Abstract
We have previously reported genetic differences between Western and Chinese prostate cancers, including different frequencies of ERG rearrangements. We investigated further ERG expression and rearrangements in prostate cancers and high-grade prostatic intraepithelial neoplasia (HGPIN) from the UK and China to determine differences between these two populations by tissue microarray based immunohistochemistry and fluorescence in situ hybridization. In keeping with our previous observation, that ERG was rearranged at a higher frequency in UK prostate cancer samples (38%, 58/155) than Chinese ones (8%, 7/93), ERG rearrangements were also found in 21% (4/19) and 0% (0/19) foci of HGPIN in UK and Chinese samples respectively. ERG nuclear expression in UK cancers (34%, 54/160) was significantly higher than that in Chinese ones (10%, 9/88) (p<0.001). ERG nuclear expression in UK HGPIN (28%, 11/39) was higher than that in Chinese HGPIN (0%, 0/9), but without statistical significance (p=0.193). ERG nuclear expression was correlated to ERG rearrangements in both UK (Kappa=0.686) and Chinese (Kappa=0.565) cancers. These data demonstrate that ERG rearrangement and expression frequencies are different in prostate cancers from UK and China as early as the precursor lesion, HGPIN. The nuclear expression is associated with ERG rearrangements which mainly occur in the Western samples. UK and Chinese prostate cancers may be the result of different genetic mechanisms.
Keywords: ERG, prostate cancer, high-grade prostatic intraepithelial neoplasia, genomic rearrangement, protein expression
Introduction
Prostate cancer is the most common cancer in Western men. The prevalence of prostate cancer in Asian countries is much less than that in Western countries [1,2]. The genetic alterations in prostate cancer cells from Western populations are well studied, but data from Asian samples are limited. To determine the similarities and/or differences in genomic alterations in prostate cancer samples from high and low incidence populations, we previously analyzed the genomic alterations in prostate cancer from China (low-incidence) and UK (high-incidence) with Affymetrix SNP Array 6.0 high-density microarrays and found specific genomic differences between cancers from China and Western countries [3]. Recently, we further demonstrated that genomic alterations of RAF genes are more common in Chinese than UK prostate cancer samples [4]. These genetic differences might underlie the regional/ethnic differences in clinical incidence and suggest different pathways of prostate carcinogenesis in these populations.
The TMPRSS2:ERG fusion gene, generated by a chromosomal internal deletion or chromosome translocation at 21q22.2-22.3, has been detected in about 50% of prostate cancer in the Western countries [5]. However, we found a low frequency of these genomic rearrangements, which are associated with the TMPRSS2:ERG fusion gene, in our Chinese samples [3]. The difference in ERG rearrangement frequency between Western and east Asian countries has also been observed in the studies of prostate cancer samples from Korea and Japan [6-8]. In this study, we evaluated the consequences of the different frequency of ERG rearrangements to protein expression level by tissue microarray (TMA)- based immunohistochemistry (IHC). We also investigated if these changes were present in earlier stages of prostate cancer. High-grade prostatic intraepithelial neoplasia (HGPIN) is well established as the only common precursor lesion associated with prostate cancer. HGPIN and prostate cancer share some genetic and molecular markers, with HGPIN representing an intermediate stage between benign epithelium and invasive carcinoma [9]. Therefore, we investigated ERG rearrangements and expression in HGPIN from these prostate cancer samples from China and UK. We observed that ERG expression is different in prostate cancers from UK and China and that the different frequency in ERG rearrangements and expression between these two populations occurs in the precursor lesion, HGPIN.
Materials and methods
Samples
Sample collection and TMA construction have been described previously [3]. Briefly, 168 prostate cancer cases from the UK (<10% were Asian and none were of Chinese origin) and 143 Chinese prostate cancer cases from mainland China were collected from radical prostatectomy specimens and manually constructed into TMAs. Ethical approval was obtained from each local ethics committee. The age of the patients ranged from 39 to 83 years with a median of 64 (UK) and from 47 to 91 years, median 73 (China). 32 (19%) UK patients had a Gleason Score of 6 or less, 90 (54%) equal to 7, and 46 (27%) greater than 7. 21 (15%) Chinese patients had Gleason Score 6 or less, 35 (25%) equal to 7, and 87 (61%) greater than 7. The Gleason Score of Chinese cases was higher than that of UK cases (p<0.001). Representative cancer areas, accompanying HGPIN and adjacent normal prostate areas were identified on H&E stained sections by histopathologists (LX, GR, LB and DB).
Fluorescence In Situ Hybridization (FISH) analysis of TMA samples
FISH had been performed in a previous study but signals were only analyzed in invasive tumors [3]. In the present study, HGPIN in these cases were identified and scored for ERG deletion and split signals. A minimum of 100 cells with clear hybridization signals were counted per core.
IHC and assessment
The standard ABC method was employed for immunostaining. Briefly, sections were cut at 4μm from the TMA blocks. After sections were dewaxed in xylene and rehydrated in alcohol and distilled water, antigen retrieval was performed by pressure oven heating (10 minutes) in low pH antigen unmasking solution. Then, sections were incubated with the primary rabbit monoclonal ERG antibody (1:3000, clone ID: EPR3864, Epitomics, San Diego, CA) for 1 hour at room temperature. This antibody has been recently validated [10] and has been used in several papers [11-14]. The bound antibody was detected with biotinylated secondary antibody for 30 minutes and AB compound (Vector Laboratories Inc, Burlingame, CA) for 20 minutes. 3,3’-diaminobenzidine (Vector Laboratories Inc, Burlingame, CA) was used as the chromogen. Finally, slides were lightly counterstained with hematoxylin. In control experiments, the primary antibody was replaced by phosphate-buffered saline (PBS).
Vascular endothelial cells, which have been consistently positively stained in previous studies [10-20], served as intrinsic positive control for the ERG IHC assay. Either cytoplasmic or nuclear localization of ERG were considered as positive, and scored separately with combined criteria of intensity and percentage of positive cells. The intensity was graded as follows: 0, negative; 1, weak; 2, moderate; 3, strong. The percentage of positive cells was scored subjectively and divided into: 0, <5%; 1, 5-25%; 2, 26-50%; 3, 51%-75%; 4, >75%. A final score was achieved by multiplication of the two scores above. Scores of 0-4 were defined as “negative expression or equivocally positive expression” (-), 5-8 as “weakly positive expression” (+), and 9-12 as “strongly positive expression” (++).
Statistical analysis
Chi-square test performed with SPSS 16.0 for Windows (SPSS, Chicago, IL, USA) was used to compare the difference of ERG expression between Chinese and UK prostate cancers as well as HGPINs, and to assess the correlations between ERG expression in prostate cancer and clinicopathological characteristics, including age and Gleason score. All of these were performed as two tailed tests. Kappa test was used to analyze the association between ERG rearrangements and ERG expression. A P value of less than 0.05 was considered statistically significant.
Results
ERG rearrangements evaluated in HGPIN using TMA based FISH
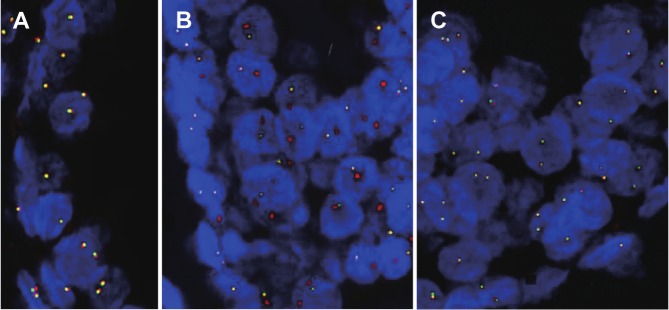
As we previously identified ERG rearrangement difference between prostate cancer samples from the UK and China [3], in this study we evaluated the differences in ERG rearrangement frequency in HGPIN lesions detected by FISH analysis (Table 1). There were 19 cases of HGPINs in UK TMAs with informative FISH data. ERG 5’ region was deleted in 21% (4/19) of these cases of HGPINs, but split signals were not detected in any cases in this cohort. No ERG rearrangements were detected in any of the 19 cases of Chinese HGPINs. Representative FISH images are shown in Figure 1.
Table 1.
ERG status in different groups
| Variable | ERG rearrangement | ERG nuclear expression | ERG cytoplasmic expression |
|---|---|---|---|
| UK cancer samples | 58/155 (37%) | 54/160 (34%) | 64/160 (40%) |
| GS <7 | 15/28 (54%) | 12/30 (40%) | 14/30 (47%) |
| GS =7 | 30/82 (37%) | 33/81 (41%) | 33/81 (41%) |
| GS >7 | 13/45 (30%) | 8/43 (19%) | 16/43 (37%) |
| UK HGPIN | 4/19 (21%) | 11/39 (28%) | 16/39 (41%) |
| Chinese cancer samples | 7/93 (8%) | 9/88 (10%) | 13/88 (15%) |
| GS <7 | 1/16 (6%) | 0/14 (0%) | 4/14 (29%) |
| GS =7 | 2/21 (10%) | 0/17 (0%) | 3/17 (18%) |
| GS >7 | 4/56 (7%) | 7/49 (14%) | 6/49 (12%) |
| Chinese HGPIN | 0/19 (0%) | 0/9(0%) | 2/9 (22%) |
GS: Gleason score; HGPIN: high-grade prostatic intraepithelial neoplasia.
Figure 1.
Representative FISH images of ERG gene status from the UK and Chinese TMAs. A. A normal prostate gland without ERG deletion (paired red and green signals); B. A HGPIN lesion from a UK case with ERG deletion (more red than green signals); C. A HGPIN lesion from a Chinese case without ERG deletion (paired red and green signals). Red signal: a probe for ERG 3’ undeleted region; Green signal: a probe for ERG 5’ region deleted in TMPRSS2:ERG fusion cases.
Expression of ERG evaluated using TMA based IHC
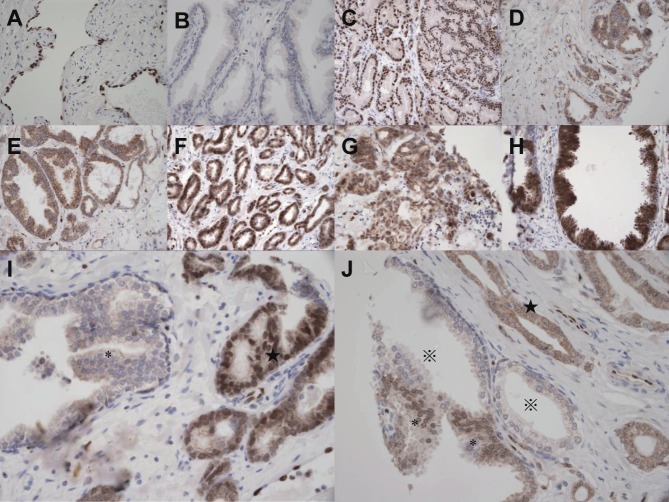
We found that endothelia of the vessels were always positive (Figure 2) and could be considered as internal positive controls. Both cytoplasmic and nuclear localization of ERG could be seen in prostate cancer and HGPIN lesions, and most of the cytoplasmic signals were weak (Figure 2). The numbers of IHC informative cancer cases were 160 and 88 in the UK and Chinese cohorts, respectively. Because the sections for the Chinese TMA blocks were deeper cutting ones, there were less cases on the IHC slides than all of those used to construct the TMAs (88 vs. 143). Scored the subcellular localization of expression signals separately, ERG showed nuclear positivity (including weak and strong positivity) in 34% (54/160) and cytoplasmic positivity in 40% (64/160) of UK cancer samples, and nuclear and cytoplasmic positivity in 10% (9/88) and 15% (13/88) of Chinese cancer samples, respectively (Table 1). Both ERG nuclear (Table 2) and cytoplasmic expressions in UK cancer samples were significantly higher than those in Chinese ones (p<0.001 for both). The numbers of IHC informative HGPIN cases were 39 (UK) and 9 (Chinese). ERG showed nuclear positivity in 28% (11/39) and cytoplasmic positivity in 41% (16/39) of UK HGPINs and nuclear and cytoplasmic positivity in 0% (0/9) and 22% (2/9) of Chinese HGPINs, respectively. ERG nuclear expression in UK HGPINs (28%, 11/39) was higher than that in Chinese HGPINs (0%, 0/9), but without statistical significance (p=0.193) (Table 3). The difference between UK and Chinese HGPINs was less for ERG cytoplasmic expression (p= 0.294) than nuclear expression (p=0.193) (Table 1).
Figure 2.
Representative IHC images of ERG expression in the UK and Chinese samples. A. Positive ERG staining in endothelium. B. Negative ERG expression in adjacent normal prostate glands. C. Nuclear positive staining in a Chinese prostate cancer sample. D. Cytoplasmic positive staining in a Chinese prostate cancer sample. E. Cytoplasmic positive staining in a Chinese HGPIN. F. Nuclear positive staining (and very weak cytoplasmic staining) in a UK prostate cancer sample. G. Cytoplasmic positive staining in a UK prostate cancer sample. H. Both cytoplasmic and nuclear positive staining in a UK HGPIN. I. Nuclear positive staining (and very weak cytoplasmic staining) in a UK prostate cancer sample (★) and negative staining in the adjacent HGPIN (*). J. Cytoplasmic positive staining in a UK prostate cancer sample (★) with nuclear positive staining in adjacent HGPIN (*) and negative staining in adjacent normal glands (※). Magnification: 400X.
Table 2.
Difference of ERG nuclear expression between UK and Chinese cancer samples
| ERG nuclear | |||||
|---|---|---|---|---|---|
|
|
|||||
| Negative or equivocal | Weakly positive | Strong positive | Total | P | |
| UK | 106 (66%) | 16 (10 %) | 38 (24 %) | 160 | |
| China | 79 (90%) | 2(2%) | 7 (8%) | 88 | <0.001 |
| Total | 185 | 18 | 45 | 248 | |
Table 3.
Difference of ERG nuclear expression between UK and Chinese high-grade prostatic intraepithelial neoplasia lesions
| ERG nuclear | |||||
|---|---|---|---|---|---|
|
|
|||||
| Negative or equivocal | Weakly positive | Strong positive | Total | P | |
| UK | 28 (72 %) | 6 (15 %) | 5 (13 %) | 39 | |
| China | 9 (100%) | 0 | 0 | 9 | 0.193 |
| Total | 37 | 6 | 5 | 48 | |
From HGPIN to invasive prostate cancer, there is a trend of increased frequency of ERG nuclear expression both in samples from UK (28% vs. 34%) and China (0% vs 10%,), although in both cohorts it was not statistically significant (p=0.259 and p=0.602 respectively). In the Chinese samples, this lack of statistical significance may be due to the small sample size. However, the frequency of ERG cytoplasmic expression in HGPINs is as high as that in invasive prostate cancer samples both in the UK (41% vs. 40%) and Chinese (22% vs. 15%,) cohorts.
Correlation of ERG rearrangements and ERG expression
In the UK cohort, ERG rearrangements were significantly correlated with ERG nuclear expression (Kappa=0.686, p<0.001). The majority (76%, 42/55) of prostate cancers with ERG rearrangements had positive nuclear expression for ERG, and the majority (91%, 81/89) of prostate cancers without ERG rearrangements had no positive nuclear expression for ERG, while 84% (42/50) with positive nuclear expression of ERG possessed ERG rearrangements (Table 4). While ERG cytoplasmic expression and ERG rearrangements were also significantly correlated (kappa=0.292, p<0.001), this correlation is much weaker than that between nuclear expression and ERG rearrangements.
Table 4.
Correlation between ERG rearrangements and ERG nuclear expression in UK prostate cancer samples
| Nuclear expression | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Total* | Negative | positive | Kappa | P | ||
| Genomics | Normal | 89 | 81(91%) | 8(9 %) | ||
| rearrangement | 55 | 13(24 %) | 42(76%) | 0.686 | <0.001 | |
| Total* | 144 | 94 | 50 | |||
Informative cases with both FISH and IHC data.
In Chinese prostate cancer samples, ERG rearrangements were also significantly correlated with ERG nuclear expression (Kappa=0.565, p<0.001). Half (50%, 3/6) of prostate cancers with ERG rearrangements had positive nuclear expression for ERG, and the majority (98%, 52/53) of prostate cancers without ERG rearrangements had no positive nuclear expression for ERG, while 75% (3/4) with positive nuclear expression of ERG possessed ERG rearrangements (Table 5). ERG cytoplasmic expression was not significantly correlated with ERG rearrangements (kappa=0.119, p=0.330).
Table 5.
Correlation between ERG rearrangements and ERG nuclear expression in Chinese prostate cancer samples
| Nuclear expression | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Total* | Negative | positive | Kappa | P | ||
| Genomics | Normal | 53 | 52(98%) | 1(2%) | ||
| rearrangement | 6 | 3(50%) | 3(50%) | 0.565 | <0.001 | |
| Total* | 55 | 4 | ||||
Informative cases with both FISH and IHC data.
Data were available both from FISH and IHC analyses for 11 UK cases of HGPINs. Only 2 cases had positive ERG expression in nucleus, and both of these two cases (100%) had ERG rearrangements. ERG rearrangements were detected in 4 cases and among them, 2 (50%) had positive ERG expression in nucleus. In the 7 cases without ERG rearrangements, no cases (0%) had nuclear expression of ERG.
Data were also available both from FISH and IHC analyses for 6 Chinese HGPIN cases. None of them have been detected with ERG rearrangement or nuclear expression.
Correlation of ERG rearrangements and ERG expression with patient age and tumor Gleason score
There were no significant correlations between ERG rearrangements and age or Gleason score in prostate cancer samples from the UK (p=0.854 and 0.103, respectively) or China (p=0.062 and 0.919, respectively). There were also no significant correlations between ERG nuclear expression and age or Gleason Score in prostate cancer samples from the UK (p=0.615 and p=0.150, respectively) or China (p=0.059 and p=0.303, respectively).
Discussion
In our previous study, we identified in Chinese cancer samples a significant reduction in the frequency of some somatic genomic alterations that were commonly found in Western prostate cancers, including the 21q22.2-22.3 rearrangements, which cause the TMPRSS2:ERG fusion gene [3]. In this study, we further evaluated this genetic change in Western and Chinese populations by FISH and IHC and found ERG expression patterns were different in UK and Chinese cohorts and that these differences, both at genomic and protein levels, were identifiable at a pre-neoplastic stage. ERG rearrangements were associated with ERG nuclear expression.
TMPRSS2:ERG is the most common gene fusion event in human cancers, occurring in around half of Western prostate cancer [5,12,21-26] and leads to over-expression of the oncogene ERG from the androgen-stimulated TMPRSS2 promoter. However, from our and the others previous studies [3,7,8,27], it is clear that the TMPRSS2:ERG fusion accounts for a much smaller proportion of prostate cancer in East Asian than Western populations. In this study, we found that the difference in ERG rearrangement frequency was already present in HGPIN, the precursor lesion of prostate cancer. TMPRSS2:ERG rearrangements have previously been observed in about 20% of HGPIN from patients from the Western countries, both using PCR-based (4/19) [28,29] and FISH-based (5/26) [28,29] techniques. In the FISH analysis, all TMPRSS2:ERG fusion genes in the HGPIN lesions were detected as 5’ region genomic deletion of ERG without any translocations [28,29]. In consistence with the previous reports, we also detected 5’ region deletion of ERG in 21% (4/19) of UK HGPIN cases, but did not find split signals in any cases. Together, these observations strongly support the hypothesis that TMPRSS2:ERG fusion through genomic deletion is an early event in prostate carcinogenesis. To our knowledge, this is the first report on ERG rearrangements in Chinese cases of HGPINs. Although the number of samples is limited, lack of ERG rearrangements in all of the 19 Chinese HGPIN samples suggests that the difference in ERG rearrangement frequency between the Western and Chinese prostate cancers may occur before the onset of the invasive cancer. This data further supports that different etiological factors and pathogenetic mechanisms contribute to the different prevalence of prostate cancer in Western countries and China and these factors/mechanisms act at a pre-neoplastic stage. We have recently demonstrated that exposure to high-dose of androgen, whose level and receptor activity are different in Western and Chinese male individuals, can induce TMPRSS2:ERG fusion gene in non-malignant prostate epithelial cells [30], which may partially explain the different frequency of TMPRSS2:ERG fusion in the HGPINs from Western and Chinese populations.
In addition to numerous reports of genomic fusions and fusion transcript expression [31], a few studies have investigated ERG expression at the protein level using IHC or immunofluorescence [10-14,18-20,32,33]. Although, as a transcription factor, ERG should fulfill its function in cell nucleus, the reports in the literature on the localization of ERG protein in prostate cancer are inconsistent. Using the same commercial anti-ERG polyclonal antibody, one study showed ERG cytoplasmic expression in both prostate cancer specimens and the TMPRSS2:ERG fusion positive prostate cancer cell line, VCaP [32], but nuclear staining was detected in other two studies [19,33]. Furusato et al. showed moderate to strong nuclear staining with cytoplasmic blush in prostate cancer using a self-made anti-ERG monoclonal antibody [18]. The rabbit anti-ERG monoclonal antibody we used, clone ID: EPR3864, has been recently validated by Park K et al. [10] and was used in several other studies [11-14]. Although cytoplasmic staining can be seen in the figures presented, only nuclear localized ERG expression was scored in those studies, which was highly predictive of ERG rearrangement status assessed by FISH or quantitative PCR, with 86-100% sensitivity and 85-99% specificity [10-12,14]. Therefore, we also mainly accessed the nuclear expression of ERG and determined its association with ERG genomic rearrangements. In our study, the ERG nuclear expression was also highly predictive of ERG rearrangement status assessed by FISH with sensitivity of 76% in UK cases and 50% in Chinese ones, and specificity of 91% and 98% in UK and Chinese cases respectively. Moreover, we showed, in consistent with ERG rearrangement data, nuclear expression of ERG was significantly more frequent in UK than Chinese cases; and this difference presented in both prostate cancer and HGPIN samples.
As we have observed both nuclear and cytoplasmic expression of ERG, we also scored the ERG cytoplasmic expression. There were more cytoplasmic than nuclear positive cases, but the reasons for frequent ERG cytoplasmic expression are currently not clear. While the difference of ERG cytoplasmic expression between UK and Chinese cancer samples is also very significant, ERG cytoplasmic expression was less strongly correlated with ERG rearrangements than nuclear expression in the UK cancer cases and ERG cytoplasmic expression is not significantly correlated with ERG rearrangements in the Chinese samples. These data further support that ERG nuclear rather than cytoplasmic expression is associated with ERG fusion status and prostate carcinogenesis.
It has been reported that up-regulation of ERG promotes prostate cell proliferation and results in the development of PIN [33]. Previous studies have investigated ERG protein expression in Western HGPIN, showing that 45.8-52% HGPIN foci had ERG nuclear positive expression [14,18]. We also observed a significant proportion of cases with nuclear positivity (29%) in UK HGPINs while no ERG nuclear expression was observed in Chinese HGPINs, demonstrating that differences in ERG expression occur in the precursor lesions, potentially caused by the difference in ERG genomic rearrangements. This also indicates that nuclear expression of ERG functionally contributes to prostate carcinogenesis and the different incidence of prostate cancer between the Western countries and China.
In the previous studies, the correlation between ERG status and Gleason score is inconsistent, and varies from a positive association [18,26,34-37], no association [28,38] to reverse association [32]. In this study, while the Gleason Score is significantly higher in the Chinese cohort than the UK one, there were no significant correlations between ERG rearrangements or expression with Gleason Score in either cohort. This is consistent with our previous genomic analysis of the association between ERG rearrangements and Gleason score [3].
Conclusions
In summary, following our previous observation that TMPRSS2:ERG fusion is more frequent in UK prostate cancer samples than Chinese ones, here we show that the difference of ERG rearrangement frequency occurs in the precursor lesion. Moreover, we demonstrate different ERG protein expression in UK and Chinese prostate cancers and that this difference is also detectable in the precursor lesions. ERG nuclear, but not the cytoplasmic, expression is closely associated with ERG rearrangements in both Western and Chinese samples. Our data from this study provide further evidence that different pathways of prostate carcinogenesis exist in Western and Chinese populations.
Acknowledgements
We thank Isabelle Bisson for sample collection and Orchid Cancer Appeal for funding support.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Gronberg H. Prostate cancer epidemiology. Lancet. 2003;361:859–864. doi: 10.1016/S0140-6736(03)12713-4. [DOI] [PubMed] [Google Scholar]
- 3.Mao X, Yu Y, Boyd LK, Ren G, Lin D, Chaplin T, Kudahetti SC, Stankiewicz E, Xue L, Beltran L, Gupta M, Oliver RT, Lemoine NR, Berney DM, Young BD, Lu YJ. Distinct genomic alterations in prostate cancers in Chinese and Western populations suggest alternative pathways of prostate carcinogenesis. Cancer Res. 2010;70:5207–5212. doi: 10.1158/0008-5472.CAN-09-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ren G, Liu X, Mao X, Zhang Y, Stankiewicz E, Hylands L, Song R, Berney DM, Clark J, Cooper C, Lu YJ. Identification of frequent BRAF copy number gain and alterations of RAF genes in chinese prostate cancer. Genes Chromosomes Cancer. 2012;51:1014–23. doi: 10.1002/gcc.21984. [DOI] [PubMed] [Google Scholar]
- 5.Clark JP, Cooper CS. ETS gene fusions in prostate cancer. Nat Rev Urol. 2009;6:429–439. doi: 10.1038/nrurol.2009.127. [DOI] [PubMed] [Google Scholar]
- 6.Magi-Galluzzi C, Tsusuki T, Elson P, Simmerman K, LaFargue C, Esgueva R, Klein E, Rubin MA, Zhou M. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate. 2011;71:489–497. doi: 10.1002/pros.21265. [DOI] [PubMed] [Google Scholar]
- 7.Lee K, Chae JY, Kwak C, Ku JH, Moon KC. TMPRSS2-ERG Gene Fusion and Clinicopathologic Characteristics of Korean Prostate Cancer Patients. Urology. 2010;76:1268.e7–e13. doi: 10.1016/j.urology.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 8.Miyagi Y, Sasaki T, Fujinami K, Sano J, Senga Y, Miura T, Kameda Y, Sakuma Y, Nakamura Y, Harada M, Tsuchiya E. ETS family-associated gene fusions in Japanese prostate cancer: analysis of 194 radical prostatectomy samples. Mod Pathol. 2010;23:1492–1498. doi: 10.1038/modpathol.2010.149. [DOI] [PubMed] [Google Scholar]
- 9.Montironi R, Mazzucchelli R, Lopez-Beltran A, Cheng L, Scarpelli M. Mechanisms of disease: high-grade prostatic intraepithelial neoplasia and other proposed preneoplastic lesions in the prostate. Nat Clin Pract Urol. 2007;4:321–332. doi: 10.1038/ncpuro0815. [DOI] [PubMed] [Google Scholar]
- 10.Park K, Tomlins SA, Mudaliar KM, Chiu YL, Esgueva R, Mehra R, Suleman K, Varambally S, Brenner JC, MacDonald T, Srivastava A, Tewari AK, Sathyanarayana U, Nagy D, Pestano G, Kunju LP, Demichelis F, Chinnaiyan AM, Rubin MA. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 12:590–598. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaux A, Albadine R, Toubaji A, Hicks J, Meeker A, Platz EA, De Marzo AM, Netto GJ. Immunohistochemistry for ERG expression as a surrogate for TMPRSS2-ERG fusion detection in prostatic adenocarcinomas. Am J Surg Pathol. 2011;35:1014–1020. doi: 10.1097/PAS.0b013e31821e8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falzarano SM, Zhou M, Carver P, Tsuzuki T, Simmerman K, He H, Magi-Galluzzi C. ERG gene rearrangement status in prostate cancer detected by immunohistochemistry. Virchows Arch. 459:441–447. doi: 10.1007/s00428-011-1128-4. [DOI] [PubMed] [Google Scholar]
- 13.He H, Magi-Galluzzi C, Li J, Carver P, Falzarano S, Smith K, Rubin MA, Zhou M. The diagnostic utility of novel immunohistochemical marker ERG in the workup of prostate biopsies with “atypical glands suspicious for cancer”. Am J Surg Pathol. 35:608–614. doi: 10.1097/PAS.0b013e31820bcd2d. [DOI] [PubMed] [Google Scholar]
- 14.van Leenders GJ, Boormans JL, Vissers CJ, Hoogland AM, Bressers AA, Furusato B, Trapman J. Antibody EPR3864 is specific for ERG genomic fusions in prostate cancer: implications for pathological practice. Mod Pathol. 24:1128–1138. doi: 10.1038/modpathol.2011.65. [DOI] [PubMed] [Google Scholar]
- 15.Baltzinger M, Mager-Heckel AM, Remy P. Xl erg: expression pattern and overexpression during development plead for a role in endothelial cell differentiation. Dev Dyn. 1999;216:420–433. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<420::AID-DVDY10>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 16.Birdsey GM, Dryden NH, Amsellem V, Gebhardt F, Sahnan K, Haskard DO, Dejana E, Mason JC, Randi AM. Transcription factor Erg regulates angiogenesis and endothelial apoptosis through VE-cadherin. Blood. 2008;111:3498–3506. doi: 10.1182/blood-2007-08-105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellett F, Kile BT, Lieschke GJ. The role of the ETS factor erg in zebrafish vasculogenesis. Mech Dev. 2009;126:220–229. doi: 10.1016/j.mod.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furusato B, Tan SH, Young D, Dobi A, Sun C, Mohamed AA, Thangapazham R, Chen Y, Mc-Master G, Sreenath T, Petrovics G, McLeod DG, Srivastava S, Sesterhenn IA. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis. 2010;13:228–237. doi: 10.1038/pcan.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gavrilov D, Kenzior O, Evans M, Calaluce R, Folk WR. Expression of urokinase plasminogen activator and receptor in conjunction with the ets family and AP-1 complex transcription factors in high grade prostate cancers. Eur J Cancer. 2001;37:1033–1040. doi: 10.1016/s0959-8049(01)00077-6. [DOI] [PubMed] [Google Scholar]
- 20.Miettinen M, Wang ZF, Paetau A, Tan SH, Dobi A, Srivastava S, Sesterhenn I. ERG transcription factor as an immunohistochemical marker for vascular endothelial tumors and prostatic carcinoma. Am J Surg Pathol. 35:432–441. doi: 10.1097/PAS.0b013e318206b67b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, Yu J, Wang L, Montie JE, Rubin MA, Pienta KJ, Roulston D, Shah RB, Varambally S, Mehra R, Chinnaiyan AM. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 22.Lapointe J, Li C, Giacomini CP, Salari K, Huang S, Wang P, Ferrari M, Hernandez-Boussard T, Brooks JD, Pollack JR. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504–8510. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
- 23.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 24.Liu W, Chang B, Sauvageot J, Dimitrov L, Gielzak M, Li T, Yan G, Sun J, Sun J, Adams TS, Turner AR, Kim JW, Meyers DA, Zheng SL, Isaacs WB, Xu J. Comprehensive assessment of DNA copy number alterations in human prostate cancers using Affymetrix 100K SNP mapping array. Genes Chromosomes Cancer. 2006;45:1018–1032. doi: 10.1002/gcc.20369. [DOI] [PubMed] [Google Scholar]
- 25.Mehra R, Tomlins SA, Yu J, Cao X, Wang L, Menon A, Rubin MA, Pienta KJ, Shah RB, Chinnaiyan AM. Characterization of TMPRSS2-ETS gene aberrations in androgen-independent metastatic prostate cancer. Cancer Res. 2008;68:3584–3590. doi: 10.1158/0008-5472.CAN-07-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins SA, Hofer MD, Pienta KG, Kuefer R, Vessella R, Sun XW, Meyerson M, Lee C, Sellers WR, Chinnaiyan AM, Rubin MA. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 27.Magi-Galluzzi C, Tsusuki T, Elson P, Simmerman K, Lafargue C, Esgueva R, Klein E, Rubin MA, Zhou M. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of caucasian, african-american and japanese patients. Prostate. 2010;71:489–97. doi: 10.1002/pros.21265. [DOI] [PubMed] [Google Scholar]
- 28.Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, Collins C, Bismar TA, Chinnaiyan AM, De Marzo AM, Rubin MA. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 29.Cerveira N, Ribeiro FR, Peixoto A, Costa V, Henrique R, Jeronimo C, Teixeira MR. TMPRSS2-ERG gene fusion causing ERG overexpression precedes chromosome copy number changes in prostate carcinomas and paired HGPIN lesions. Neoplasia. 2006;8:826–832. doi: 10.1593/neo.06427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bastus NC, Boyd LK, Mao X, Stankiewicz E, Kudahetti SC, Oliver RT, Berney DM, Lu YJ. Androgen-induced TMPRSS2:ERG fusion in nonmalignant prostate epithelial cells. Cancer Res. 70:9544–9548. doi: 10.1158/0008-5472.CAN-10-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrovics G, Liu A, Shaheduzzaman S, Furusato B, Sun C, Chen Y, Nau M, Ravindranath L, Chen Y, Dobi A, Srikantan V, Sesterhenn IA, McLeod DG, Vahey M, Moul JW, Srivastava S. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847–3852. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
- 32.Bonaccorsi L, Nesi G, Nuti F, Paglierani M, Krausz C, Masieri L, Serni S, Proietti-Pannunzi L, Fang Y, Jhanwar SC, Orlando C, Carini M, Forti G, Baldi E, Luzzatto L. Persistence of expression of the TMPRSS2:ERG fusion gene after pre-surgery androgen ablation may be associated with early prostate specific antigen relapse of prostate cancer: preliminary results. J Endocrinol Invest. 2009;32:590–596. doi: 10.1007/BF03346514. [DOI] [PubMed] [Google Scholar]
- 33.Klezovitch O, Risk M, Coleman I, Lucas JM, Null M, True LD, Nelson PS, Vasioukhin V. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci U S A. 2008;105:2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajput AB, Miller MA, De Luca A, Boyd N, Leung S, Hurtado-Coll A, Fazli L, Jones EC, Palmer JB, Gleave ME, Cox ME, Huntsman DG. Frequency of the TMPRSS2:ERG gene fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007;60:1238–1243. doi: 10.1136/jcp.2006.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, Palanisamy N, Tomlins SA, Chinnaiyan AM, Shah RB. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol. 2009;22:1083–1093. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reynolds MA. Molecular alterations in prostate cancer. Cancer Lett. 2008;271:13–24. doi: 10.1016/j.canlet.2008.04.047. [DOI] [PubMed] [Google Scholar]
- 37.Majumder PK, Sellers WR. Akt-regulated pathways in prostate cancer. Oncogene. 2005;24:7465–7474. doi: 10.1038/sj.onc.1209096. [DOI] [PubMed] [Google Scholar]
- 38.Lapointe J, Kim YH, Miller MA, Li C, Kaygusuz G, van de Rijn M, Huntsman DG, Brooks JD, Pollack JR. A variant TMPRSS2 isoform and ERG fusion product in prostate cancer with implications for molecular diagnosis. Mod Pathol. 2007;20:467–473. doi: 10.1038/modpathol.3800759. [DOI] [PubMed] [Google Scholar]