well. Furthermore, for both low- and high-heritability traits, the prediction accuracies were consistent across the three data sets.
Figure 3 .
Effect of error correlation (−0.2, 0, and 0.2 for columns I, II, and III) on genetic parameter estimation and prediction accuracy using MT-BayesCπ. True parameter values are shown with dashed lines. (A) Genetic correlation; (B) error correlation; (C) heritability for low-heritability trait (shaded bar; h2 = 0.1) and high-heritability trait (solid bar; h2 = 0.5); (D) prediction accuracy low-heritability trait (shaded; h2 = 0.1) and high-heritability trait (solid; h2 = 0.5).
Real pine breeding data analysis using multi-trait GS
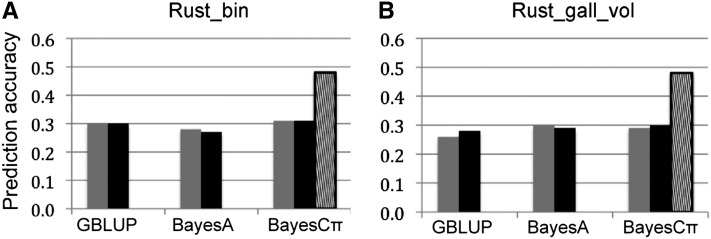
The MT-GS models were applied to two disease-resistance traits in published pine breeding data (Resende et al. 2012) using a two-step analysis that reduced marker numbers by selecting on the rank of marker effect (see Materials and Methods) (Figure 4). Compared to prediction in the original publication (Resende et al. 2012), the ST-GS models in this study showed similar results for all models (GBLUP, BayesA, and BayesCπ). This result suggests that the two-step analysis may be a useful variable selection method when millions of SNP markers from new sequencing technologies are used in genomic selection.
Figure 4 .
Comparison of ST-GS (shaded) and MT-GS (solid) for two disease-resistance traits of pine tree: Rust_bin (A) and Rust_gall_vol (B). The striped bars show prediction accuracy for MT-BayesCπ when the phenotype for the focal trait was unknown, but that for the other trait was observed.
The phenotype and genotype data used for ST-GS analysis were also fit with three MT-GS models. Within each of GBLUP, BayesA, and BayesCπ, the MT-GS exhibited similar prediction capability to the ST-GS models (Figure 4). This prediction pattern was similar to the pattern for the polygenic genetic architecture in the simulation study. With MT-GS models it is also possible to predict a trait when individuals have been measured for other traits. For example, by setting each 10% of the Rust_gall_vol values to missing (similar to 10-fold cross-validation) and using both marker and Rust_bin data to predict these values, MT-BayesCπ had a prediction accuracy of 0.48 (Figure 4), which was a 60% increase relative to the ST–GS method (0.30).
Discussion
Hyperprior optimization of Bayesian model for ST-GS and MT-GS methods
The conventional, fixed hyperparameter BayesA model allows locus-specific marker variances for markers in the model. This is a natural way to model the assumption that some markers are in strong LD with important QTL while others are not (Meuwissen et al. 2001). BayesA is easy to implement using conjugate priors through Gibbs sampling and has at times been shown, in both simulated and empirical data, to achieve higher prediction accuracy than ridge regression (Hayes et al. 2010; Meuwissen et al. 2001). In BayesA, the hyperprior for the marker-specific variance is a scaled inverse-χ2 distribution with two parameters, degree of freedom ν, and scale s. Because most markers, in particular SNPs, are biallelic, we estimate only a single marker-substitution effect per locus and the posterior and prior distributions differ by only a single degree of freedom (Gianola et al. 2009; although note that in the original publication, BayesA was applied not to biallelic markers but to multiallelic marker haplotypes, Meuwissen et al. 2001). Consequently, the scale parameter s in the prior has a strong effect on the shrinkage of marker effects. To address this drawback, Habier et al. (2011) developed BayesDπ that treated the scale parameter s as a random variable to be estimated but still treated the degrees of freedom as known although this parameter strongly affects the shape of distribution. Thus BayesDπ reduced the problems of BayesA but did not solve the dominance of the prior over the posterior distribution. Gianola et al. (2009) suggested several possible solutions including development of a full hierarchical approach to estimating the optimal priors from the data instead of assigning fixed values. In this study, both the degrees of freedom and the scale s parameter were given a flat prior and estimated using Metropolis sampling (Appendix). Under a simulated polygenic architecture, the full hierarchical BayesA model performed significantly better than the conventional fixed prior BayesA, and the difference was more important for multi- than single-trait analyses. Given that the genetic architecture of traits of interest is unknown in practice use of the full hierarchical BayesA appears prudent.
Comparison of single-trait and multi-trait GS models
Daetwyler et al. (2010) investigated the impact of genetic architecture on the prediction accuracy of genomic selection. They found that the GBLUP linear method showed relatively constant performance across different genetic architectures while the Bayesian variable selection method (BayesB) gave a higher accuracy compared to GBLUP when the traits were controlled by few QTL. This observation derived from simulation was also confirmed in real breeding data from different traits of Holstein cattle (Hayes et al. 2010). In a previous MT-GS study (Calus and Veerkamp 2011), different MT-GS methods were compared with each other and with the corresponding ST-GS methods with simulated data under a single genetic architecture. In our study, genetic architecture affected the relative superiority of MT-GS over ST-GS. Under a major QTL genetic architecture, the Bayesian models performed better than GBLUP in both single- and multi-trait models, and the multi-trait analysis was strongly beneficial. Under the polygenic genetic architecture, however, GBLUP was equal to the Bayesian models and multi-trait analysis provided a slight improvement at best. This observation suggests that MT-GS can capture the genetic correlation between traits when major QTL are present more efficiently than when they are not. In addition, if other phenotypes are available on individuals that have missing data, phenotype imputation with MT-GS methods can be very useful (Calus and Veerkamp 2011), which was shown in the MT-BayesCπ analysis of real pine data.
Genetic correlation between traits is the basis for the benefit of MT-GS models. Among traits measured by breeders, not all traits are genetically correlated with other traits. For two traits simulated without genetic correlation, we found that MT-GS was inferior to ST-GS (data not shown). The decreased accuracy presumably arises because sampling leads to nonzero estimates of correlation in the training population and then to erroneous information sharing across traits in the validation population. To avoid the application of MT-GS on traits that are not genetically correlated, we can estimate that correlation between traits using the GEBVs derived from ST-GS models and apply MT-GS only where it is likely to be beneficial.
Low-heritability traits benefit from correlated high-heritability traits
Genetic correlation between traits has previously been exploited to improve the statistical power to detect QTL controlling traits of interest (Jiang and Zeng 1995; Fernie et al. 2004; Chesler et al. 2005; Banerjee et al. 2008; Breitling et al. 2008; Xue et al. 2008; Xu et al. 2009). In genomic prediction rather than QTL identification, we have found that low-heritability traits can borrow information from correlated high-heritability traits and consequently achieve higher prediction accuracy. This improvement was not observed, however, for the high-heritability trait. This characteristic of MT-GS could be very important in plant breeding since many traits of interest have low heritability. In addition, plant breeders often want to reduce the undesirable genetic correlation between traits (Chen and Lubberstedt 2010). It is important to note that MT-GS is modeled by directly taking advantage of such genetic correlation, whether it is favorable or unfavorable, and is not designed to break the undesirable genetic correlation.
Supplementary Material
Acknowledgments
We thank Mark Sorrells for valuable feedback on the manuscript. Partial funding for this research was provided by U.S. Department of Agriculture, National Institute of Food and Agriculture, Agriculture and Food Research Initiative grants, award numbers 2009-65300-05661 and 2011-68002-30029.
Appendix
Metropolis Algorithm for Single-Trait BayesA Model
The joint posterior probability used for sampling the ν and s parameters is
where and are normal distributions, is a scaled inverse-χ2 distribution and is an improper constant prior. The symmetrical jumping distribution to sample the candidates of ν or s was normal with the existing value of (ν, s) as mean and variance 0.2. To avoid the negative values sampled from the normal distribution, the absolute sampled values were used as the candidates. The usual Metropolis rule was used: if the posterior density of the candidate values was higher than that of the existing values, the candidate values were accepted. If not, the candidates were accepted with probability equal to the ratio of the candidate to the existing density.
Metropolis Algorithm for Multi-Trait BayesA Model
The joint posterior probability used for sampling the ν and parameters is
where and were multivariate (m × m) normal distributions, was a scaled inverse Wishart distribution and was constant. The jumping distribution to sample the candidate of ν is the normal distribution with the existing value of ν as mean and variance equal to 0.2. The jumping distribution to sample the candidate scale matrix S*m×m was scaled-inversed-Wishart(100, Sm×m).
Variable Selection Procedure and Posterior Distributions for Single-Trait BayesCπ
The posterior distribution of is
where and are all marker effects and indicator variables except for marker j, respectively, equals xjT(xjαj + e), and xj is the genotype vector for marker j.
In addition, is proportional to , where can be two possible values, v0 or v1, depending whether the marker is in the model or not,
Then if the is larger than the value sampled from a unit uniform distribution, the marker is included in the model. For markers in the model, the posterior distribution of marker effect, αj, is a normal distribution,
where x−j and α−j are the marker genotype and effect excluding marker j, , is the common variance shared by all the markers in the model. For the markers not in the model, the marker effect is equal to zero. The posterior distribution of overall population mean μ and error variance is the same as in ST-BayesA. Full conditional distributions used for Gibbs sampling for parameters were as follows.
For the common variance of marker effect, , a scaled inverse-χ2 distribution,
where ν, the degree of freedom in the prior, was assigned a value of 3, κ is the number of markers included in the model, and s, the scale parameter in the prior, is 0.01. For the probability of marker having a zero effect, π, a beta distribution:
Variable Selection Procedure and Posterior Distributions for Multi-Trait BayesCπ
The posterior distribution of is similar to the ST-BayesCπ except several parameters become matrices,
where is equal to xjT(xjαj + e), is proportional to
where can be two possible values, v0 or v1, depending whether the marker is in the model,
The posterior distribution for π in MT-BayesCπ is a beta distribution as in the ST-BayesCπ. The prior of and common variance–covariance across markers between traits were inv-Wishart(ν, Sm×m), where ν was the number of traits plus 1 and Sm×m is a diagonal matrix with size equal to number of traits and 0.01 on the diagonal. Full conditional distributions used for Gibbs sampling for parameters were as follows:
For the common variance of marker, , a scaled inverse Wishart distribution
where κ was the number of markers in the model after the previous variable selection procedure and a was the matrix of estimated marker effects. For the error variance, , a scaled inverse Wishart distribution
where n was the number of individuals in the training population. Given the error variance and marker effect a, the overall population mean vector is sampled from the multinormal distribution,
The posterior distribution for aj is a multinormal distribution,
Footnotes
Communicating editor: D. J. de Koning
Literature Cited
- Banerjee S., Yandell B. S., Yi N., 2008. Bayesian quantitative trait loci mapping for multiple traits. Genetics 179: 2275–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R., Li Y., Tesson B. M., Fu J., Wu C., et al. , 2008. Genetical genomics: spotlight on QTL hotspots. PLoS Genet. 4: e1000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calus M. P., Veerkamp R. F., 2011. Accuracy of multi-trait genomic selection using different methods. Genet. Sel. Evol. 43: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Lubberstedt T., 2010. Molecular basis of trait correlations. Trends Plant Sci. 15: 454–461 [DOI] [PubMed] [Google Scholar]
- Chesler E. J., Lu L., Shou S., Qu Y., Gu J., et al. , 2005. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat. Genet. 37: 233–242 [DOI] [PubMed] [Google Scholar]
- Daetwyler H. D., Pong-Wong R., Villanueva B., Woolliams J. A., 2010. The impact of genetic architecture on genome-wide evaluation methods. Genetics 185: 1021–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie A. R., Trethewey R. N., Krotzky A. J., Willmitzer L., 2004. Metabolite profiling: from diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 5: 763–769 [DOI] [PubMed] [Google Scholar]
- Gianola D., de los Campos G., Hill W. G., Manfredi E., Fernando R., 2009. Additive genetic variability and the Bayesian alphabet. Genetics 183: 347–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour A. R., Gogel B. J., Cullis B. R., Thompson R., 2009. 2009 ASReml User Guide, release 3.0. VSN Intl., Hemel Hempstead, UK [Google Scholar]
- Habier D., Fernando R. L., Kizilkaya K., Garrick D. J., 2011. Extension of the Bayesian alphabet for genomic selection. BMC Bioinformatics 12: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B. J., Pryce J., Chamberlain A. J., Bowman P. J., Goddard M. E., 2010. Genetic architecture of complex traits and accuracy of genomic prediction: coat colour, milk-fat percentage, and type in Holstein cattle as contrasting model traits. PLoS Genet. 6: e1001139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Zeng Z. B., 1995. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140: 1111–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Zollner S., Abecasis G. R., 2007. GENOME: a rapid coalescent-based whole genome simulator. Bioinformatics 23: 1565–1567 [DOI] [PubMed] [Google Scholar]
- Meuwissen T. H., Hayes B. J., Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende M. F., Jr, Munoz P., Resende M. D., Garrick D. J., Fernando R. L., et al. , 2012. Accuracy of genomic selection methods in a standard data set of loblolly pine (Pinus taeda L.). Genetics 190: 1503–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Wang X., Li Z., Xu S., 2009. Mapping QTL for multiple traits using Bayesian statistics. Genet. Res. 91: 23–37 [DOI] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., et al. , 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40: 761–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.